Abstract
PRDM1/Blimp-1, a master regulator in terminal B-cell differentiation, has been recently identified as a tumor suppressor target for mutational inactivation in diffuse large B-cell lymphomas of the activated B-cell type. Our studies here demonstrate that PRDM1/blimp-1 is also a target for microRNA (miRNA)-mediated down-regulation by miR-9 and let-7a in Hodgkin/Reed-Sternberg (HRS) cells of Hodgkin lymphoma (HL). MiRNA expression profiling by direct miRNA cloning demonstrated that both of these miRNAs are among the most highly expressed in cultured HRS cells. These miRNAs functionally targeted specific binding sites in the 3′ untranslated region of PRDM1/blimp-1 mRNA and repressed luciferase reporter activities through repression of translation. In addition, high levels of miR-9 and let-7a in HL cell lines correlated with low levels of PRDM1/Blimp-1. Similar to their in vitro counterparts, the majority of HRS cells in primary HL cases showed weak or no PRDM1/Blimp-1 expression. Over-expression of miR-9 or let-7a reduced PRDM1/Blimp-1 levels in U266 cells by 30% to 50%, whereas simultaneous inhibition of their activities in L428 cells resulted in an approximately 2.6-fold induction in PRDM1/Blimp-1. MiRNA-mediated down-regulation of PRDM1/Blimp-1 may contribute to the phenotype maintenance and pathogenesis of HRS cells by interfering with normal B-cell terminal differentiation, thus representing a novel molecular lesion, as well as a potential therapeutic target in HL.
PRDM1, also known as Blimp-1, belongs to the PRDM gene family of transcription repressors containing Kruppel-type zinc fingers and the SET-related PR (PRDI-BF1-RIZ) domain. PRDM1 is expressed as two isoforms, α and β, as a result of alternative promoter usage. They differ in that the latter lacks the amino-terminal acidic domain and part of the PR domain, and is functionally impaired.1 PRDM1 plays a critical role in terminal differentiation of lymphocytes2 and epidermal cells,3 as well as cell fate specification of primordial germ cells4 and other cell types.5 In B cells, PRDM1 is a key differentiation factor in post-germinal center (GC) cells and is regarded as a master regulator for plasma cell differentiation.6 In normal lymphoid tissues, PRDM1 is co-expressed with interferon regulatory factor-4 (IRF4) in plasma cells and in a subset of GC cells that demonstrate evidence of plasma cell commitment and differentiation.7,8,9 Both of these transcription factors are required for plasma cell differentiation.9 Microarray profiling demonstrates that PRDM1/Blimp-1 orchestrates plasma cell differentiation by repressing genetic programs associated with activated B cells and/or GC B cells, including those that control cell proliferation, and by activating genetic programs associated with plasma cell functions, including apoptosis.10
Recent published data supported a role for interference of PRDM1 functions in the pathogenesis of human lymphomas. PRDM1 has been shown to be inactivated by a classic mechanism for tumor suppressor genes in large B-cell lymphoma diffuse (DLBCL), specifically those of the non-GCB type, strongly suggesting that inhibition of terminal B-cell differentiation may play a role in their pathogenesis.11,12 In addition, BCL-6-mediated transcription repression of PRDM1 causes blockade of terminal differentiation in GC-type DLBCL.13
The neoplastic cells in classical Hodgkin lymphoma (HL), the Hodgkin/Reed-Sternberg (HRS) cells, resemble post-GC cells immunophenotypically and genetically, despite their putative origin from preapoptotic GC cells.14,15 They frequently lack BCL-6 and consistently express IRF4.16,17,18 They harbor somatic mutations in their immunoglobulin genes but show no evidence of ongoing somatic hypermutation.15 In addition, HL cell lines have a similar gene expression pattern as that of Epstein Barr Virus-transformed B cells and DLBCL cell lines showing features of in vitro-activated B cells.19 Since HRS cells and non-GCB DLBCL appear to be in similar differentiation stages, we hypothesize a role of interference of PRDM1 functions in HRS cell pathogenesis. Sequence analysis has not identified inactivating mutation of PRDM1 in HL cell lines (unpublished data). Decrease in accumulation of PRDM1, however, may occur through quantitative changes in its synthesis or stability. One way by which PRDM1 synthesis can be altered is via regulation by miRNA. In fact, deregulation of target protein production by altered expression of miRNA is a well documented mechanism of oncogenesis.20 In this report, we provide experimental evidence that PRDM1 is a target for down-regulation by miRNAs in HRS cells.
Materials and Methods
Cell Lines
The GC-like DLBCL cell lines SUDHL6 and OCI-Ly1, the myeloma cell line U266, the primary effusion lymphoma (PEL) cell lines BC1, BC2, BC3, and BCBL1, and the HL cell lines L428, KMH2, and L1236, were cultured in RPMI medium 1640 with 10% heat-inactivated fetal calf serum (Invitrogen, Carlsbad, CA). 293T cells were maintained in Dulbecco’s modified Eagle’s medium with 10% heat-inactivated fetal bovine serum (Invitrogen).
Patient Tissue Samples
Formalin-fixed, paraffin-embedded archival tissue of classical HL cases were obtained according to the protocols approved by the Institutional Review Board. All samples were reviewed and classified according to the World Health Organization criteria.
Antibody
For Western blots and immunohistochemistry, a monoclonal antibody against human PRDM1 (ROS) was used. This antibody recognizes both PRDM1α and PRDM1β.21
Western Blotting and Immunohistochemistry
Immunoblotting and immunoperoxidase staining on paraffin tissue sections were performed as previously described.21 Quantitation of PRDM1α expression in Western blots was done by densitometry and normalized with loading controls (lamin B or β-actin).
Reverse Transcription and Real-Time PCR
Total RNA was extracted from cell lines and treated with RNase-free DNase I. For quantitative detection of PRDM1α mRNA, cDNA was synthesized from 1 μg of total RNA using random primers. Monoplex real-time PCR was conducted using ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). The PCR reaction was done using 50 ng of cDNA template according to the manufacturer’s protocol using the following PCR conditions: 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, 58°C for 30 seconds, and 72°C for 1 minute. A standard curve, consisting of serially diluted U266 RNA, was included on each 96-well reaction plate for each run. Each cDNA template was assayed in duplicate, and each sample was run three independent times. Relative quantities of PRDM1α mRNA were calculated using the standard curve method, normalized, and expressed relative to U266 (set as one). For normalization, the 6 most stable reference genes identified using the freely distributed MicroSoft Excel application geNorm22 among 11 candidate genes described previously23 were used. These six genes are: glyceraldehyde-3-phosphate dehydrogenase, protein kinase cGMP-dependent, type 1, hypoxanthine phosphoribosyltransferase 1, TATA box binding protein, large ribosomal phospoprotein PO, and β-glucoronidase.
Primer and probe sequences for real-time detection of PRDM1α mRNA are as follows: Forward: 5′-TCCAGCACTGTGAGGTTTCA-3′; Reverse: 5′-TCAAACTCAGCCTCTGTCCA-3′; Probe: FAM-5′-ATGGACATGGAGGATGCGGATATG-3′-TAMRA. Real-time quantification of the endogenous control genes was performed using the Human Taqman predeveloped assays reagents endogenous controls (Applied Biosystems).
For quantitative measurement of luciferase reporter mRNA in cell transfectants, cDNA was generated and real-time PCR was performed as described above using SYBR Green PCR Master Mix (Applied Biosystems). The relative Renilla luciferase mRNA levels (negative control transfectant set as 100) were calculated by the ΔΔCt method using firefly luciferase mRNA expression as normalization. Primer sequences are as follows: Renilla luciferase: Forward: 5′-AAGAGCGAAGAGGGCGAGAA-3′, Reverse: 5′-TGCGGACAATCTGGACGAC-3′; Firefly luciferase: Forward: 5′-CGTGCCAGAGTCTTTCGACA-3′, Reverse: 5′-ACAGGCGGTGCGATGAG-3′.
MicroRNA Quantitation
Let-7a and miR-9 levels in cell lines were determined using TaqMan MicroRNA Assays (Applied Biosystems) following the protocols recommended by the manufacturer. Five ng of total RNA was used per 15 μl reaction during reverse transcription, and 1 μl of reverse transcription product was used for subsequent real-time PCR reactions. For each sample, three independent reverse transcription reactions were performed and each reaction was assayed in triplicate for real-time PCR. The levels of miRNAs were normalized with 5S RNA and the relative levels were calculated using the ΔΔCt method. Direct cloning of miRNAs was performed as previously described.24
Computer Prediction of MiRNA Target Sites in the 3′ Untranslated Region of PRDM1 mRNA
Putative target sites for miRNA in the 3′ untranslated region (UTR) of human, mouse, rat, and chicken PRDM1 mRNA were identified using the MiRANDA Human miRNA Targets web site (http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl) and the Targetscan (version 4.1) web site (http://genes.mit.edu/targetscan) based on the target prediction algorithms developed by John et al25 and Lewis et al,26 respectively. The binding of chicken miR-9 to its target sequences is not depicted in either of these web sites, and was deduced based on the available chicken sequence database and with the assistance of the mFOLD program (version 3) available at the website http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi.27
Constructs
The miR-155/BIC expression plasmid has been previously described.28 To generate the miR-9-expressing plasmid pcDNA3.miR-9-1, a 515-bp DNA fragment encompassing pre-miR-9-1 was amplified by PCR using genomic DNA template with sense and antisense primers flanked at the 5′ end by EcoRI and BamHI sites, respectively. The sequences of the primers were 5′-ATATAGAATTCCCAAGCAGTGACCCAGA-3′ (sense) and 5′-TAATAGGATCCTTCCCTCCTACTCCCGCTGA-3′ (antisense). The PCR-generated fragment were digested with EcoRI and BamHI and subcloned into pcDNA3(+) (Invitrogen) between the EcoRI and BamHI sites.
Construction of pSIC.PRDM1.3′UTR.538–2419 is as follows: A genomic fragment encompassing nucleotides 1 to 2419 of the PRDM1 3′UTR was PCR-amplified using primers flanked by XhoI and NotI sites, and cloned into pGEM-T-Easy (Promega, Madison, WI). The sequences of the primers were: 5′-CTCGAGGATTTTCAGAAAACACTTATTTTGTTTC-3′ (sense) and 5′-GCGGCCGCACATTTTGACAATTTGCACATAAATAAC-3′ (antisense). Sequence of the insert was confirmed by double-stranded sequencing. Plasmid DNA was isolated from recombinant clones and digested with XhoI and NotI. The insert was gel purified and cloned into psiCHECK-2 (Promega) downstream of the Renilla luciferase coding region between the XhoI and NotI sites. The resulting plasmid was then digested with XhoI and PshAI, followed by fill-in. The larger product was then gel purified and self-ligated to give the final product.
The mutant reporter constructs were generated by using pSIC.PRDM1.3′UTR.538-2419 as a template and mutating the fifth and sixth positions (from the 3′end) of the putative miR-9 or let-7a binding sites using the GeneEditor in vitro Site-Directed Mutagenesis System (Promega). Mutations were generated in one or more of the three putative miR-9 binding sites, designated as Mut1, Mut2, Mut3, Mut(2 + 3), and Mut(1 + 2 + 3). The numbers 1 to 3 denote positions from the proximal to distal end. Sequence of the mutant report constructs thus obtained were confirmed by double-stranded sequencing.
The mutagenic primers used (mutant nucleotides in italics) were as follows: miR-9.Mut1: 5′-CTTTTATTCTGCTAAGCCGTAAGATTACATGTTGG-3′; miR-9.Mut2: 5′-CTGAAGGTAAACGTAAGCATCACGTTGAC-3′; miR-9.Mut3: 5′-CAAAGTTAAAACTGACGTAAGTTACTGGCTTTTTAC-3′; and, let-7a: 5′-AGTTGTTCAACAACAGTTTGCTCATTGAGTGTGTCC-3′.
Transfections and Luciferase Reporter Assays
293T cells were transfected with 75 ng of miRNA expression plasmids (miR-9 or miR-155) or pcDNA3 (negative plasmid control) in 96-well plates using Effectene (Qiagen, Valencia, CA), or with 20 nmol/L of miRNA precursor molecules (let-7a or negative miRNA control) purchased from Ambion (Austin, TX) in 24-well plates using Lipofectin (Invitrogen). For experiments evaluating the cooperative effects of miR-9 and let-7a, 50 nmol/L of either miRNA was transfected. These plasmids and miRNA analogs were co-transfected with 25 ng of pSIC.PRDM1.3′UTR.538–2419 (wild type or mutants), respectively. Twenty-four hours after transfection, Renilla and firefly luciferase activities were determined using the Dual-Glo Luciferase Assay System (Promega). The Renilla luciferase activities were normalized by firefly luciferase activities, which served as internal controls. The normalized values were compared with that of the negative controls to determine percentage change. The mean values (±SE) from three or four independent experiments are shown.
Transfection of Suspension Cell Lines with miRNAs or Anti-miRNA Inhibitors
Optimized nucleofection protocols generated by Amaxa (Cologne, Germany) were followed for the transfection of U266 (Nucleofector Kit C, Program X-005) and L428 (Nucleofector Kit L, Program X-001) with miRNA precursors (Ambion) or anti-miRNA inhibitors (Ambion), respectively. A total of 100 nmol/L was used for all transfections. For co-transfections of anti-miR-9 and anti-let-7a inhibitors, 50 nmol/L each was added. Cells were collected at 24 to 48 hours after electroporation and subject to Western blot analysis for determination of PRDM1 expression. These transfection experiments were repeated four independent times.
Statistical Analysis
P values were calculated by Student’s t-test using the StatView software (SAS Institute Inc., Cary, NC).
Results
Direct Cloning and Computer Prediction Identified miRNAs that Potentially Interact with PRDM1 mRNA
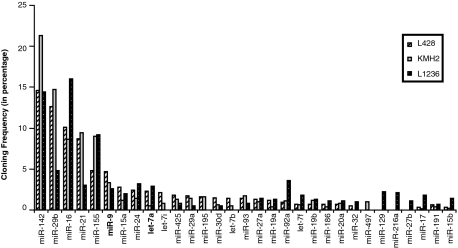
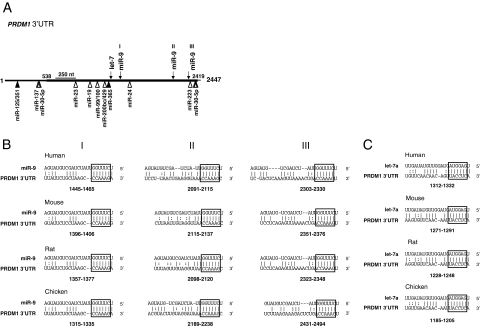
As a first step to identify miRNAs that may regulate PRDM1 expression, miRNA profiles for HL cell lines L428, KMH2, and L1236 generated by direct cloning29 were examined (Figure 1). Based on these expression profiles, we selected for further analysis two miRNAs, miR-9 and let-7a, with high potential to functionally interact with PRDM1 mRNA. Both these miRNAs are among the 10 most abundant miRNAs in HL cell lines and constitute on average about 3.5% and 1.9% of the total miRNA population, respectively. These expression levels rank close to miR-155, a highly expressed miRNA in HRS cells.30,31 In addition, putative binding sites for these miRNAs, including three for miR-9 and one for let-7, are predicted in the PRDM1 3′UTR independently by two computer algorithms: miRanda and Targetscan (release 4.1, January 2008).25,32,33 The pairing between these miRNAs and their target sequences are evolutionarily conserved, with absolute conservation between the 6bp “seed” region32 at the 5′ end of the miRNA and the complementary sequence of the target site (Figure 2). Conserved binding sites for two other miRNAs, miR-125/351 and miR-365, are also predicted in the PRDM1 3′UTR by both miRanda and Targetscan; however, these miRNAs are present at very low amount in HL cell lines based on miRNA cloning (<0.5% of the total miRNA population) and therefore not selected for further analysis.
Figure 1.
Profiles of miRNA expression in HL cell lines. The cloning frequency of each miRNA is shown as a percentage of total miRNA clones isolated for each HL cell line sample. Only those miRNAs representing greater than 0.5% of the entire miRNA population (based on average of the three cell lines) are included. MiRNAs miR-9 and let-7a are indicated in bold.
Figure 2.
PRDM1 3′UTR harbors putative target sites for miR-9 and let-7a. A: A schematic representation of the PRDM1 3′ UTR is shown. The positions of the putative miR-9 and let-7a sites are indicated by arrows, and other conserved miRNA binding sites predicted by TargetScan (v.4) only or by both TargetScan and MiRANDA are marked by open and filled triangles, respectively. The portion of the PRDM1 3′ UTR that is cloned and used for the luciferase reporter assays is marked in bold line. B and C: Complementarity between miR-9 or let-7a (top strand) and its conserved putative binding sites (bottom strand) in the PRDM1 3′ UTR is shown for different species. A Watson-Crick bp is indicated by a vertical line, and the U:G wobble base is marked by double dots. The highly conserved 6-bp “seed” pairing is highlighted. The numbers indicate the locations of the putative binding sites in the PRDM1 3′ UTRs downstream from the PRDM1 stop codon.
miR-9 and let-7a Target the 3′UTR of PRDM1 mRNA
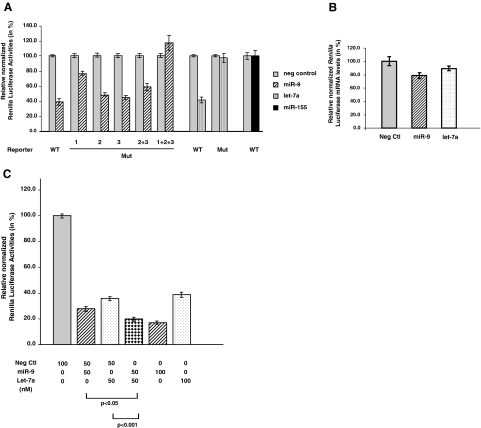
Luciferase reporter assays were performed to determine whether miR-9 and let-7a can target the 3′UTR of PRDM1 mRNA. The reporter plasmid, psiC.PRDM1.3′UTR.538–2419, contains the majority of the PRDM1 3′UTR that harbors all of the putative binding sites for the two miRNAs (Figure 2A). This plasmid was cotransfected into 293T cells with miR-9, let-7a, miR-155, or negative miR controls. No mir-155 binding sites were predicted in the PRDM1 3′UTR. MiR-9 and let-7a decreased normalized Renilla luciferase activities to 39.2% ± 4.6%, and 41.7% ± 3.8% of negative controls, respectively, whereas cotransfection with miR-155 showed no significant difference (Figure 3A). Reduction in luciferase activities by miR-9 and let-7a is mediated mainly by translation repression. No significant difference in Renilla luciferase mRNA levels was detected among the negative control, miR-9 and let-7a transfectants after normalization, although miR-9 transfectants may tend to show a slight reduction (Figure 3B).
Figure 3.
MiR-9 and let-7a target PRDM1 3′UTR and inhibit reporter activity. A: 293T cells were transfected with wild-type psiC.PRDM1/3′UTR.538-2419 or one of the mutant reporter plasmids harboring point mutations in the target sites for miR-9 or let-7a. For miR-9 binding sites, mutation is present in either one of the three sites (Mut 1, 2, or 3), two of the three sites (Mut [2 + 3]) or all three sites (Mut [1 + 2 + 3]). These reporter plasmids are co-transfected with miRNA expression plasmids (75 ng) pcDNA3.miR-9–1 (miR-9) or pcDNA3.BIC/miR-155 (miR-155), or with precursor let-7a (20 nmol/L) obtained from Ambion. pcDNA3 plasmid or miRNA negative control oligonucleotides (Ambion) were used as negative controls. Luciferase activities (in triplicates) were measured 24 hours after transfection. Renilla luciferase activities were normalized against firefly luciferase activities, and mean normalized Renilla luciferase activities (±SE) from three or four independent experiments were determined and expressed relative to control values. B: Levels of Renilla luciferase mRNA in 293T cells transfected with wild-type psiC.PRDM1/3′UTR.538-2419 and either miRNA Negative Control oligonucleotides pcDNA3.miR-9-1, or precursor let-7a (see above) were measured by quantitative real time PCR and normalized against firefly luciferase mRNA expression. The mean (±SE) from three independent experiments were shown. There is no significant difference in the miR-9 (P = 0.06) or let-7a (P = 0.24) transfectants compared with the negative control. C: 293T cells were transfected with psiC.PRDM1/3′UTR.538-2419 and precursor miRNAs (Ambion) as indicated. Relative luciferase activities were calculated as in (A). Transfection of both miR-9 and let-7a (each at 50 nmol/L) resulted in significantly greater reduction of luciferase activities compared with transfection of either miR-9 or let-7a (at 50 nmol/L) with negative miRNA control (at 50 nmol/L). Transfection of miR-9 at 100 nmol/L leads to further repression in luciferase activities, whereas let-7a at 100 nmol/L results in no further decrease.
To demonstrate that both miRNAs interact specifically with their predicted target sequences, additional reporter constructs harboring mutations in the “seed pairing” sequences of the putative binding sites were generated using site-directed mutagenesis. These mutated reporter constructs were transfected into 293T cells with the corresponding miRNAs as described above and luciferase reporter assays were measured. Mutations in the predicted target sites for each of these miRNAs relieve repression of luciferase activities (Figure 3A). These results indicate that miR-9 and let-7 can repress translation by direct and specific interaction with the PRDM1 3′UTR.
Interestingly for miR-9, the extent of de-repression depends on which and how many miR-9 binding sites are mutated. Mutations in the most proximal site (Mut 1) resulted in partial relief in repression (77.0% ± 2.5% of negative control), whereas mutations in each of the other two miR-9 binding sites (Mut 2 and Mut 3) do not have significant effects compared with wild type (48.6% ± 2.4% and 45.2% ± 2.6%). Mutations in the middle and distal sites (Mut[2 + 3]) resulted in a slight de-repression (59.1% ± 4.7% of negative control), whereas mutations in all three miR-9 binding sites (Mut[1 + 2 + 3]) led to a complete de-repression (117.5% ± 9.9%). These results suggest a differential, as well as cooperative, repression effect for the three miR-9 target sites in the PRDM1 3′ UTR.
We tested whether interaction of both miR-9 and let-7a with the PRDM1 3′UTR may increase the repression effects relative to either miRNA alone by cotransfecting psiC.PRDM1.3′UTR.538–2409 with miR-9 and let-7a. Cotransfection of miR-9 and let-7a (each at 50 nmol/L) reduced luciferase activity to 19.7% ± 1.5% of negative control, significantly different from the effects of miR-9 or let-7a alone (P < 0.05 and P < 0.001, respectively) (Figure 3B). This increase in repression is likely due to additive effects of miR-9 and let-7a, because similar repression was obtained when miR-9 concentration was increased to 100 nmol/L. Let-7a at 50 nmol/L or 100 nmol/L shows no significant difference in repression, suggesting that it has already reached saturation at these concentrations.
High miR-9 and let-7 Levels Are Associated with Low Levels of PRDM1 Expression in HRS Cells
If miR-9 and let-7a functionally interact with endogenous PRDM1 mRNA to down-regulate PRDM1 protein expression in HRS cells, we should expect association of high levels of miR-9 and let-7a with relatively lower levels of PRDM1 in these cells. For comparisons, we selected cell lines that exhibit plasmablastic/plasmacytic differentiation such as PEL and myeloma cell lines, and cell lines that correspond to the GC differentiation stage such as the GC-DLBCL cell lines.
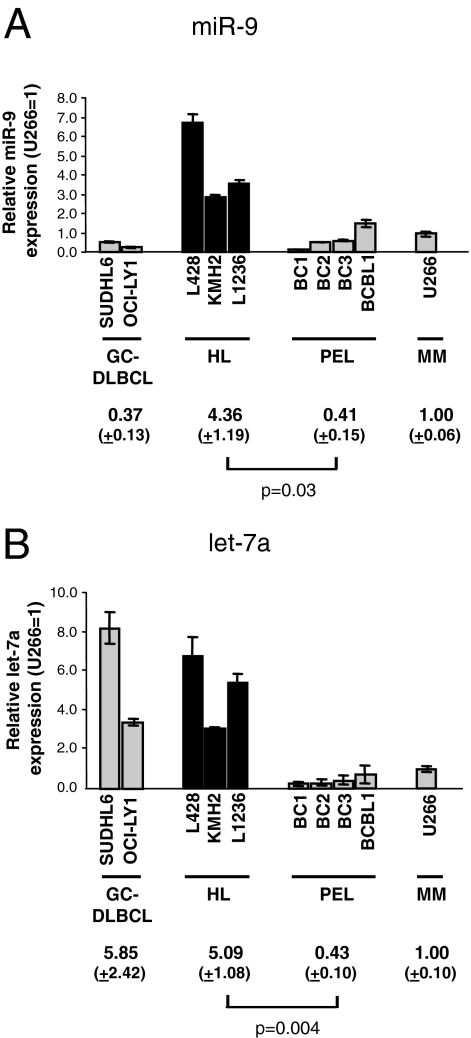
Real-time RT-PCR analysis confirmed the miRNA cloning data and showed that miR-9 expression in HL cell lines is distinctly higher: ∼10-fold of PEL lines (P = 0.03), ∼4-fold of U266, and ∼12-fold of GC-DLBCL lines. Let-7a expression levels in HL cell lines are similar to GC-DLBCL lines, but ∼11-fold that of PEL lines (P = 0.004) and ∼5 fold of U266 (Figure 4).
Figure 4.
HL cell lines express relatively high levels of miR-9 and let-7a. A and B: The relative miR-9 and let-7a levels in GC-DLBCL, HL, PEL, and myeloma cell lines determined by quantitative real-time PCR were shown. The level in U266 was arbitrarily set as 1.
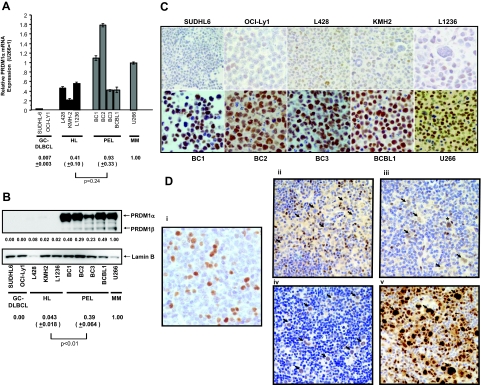
As a group, the steady-state levels of PRDM1α mRNA levels in HL cell lines are approximately half of those in PEL cell lines and U266, but did not reach statistical significance (P = 0.24) (Figure 5A). On the other hand, PRDM1α is present at much lower levels (∼9 to 25-fold lower) in the HL cell lines compared with PEL lines (P = 0.007) and U266 (Figure 5B). Further comparison of individual cell lines from the HL and PEL groups further illustrates the discordance in PRDM1 mRNA and protein levels in HL cell lines. For example, whereas the HL cell line L1236 has similar levels of PRDM1α mRNA as the PEL cell lines BC3 and BCBL1, PRDM1 expression in L1236 is ∼10 to 20-fold lower. PRDM1 is undetectable in GC-DLBCL lines, as expected from their low levels of transcript expression.
Figure 5.
HRS cells show evidence of PRDM1 mRNA induction but express low levels of PRDM1. A: Real-time PCR analysis of PRDM1α mRNA in GC-DLBCL, HL, PEL, and myeloma cell lines using specific primers as described in Materials and Methods. The actual relative expression levels are shown (U266 as 1). B: Western blot analysis of nuclear extracts prepared from the above cell lines using the monoclonal antibodies (ROS clone) against PRDM1. Both PRDM1α (∼90 kDa) and PRDM1β (∼80 kDa) were indicated. The same blot was reprobed with anti-lamin B polyclonal antibodies for normalization of nuclear protein quantity. The relative protein amount of PRDM1α after densitometric measurement and normalization was indicated for each cell line (U266 = 1). C: Immunoperoxidase staining on formalin-fixed, paraffin-embedded cell blocks using the anti-PRDM1 monoclonal antibodies (ROS clone). Though scattered PRDM1-positive cells were detected in the HL cell lines, PRDM expression in PEL and U266 cells is much stronger and more uniform. D: Immunohistochemistry performed on formalin-fixed, paraffin-embedded tissue sections of clinical HL cases with the monoclonal anti-PRDM1 antibodies (ROS clone). Representative results are shown. Normal tonsils were stained as a control. i: A subset of PRDM1(+) cells within a reactive GC in a hyperplastic tonsil. ii: PRDM1-negative HRS cells (arrows). iii: HRS cells weakly positive for PRDM1 (arrows). Note their relatively weaker staining compared with the surrounding scattered reactive cells. A HRS cell that has relatively strong (2+) staining intensity is also seen (asterisk). iv: A rare case in which most HRS cells show strong reactivity toward PRDM1 (arrows). Their staining intensity is similar to the surrounding reactive cells. v: HRS cells strongly express IRF4/MUM1 (arrows). C and D, i–v are shown at original magnification = ×400).
In keeping with the Western blotting results, immunohistochemical staining on formalin-fixed, paraffin-embedded cell blocks prepared from the cell lines using the ROS monoclonal antibody showed strong and uniform reactivity in the PEL and U266 cells. In contrast, only a small fraction of relatively weaker positive cells were detected in the HL cell lines. SUDHL6 and OCI-Ly1 cells show few positive cells (Figure 5C).
PRDM1 expression in primary HRS cells reflects their in vitro counterparts (Table 1; Figure 5D). In 13 of the 21 HL cases (62%) examined, positivity for PRDM1 was detected in 20% or fewer of the HRS cells. In 8/21 of the cases, PRDM1 is expressed in more than 20% of the HRS cells, although the majority of them exhibit only weak positivity. In one case (#13), strong PRDM1 expression was observed in the majority of HRS cells. A variable fraction (10 to 70%) of reactive lymphocytes and plasma cells are also positive for PRDM1 in each case. Thus, PRDM1 is either absent or expressed at low levels in the majority of primary HRS cells, a finding similar to previously published results.18,21 In addition, PRDM1 expression in primary HRS cells is independent of Epstein Barr Virus infection status. In contrast, HRS cells consistently and strongly express IRF4/MUM1 (Figure 5D). Because PRDM1 and IRF4/MUM1 are normally co-expressed in GC centrocytes that are committed to the plasma cell differentiation pathway and both factors are required for the generation of competent plasma cells,9 the absence or low level expression of PRDM1 in HRS cells implies a pathogenetic dissociation between PRDM1 and IRF4/MUM1 expression.
Table 1.
PRDM1 Expression in HRS Cells in Primary Hodgkin Lymphoma Cases
| Cases | EBV | PRDM1 (% H/RS cells)
|
||
|---|---|---|---|---|
| 0 | 1+ (weak) | 2+ (strong) | ||
| 1 | N | 100 | 0 | 0 |
| 2 | P | 100 | 0 | 0 |
| 3 | N | 30 | 40 | 30 |
| 4 | N | 70 | 30 | 0 |
| 5 | P | 95 | 5 | 0 |
| 6 | N | 95 | 5 | 0 |
| 7 | N | 35 | 65 | 5 |
| 8 | N | 30 | 60 | 10 |
| 9 | P | 80 | 20 | 0 |
| 10 | N | 100 | 0 | 0 |
| 11 | N | 95 | 5 | 0 |
| 12 | N | 100 | 0 | 0 |
| 13 | N | 20 | 10 | 70 |
| 14 | N | 90 | 10 | 0 |
| 15 | N | 45 | 50 | 5 |
| 16 | N | 95 | 5 | 0 |
| 17 | P | 100 | 0 | 0 |
| 18 | P | 95 | 5 | 0 |
| 20 | P | 20 | 75 | 5 |
| 21 | P | 95 | 5 | 0 |
| 22 | P | 20 | 80 | 0 |
| Average | 72 | 22 | 6 | |
| SE | 7 | 6 | 4 | |
P, positive; N, negative; SE, standard error.
Our results indicate that cultured HRS cells, as well as HRS cells of primary HL cases, harbor low levels of PRDM1 that correlate better with miR-9 and let-7a expression than PRDM1 transcript levels. This data provides circumstantial evidence for down-regulation of PRDM1 expression in HRS cells by miR-9 and let-7a. In contrast, the low levels of PRDM1 in GC-DLBCL cells appear to be mediated by transcriptional repression.
Manipulation of miR-9 or let-7a Levels Alters Endogenous PRDM1 Expression
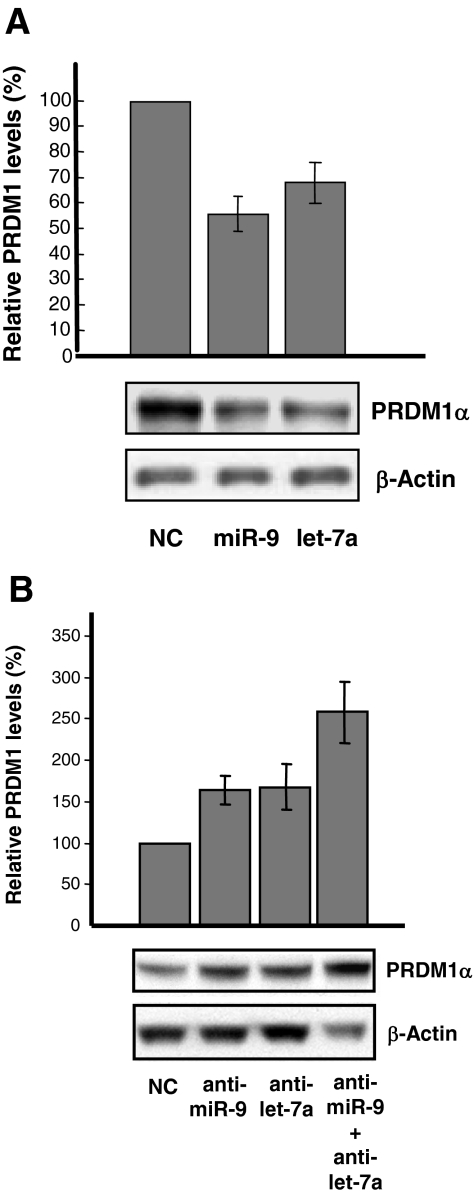
To confirm further that PRDM1 is an in vivo target for miR-9 and let-7a, U266 cells were transfected with miR-9, let-7a or negative miRNA controls. The levels of PRDM1 were determined by Western blotting 24 to 48 hours after transfection. MiR-9 or let-7a transfected at a concentration of 100 nmol/L reduced endogenous PRDM1 expression in U266 cells by ∼30 to 50% relative to negative controls (Figure 6A).
Figure 6.
Changes in miR-9 or let-7a levels alter endogenous PRDM1 expression in U266 and L428 cells. A: U266 cells were transfected with miR-Control (NC), miR-9 or let-7a (each at 100 nmol/L). Total cell lysates were isolated from the transfected cells after 24 to 48 hours and PRDM1 levels were measured by Western blot analysis using the anti-PRDM1 monoclonal antibody (ROS). β-actin was used as a loading control. Quantitation of PRDM1α expression was determined by densitometry, normalized with β-actin, and expressed as a percent relative to the PRDM1 expression level in cells transfected with miR-Control. The histogram was derived from four independent transfection experiments. A representative Western blot was also shown. B: L428 cells were transfected with anti-miRNA negative control (NC), anti-miR-9, and/or anti-let-7a at a total concentration of 100 nmol/L. Western blot analysis and quantitation were performed as described in (A) on total cell lysates prepared 24 to 48 hours after transfection. The histogram was derived from four independent transfection experiments. A representative Western blot was also shown. P = 0.05 (anti-miR-9 vs. anti-miR-9/anti-let-7a); P = 0.10 (anti-let-7a versus anti-miR-9/anti-let-7a).
To directly demonstrate that PRDM1 expression in HL cell lines is negatively regulated by endogenous levels of miR-9 and/or let-7a, L428 cells were transfected with miR-9 and/or let-7a antisense RNA oligonucleotides to reduce their endogenous levels, and PRDM1 expression was determined by Western blotting 24 to 48 hours after transfection. Cells transfected with anti-miR-9 or anti-let-7a showed an increase in PRDM1 by 64.5% ± 17.4% and 67.6% ± 27.4%, respectively, compared with negative control anti-miRNA-transfected cells. Inhibition of both miR-9 and let-7a resulted in a greater increase in PRDM1 expression, up to 2.6 ± 0.37-fold that of negative control (Figure 6B). This increase is significantly different from that observed for anti-miR-9 alone (P = 0.05). These results suggest that both miR-9 and let-7a contribute to down-regulate PRDM1 expression in L428 cells and are in line with the additive regulatory effects observed between miR-9 and let-7a in the luciferase reporter assays (Figure 3B).
Discussion
PRDM1 as a Target of miR-9 and let-7a
Our study proposed an epigenetic mechanism that may account for the low-level PRDM1 expression in HRS cells of HL cell lines and primary HL cases. Several lines of evidence presented in this report strongly suggest that PRDM1 expression in HRS cells can be negatively regulated by two endogenous miRNAs, miR-9 and let-7a, through direct interaction with the 3′UTR of PRDM1 mRNA. First, the PRDM1 3′UTR contains complementary binding sties for these two miRNAs, which are among the major miRNAs in HL cell lines. Second, these miRNAs can translationally repress expression of a reporter gene through specific interactions with their target sites in the PRDM1 3′UTR. Interestingly, although all three putative miR-9 binding sites predicted in the PRDM1 3′UTR show conserved pairing with the miR-9 seed sequence, their regulatory effects differ. The most proximal target site appears to be the major effector, whereas the other two have minor contributory roles. All three sites cooperate to produce a maximal effect. Thus, the functionality of a miRNA binding site may not be determined solely by its seed pairing sequence but also by its surrounding context.33 In addition, miR-9 and let-7a can cooperate in their repressive effects. Third, a reciprocal relationship can be demonstrated between PRDM1 and miR-9/let-7a expression in HL cell lines (PRDM1 low, miR-9/let-7a high) and in PEL or U266 cell lines (PRDM1 high, miR-9/let-7a low). Fourth, enforced expression of these miRNAs in U266 cells caused a reduction in PRDM1 levels. Moreover, we observed an increase in endogenous PRDM1 expression on inhibition of miR-9 and/or let-7a activity in the HL cell line L428, providing evidence that these two miRNAs physiologically down-regulate PRDM1 expression in HRS cells. However, the actual extent by which these miRNAs down-regulate PRDM1 expression in vivo is unclear. We are able to achieve a maximum of about 50% inhibition of miR-9 and let-7a activity using the miR-9/let-7a antagomirs in L428 cells based on estimation from the reduction in endogenous miRNA levels34 (data not shown). Considering this degree of knock-down, it is possible that miR-9 and let-7a have greater in vivo effects than what is shown in our experiments. However, we cannot exclude the possibility that other mechanisms unrelated to miRNA may also contribute in lowering PRDM1 levels in HRS cells.
The mechanism(s) responsible for the high levels of miR-9 and let-7 in HL cell lines remain(s) to be elucidated. Though the expression patterns of miR-9 in different cell types and tissues have not been studied in detail, it appears to be preferentially expressed in neural tissues.35,36 Deregulation of miR-9 expression has been observed in tumors of epithelial cell origin.37,38 Recently, miR-9, along with miR-17-92 cluster and several other miRNAs, are found to be differentially expressed in some hematological cell lines relative to normal lymphocyte populations based on microarray profiling.39 In addition, miR-9 may play a role in B cell activation, being up-regulated during in vitro activation of B cells by interleukin (IL)-2 or IgM.39 Thus, it is conceivable that the high levels of miR-9 observed in HRS cells reflect an exaggerated response to signals related to B cell activation. Let-7 has been implicated as a tumor suppressor.40 However, an oncogenic potential for let-7a has also been suggested.41 The ability of let-7a to down-regulate PRDM1 suggests that it can function as an oncogene in HRS cells. Our expression analysis on let-7a, which shows high let-7a expression in GC-DLBCL and HL cell lines and much lower let-7a expression in PEL and myeloma cell lines, suggests let-7a down-regulation on plasmablastic differentiation. Down-regulation of pre-let-7a in PEL cell lines has also been observed in a recent microarray profiling study.42 Whether the high let-7a observed in cultured HRS cells is due to deregulation as a result of genetic or epigenetic abnormalities or is simply a reflection of the differentiation stage needs to be determined by further experiments.
Translation Repression of PRDM1 by miRNAs as a Potential Novel Molecular Lesion in HL
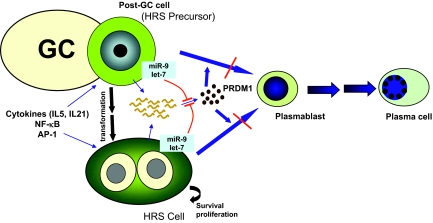
Direct demonstration of the biological consequences of miRNA-mediated down-regulation of PRDM1 in HRS cells will probably require more stable inhibition of miRNA function in selected HRS cell clones. However, a hypothetical model can be inferred based on the known biological functions of PRDM1 as a master regulator of plasma cell differentiation and as a tumor suppressor (Figure 7). Full differentiation of an activated B cell to a mature plasma cell proceeds through different developmental stages associated with a gradual quantitative increase in PRDM1 expression,43 beginning with a PRDM1/Blimp-1-independent stage followed by a PRDM1/Blimp-1-dependent stage.44 HRS cells are constantly exposed to an environment containing cytokines that may induce PRDM1 expression, for example, IL-5 and IL-21.45,46,47,48 In addition, a constitutively activated nuclear factor-κB14 or AP-149 in HRS cells may up-regulate PRDM1 expression.50 As suggested by the induction of PRDM1 mRNA in HL cell lines, plasma cell differentiation appears to have been initiated in HRS cells. However, it is conceivable that the dampening effect of miRNA on PRDM1 production prevents PRDM1 from reaching a critical level in HRS cells, resulting in abortive terminal differentiation and arrest in the PRDM1-independent, pre-plasmablastic stage (Figure 7). This interference is advantageous for the continued survival and growth of HRS cells and may also be pathogenetic in the transformation of postgerminal center B cells to HRS cells. Additional experiments involving more long-term, regulatable manipulation of miRNAs and PRDM1 expression will help address the above hypothesis.
Figure 7.
A hypothetical model of microRNA-mediated PRDM1 inactivation in pathogenesis of HRS cells. PRDM1 mRNA may be induced in HRS cells and precursors by certain cytokines (eg, IL-5, IL-21) and by a constitutively active nuclear factor-κB or AP1, which may drive the cells toward plasma cells differentiation (blue arrows). However, they may fail to accumulate sufficiently high levels of PRDM1 to complete the terminal differentiation process despite induction of PRDM1 transcripts because of translation repression by miRNAs such as miR-9 and let-7a (red lines). As a result, the plasma cell differentiation program is initiated but aborted, possibly at the pre-plasmablastic stage. This microRNA-mediated down-regulation of PRDM1 may contribute to the pathogenesis and phenotype maintenance of HRS cells.
In view of the recent findings of PRDM1 as a target for mutational inactivation in activated B cells-DLBCL11,12 and transcription repression in nm-GCB-DLBCL,13 our data suggests that disruption of normal plasma cell differentiation through functional inactivation of PRDM1 is a common event in the pathogenesis of lymphomas derived from GC B cells. The normal functions of this tumor suppressor gene can be inactivated by genetic mutations, transcriptional repression or epigenetic alterations such as miRNA-mediated down-regulation.
Role of the miRNA Milieu in HL Pathogenesis
Ours studies provide supporting evidence that the miRNA milieu present in HRS cells has the potential to contribute to their pathogenesis and maintenance by down-regulating genes important for B-cell development and differentiation. It is likely that the pathobiological functions of miR-9 and let-7 in HRS cells are mediated not only through PRDM1, but also through multiple other targets. Our miRNA profiling data identified other miRNAs that are highly expressed in HL cell lines, for example, miR-155. Its functions in HRS cells are not yet clear, but may be related to its normal role in B-cell immunity. MiR-155 plays a critical role in adaptive immune responses.51,52 MiR-155 deficient mice demonstrated an intrinsic defect in the generation of high-affinity, class-switched post-GC plasma cells and memory B cells,53 suggesting that miR-155 regulates selection of GC B cells and the formation and/or maintenance of a common precursor before final differentiation in the GC.54 The high miR-155 in HRS cells may facilitate processes related to GC cell selection, such as functioning of cytokine receptors and response to T cells. Identification of miR-155 target genes in HRS cells might explain how this miRNA contributes to their pathogenesis of HL. Future research is necessary to better define the role of the miRNA environment that modulates expression of protein factors critical for the pathogenesis of HL.
Acknowledgments
We thank Dr. Ethel Cesarman for providing the PEL cell lines.
Footnotes
Address reprint requests to Wayne Tam, Department of Pathology & Laboratory Medicine, Weill Medical College of Cornell University, 525 East 68th St., ST 711A, New York, NY 10021. E-mail: wtam@med.cornell.edu.
Supported by departmental funds for translational research from the Department of Pathology and Laboratory Medicine at Weill Cornell (to W.T.).
Current address for P.L.: Children’s Hospital of the Heinrich-Heine University Düsseldorf, Germany.
Current address for J.-F. G.: Dako Diagnosticos S.A., Barcelona, Spain.
References
- Gyory I, Fejer G, Ghosh N, Seto E, Wright KL. Identification of a functionally impaired positive regulatory domain I binding factor 1 transcription repressor in myeloma cell lines. J Immunol. 2003;170:3125–3133. doi: 10.4049/jimmunol.170.6.3125. [DOI] [PubMed] [Google Scholar]
- Kallies A, Nutt SL. Terminal differentiation of lymphocytes depends on Blimp-1. Curr Opin Immunol. 2007;19:156–162. doi: 10.1016/j.coi.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Magnusdottir E, Kalachikov S, Mizukoshi K, Savitsky D, Ishida-Yamamoto A, Panteleyev AA, Calame K. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc Natl Acad Sci USA. 2007;104:14988–14993. doi: 10.1073/pnas.0707323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, de Sousa Lopes SM, Surani MA. Germ cell specification in mice. Science. 2007;316:394–396. doi: 10.1126/science.1137545. [DOI] [PubMed] [Google Scholar]
- Robertson EJ, Charatsi I, Joyner CJ, Koonce CH, Morgan M, Islam A, Paterson C, Lejsek E, Arnold SJ, Kallies A, Nutt SL, Bikoff EK. Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice. Development. 2007;134:4335–4345. doi: 10.1242/dev.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Shapiro-Shelef M, Tunyaplin C, Calame K. Regulatory events in early and late B-cell differentiation. Mol Immunol. 2005;42:749–761. doi: 10.1016/j.molimm.2004.06.039. [DOI] [PubMed] [Google Scholar]
- Angelin-Duclos C, Cattoretti G, Lin KI, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J Immunol. 2000;165:5462–5471. doi: 10.4049/jimmunol.165.10.5462. [DOI] [PubMed] [Google Scholar]
- Cattoretti G, Angelin-Duclos C, Shaknovich R, Zhou H, Wang D, Alobeid B. PRDM1/Blimp-1 is expressed in human B-lymphocytes committed to the plasma cell lineage. J Pathol. 2005;206:76–86. doi: 10.1002/path.1752. [DOI] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- Tam W, Gomez M, Chadburn A, Lee JW, Chan WC, Knowles DM. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:4090–4100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, Aster JC, Murty VV, Shipp MA, Dalla-Favera R. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh S, Polo JM, Shaknovich R, Juszczynski P, Lev P, Ranuncolo SM, Yin Y, Klein U, Cattoretti G, Dalla Favera R, Shipp MA, Melnick A. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood. 2007;110:2067–2074. doi: 10.1182/blood-2007-01-069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt LM. The molecular and cellular origins of Hodgkin’s disease. J Exp Med. 2000;191:207–212. doi: 10.1084/jem.191.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppers R, Hansmann ML. The Hodgkin and Reed/Sternberg cell. Int J Biochem Cell Biol. 2005;37:511–517. doi: 10.1016/j.biocel.2003.10.025. [DOI] [PubMed] [Google Scholar]
- Carbone A, Gloghini A, Aldinucci D, Gattei V, Dalla-Favera R, Gaidano G. Expression pattern of MUM1/IRF4 in the spectrum of pathology of Hodgkin’s disease. Br J Haematol. 2002;117:366–372. doi: 10.1046/j.1365-2141.2002.03456.x. [DOI] [PubMed] [Google Scholar]
- Carbone A, Gloghini A, Gaidano G, Franceschi S, Capello D, Drexler HG, Falini B, Dalla-Favera R. Expression status of BCL-6 and syndecan-1 identifies distinct histogenetic subtypes of Hodgkin’s disease. Blood. 1998;92:2220–2228. [PubMed] [Google Scholar]
- Buettner M, Greiner A, Avramidou A, Jack HM, Niedobitek G. Evidence of abortive plasma cell differentiation in Hodgkin and Reed-Sternberg cells of classical Hodgkin lymphoma. Hematol Oncol. 2005;23:127–132. doi: 10.1002/hon.764. [DOI] [PubMed] [Google Scholar]
- Kuppers R, Klein U, Schwering I, Distler V, Brauninger A, Cattoretti G, Tu Y, Stolovitzky GA, Califano A, Hansmann ML, Dalla-Favera R. Identification of Hodgkin and Reed-Sternberg cell-specific genes by gene expression profiling. J Clin Invest. 2003;111:529–537. doi: 10.1172/JCI16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: a primer. Am J Pathol. 2007;171:728–738. doi: 10.2353/ajpath.2007.070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JF, Roncador G, Sanz AI, Maestre L, Lucas E, Montes-Moreno S, Fernandez Victoria R, Martinez-Torrecuadrara JL, Marafioti T, Mason DY, Piris MA. PRDM1/BLIMP-1 expression in multiple B and T-cell lymphoma. Haematologica. 2006;91:467–474. [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. Epub 2002 Jun 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossos IS, Czerwinski DK, Wechser MA, Levy R. Optimization of quantitative real-time RT-PCR parameters for the study of lymphoid malignancies. Leukemia. 2003;17:789–795. doi: 10.1038/sj.leu.2402880. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- Tam W, Dahlberg JE. miR-155/BIC as an oncogenic microRNA. Genes Chromosomes Cancer. 2006;45:211–212. doi: 10.1002/gcc.20282. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Saunders NJ, Soneji S, Palazzo S, Dunlop HM, Cooper CD, Brown PJ, Troussard X, Mossafa H, Enver T, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS. MicroRNA expression in lymphocyte development and malignancy. Leukemia. 2008 doi: 10.1038/sj.leu.2405083. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, Sultmann H, Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- O'Hara A, Vahrson W, Dittmer DP. Gene alteration and precursor and mature microRNA transcription changes contribute to the miRNA signature of primary effusion lymphoma. Blood. 2008;111:2347–2353. doi: 10.1182/blood-2007-08-104463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Hasbold J, Tarlinton DM, Dietrich W, Corcoran LM, Hodgkin PD, Nutt SL. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J Exp Med. 2004;200:967–977. doi: 10.1084/jem.20040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Hasbold J, Fairfax K, Pridans C, Emslie D, McKenzie BS, Lew AM, Corcoran LM, Hodgkin PD, Tarlinton DM, Nutt SL. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 2007;26:555–566. doi: 10.1016/j.immuni.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Maggio E, van den Berg A, Diepstra A, Kluiver J, Visser L, Poppema S. Chemokines, cytokines and their receptors in Hodgkin’s lymphoma cell lines and tissues. Ann Oncol. 2002;13 Suppl 1:52–56. doi: 10.1093/annonc/13.s1.52. [DOI] [PubMed] [Google Scholar]
- Horikawa K, Takatsu K. Interleukin-5 regulates genes involved in B-cell terminal maturation. Immunology. 2006;118:497–508. doi: 10.1111/j.1365-2567.2006.02382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K, Spolski R, Ettinger R, Kim HP, Wang G, Qi CF, Hwu P, Shaffer DJ, Akilesh S, Roopenian DC, Morse HC, 3rd, Lipsky PE, Leonard WJ. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J Immunol. 2004;173:5361–5371. doi: 10.4049/jimmunol.173.9.5361. [DOI] [PubMed] [Google Scholar]
- Ettinger R, Sims GP, Fairhurst AM, Robbins R, da Silva YS, Spolski R, Leonard WJ, Lipsky PE. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. J Immunol. 2005;175:7867–7879. doi: 10.4049/jimmunol.175.12.7867. [DOI] [PubMed] [Google Scholar]
- Mathas S, Hinz M, Anagnostopoulos I, Krappmann D, Lietz A, Jundt F, Bommert K, Mechta-Grigoriou F, Stein H, Dorken B, Scheidereit C. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappa B. EMBO J. 2002;21:4104–4113. doi: 10.1093/emboj/cdf389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo Y, Arima M, Arguni E, Okada S, Yamashita K, Asari S, Obata S, Sakamoto A, Hatano M, O-Wang J, Ebara M, Saisho H, Tokuhisa T. A role for c-fos/activator protein 1 in B lymphocyte terminal differentiation. J Immunol. 2005;174:7703–7710. doi: 10.4049/jimmunol.174.12.7703. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calame K. MicroRNA-155 function in B cells. Immunity. 2007;27:825–827. doi: 10.1016/j.immuni.2007.11.010. [DOI] [PubMed] [Google Scholar]