Abstract
DinB, a Y-family DNA polymerase, is conserved among all domains of life; however, its endogenous substrates have not been identified. DinB is known to synthesize accurately across a number of N2-dG lesions. Methylglyoxal (MG) is a common byproduct of the ubiquitous glycolysis pathway and induces the formation of N2-(1-carboxyethyl)-2′-deoxyguanosine (N2-CEdG) as the major stable DNA adduct. Here, we found that N2-CEdG could be detected at a frequency of one lesion per 107 nucleosides in WM-266-4 human melanoma cells, and treatment of these cells with MG or glucose led to a dose-responsive increase in N2-CEdG formation. We further constructed single-stranded M13 shuttle vectors harboring individual diastereomers of N2-CEdG at a specific site and assessed the cytotoxic and mutagenic properties of the lesion in wild-type and bypass polymerase-deficient Escherichia coli cells. Our results revealed that N2-CEdG is weakly mutagenic, and DinB (i.e., polymerase IV) is the major DNA polymerase responsible for bypassing the lesion in vivo. Moreover, steady-state kinetic measurements showed that nucleotide insertion, catalyzed by E. coli pol IV or its human counterpart (i.e., polymerase κ), opposite the N2-CEdG is both accurate and efficient. Taken together, our data support that N2-CEdG, a minor-groove DNA adduct arising from MG, is an important endogenous substrate for DinB DNA polymerase.
Keywords: glycolysis, mutagenesis, polymerase κ, translesion synthesis
Living cells are constantly exposed to environmental and endogenous agents, which can damage DNA (1). To counteract the deleterious effects of DNA lesions, cells have evolved an intricate DNA repair system (2). When DNA repair is not efficient enough, the presence of unrepaired DNA lesions in replicating DNA may lead to replication fork stalling, thereby inducing cell death. It was proposed that, when a high-fidelity replication fork is arrested at the lesion site, translesion synthesis polymerases can take over from replicative polymerases temporarily to bypass synthetically the lesion residing in the template (2, 3). Among the Y-family DNA polymerases, DinB [also known as polymerase (pol) IV in Escherichia coli and pol κ in mammalian cells] is conserved among all domains of life; however, the basis for this marked conservation remains unclear (4). In particular, the physiological substrates for this polymerase remain to be identified.
Similar to eukaryotic pol η, which is specialized in bypassing efficiently and accurately TT cyclobutane pyrimidine dimer (5–7), E. coli pol IV and human pol κ can insert preferentially the correct nucleotide, dCMP, opposite a number of N2-dG lesions (8, 9). In addition, DinB polymerase is capable of bypassing the N2-dG adduct induced by benzo[a]pyrene-7,8-diol-9,10-epoxide (BPDE) (10–14).
Aside from the difference in substrate specificity, E. coli pol IV distinguishes from pol V in expression levels (4). In uninduced cells, pol V is not detectable by Western analysis, whereas pol IV can be detected at a level of ≈250 copies per cell. Upon SOS induction, there are only ≈15 molecules of pol V per cell; however, the level of pol IV reaches ≈2,500 copies per cell (4). The high level of expression of pol IV suggests there is an important, basic, and yet-to-be-discovered function of pol IV in general metabolism (4).
In addition to reactive oxygen species, which constitutes a major endogenous source of DNA damage (1, 15), genomic DNA in living cells is susceptible to damage from exposure to reactive carbonyl species, and methylglyoxal (MG) is one of them. In this respect, MG can be produced endogenously in all cells and all organisms from the nonenzymatic fragmentation of triose phosphates, which include glyceraldehyde-3-phosphate and dihydroxyacetone phosphate, and are produced as metabolites of the highly conserved glycolysis pathway (16–18). In this regard, treatment of human red blood cells with increasing concentrations of glucose in vitro can result in increases of intracellular MG concentration (19). The MG concentration was also found to be elevated in the kidney (cortex and medulla), lens, and blood of streptozotocin-induced diabetic rats (20) and in blood samples of diabetic patients (21).
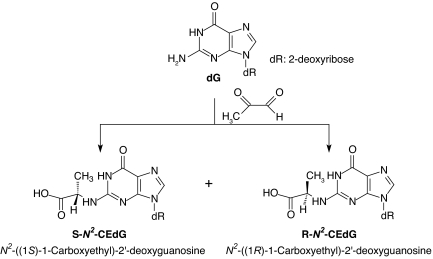
N2-(1-carboxyethyl)-2′-deoxyguanosine (N2-CEdG) was the major stable adduct formed in calf thymus DNA upon exposure to MG at physiological concentration and temperature (Fig. 1) (22). A competitive enzyme-linked immunosorbant assay showed that the adduct can be detected in urine samples of 121 healthy human subjects at levels ranging from 1.2- to 117-ng N2-CEdG equivalent per milligram of creatinine (23). Immunohistochemistry using a monoclonal antibody against N2-CEdG revealed that the level of the lesion is enhanced in the kidney and aorta of patients with diabetic nephropathy and uremic atherosclerosis, respectively (24).
Fig. 1.
Formation of N2-CEdG.
DNA adducts arising from MG could induce mutations in E. coli cells and G → C and G → T transversions in supF gene in mammalian cells (25, 26). However, the lesion-containing DNA substrates used in these previous mutagenesis experiments were prepared by the direct treatment of undamaged DNA with dihydroxyacetone or MG (25, 26); thus, the identity and homogeneity of the DNA adducts were not carefully assessed. In light of the previous findings that DinB is capable of bypassing, in an error-free fashion, a number of N2-dG lesions (8–14), we reason that this polymerase may also be involved in the bypass of N2-CEdG.
In the present study, we prepared single-stranded M13 genomes carrying individual diastereomers of N2-CEdG and assessed the cytotoxic and mutagenic properties of the lesion in E. coli cells. Our results showed that pol IV was the major polymerase responsible for the error-free bypass of N2-CEdG in vivo. Steady-state kinetic measurements revealed that the nucleotide incorporation, catalyzed by E. coli pol IV or its human homolog (i.e., pol κ), is both accurate and efficient. In addition, N2-CEdG could be detected in untreated human cells, and exposure of cells to glucose or MG enhanced the formation of N2-CEdG. These observations support that N2-CEdG is an important endogenous substrate for DinB DNA polymerase.
Results
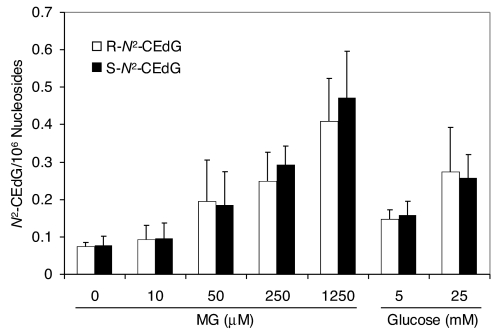
To explore the possibility that N2-CEdG might constitute an endogenous substrate for DinB DNA polymerase, we first assessed the formation of N2-CEdG in WM-266-4 human melanoma cells that are either untreated or treated with glucose or MG. LC-MS/MS analysis using the accurate isotope-dilution method revealed that both diastereomers of N2-CEdG could be detected in untreated cells at a level of one lesion per 107 nucleosides [Fig. 2; LC-MS/MS data and calibration curves are shown in supporting information (SI) Figs. S1 and S2]. In addition, incubation of the melanoma cells with MG led to a dose-dependent increase in the level of N2-CEdG (Fig. 2). Moreover, culturing of these cells in media containing 5 and 25 mM glucose for 5 days resulted in the increase of the levels of the lesions to approximately 2.5 and 4 lesions per 107 nucleosides (Fig. 2). In this context, it is worth noting that, in response to change in blood glucose level, glucose transporters are regulated in some types of cells (27). Therefore, the intracellular glucose concentration in WM-266-4 cells may not increase proportionally with the glucose concentration in the culture media, which may explain why the levels of N2-CEdG lesions induced in glucose-treated cells are not proportional to the applied glucose dose. Taken together, both diastereomers of N2-CEdG can be induced endogenously in WM-266-4 human melanoma cells, and the exposure of these cells to MG or glucose further enhances the formation of N2-CEdG. In this context, it is worth noting that the two diastereomers of N2-CEdG can also be detected readily in HeLa-S3 cells (data not shown).
Fig. 2.
The dose-dependent formation of N2-CEdG in WM-266-4 cells, either untreated or treated with MG for 3 h or d-glucose for 5 days. The concentrations of MG and d-glucose are indicated. The data represent the mean and standard deviations of results from three independent cell culture and treatments.
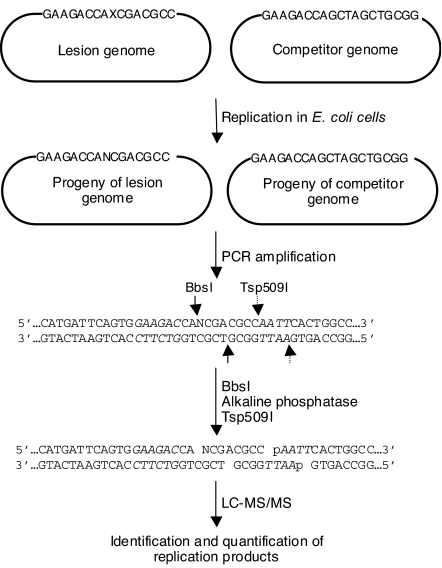
We next asked how the presence of N2-CEdG compromises DNA replication, and which SOS-induced DNA polymerase is involved in bypassing the lesion in E. coli cells. To this end, we synthesized oligodeoxyribonucleotides (ODNs) harboring a site-specifically incorporated S- or R-N2-CEdG, following our procedures (28). We then ligated the ODNs into single-stranded M13 genome and assessed the bypass efficiencies and mutation frequencies of the two diastereomers by using the competitive replication and adduct bypass (CRAB) and restriction endonuclease and postlabeling (REAP) assays introduced by Essigmann and coworkers (Fig. 3) (29–31).
Fig. 3.
Method for the determination of the cytotoxicity and mutagenicity of N2-CEdG in E. coli cells. “X” in the 16-mer ODN represents S-N2-CEdG, R-N2-CEdG or unmodified dG. “N” in the progeny of lesion genome represents the nucleoside inserted at the initial lesion site. BbsI and Tsp509I restriction endonuclease recognition sites are indicated in italic, and the cleavage sites induced by the two enzymes are designated by solid and broken arrows. Only partial sequences of the PCR products for the lesion genome are shown, and the PCR products of competitor genome are not depicted.
If there is no deletion mutation, restriction digestion of the PCR products of the progeny M13 genome arising from the replication of the lesion-carrying vector affords a 8-mer fragment harboring the site where the N2-CEdG was initially incorporated. The corresponding digestion of PCR products of the progeny of the competitor genome gives an 11-mer fragment (Fig. 3). The failure to detect any radiolabeled fragments with lengths shorter than 8 mer supports that neither diastereomer gives rise to deletion mutations (Fig. S3). The bypass efficiency can be calculated from the ratio of the 8- over the 11-mer product with the consideration of the genome ratio used in the initial transfection experiment. The bypass efficiency for the lesion-carrying genome is then normalized against that for the control lesion-free genome (Fig. 4A).
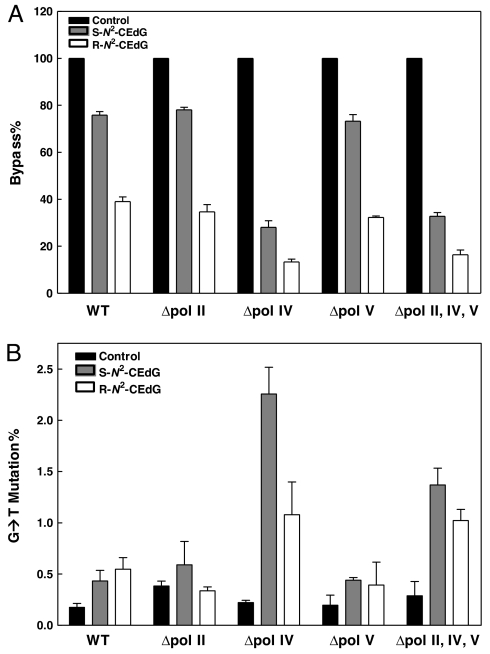
Fig. 4.
Bypass efficiencies (A) and mutation frequencies (B) of dG, S-N2-CEdG, and R-N2-CEdG lesions in wild-type; pol II-, pol IV-, and pol V-deficient; and triple-knockout AB1157 E. coli cells. Black, gray, and white columns represent the results for substrates carrying dG, S-N2-CEdG and R-N2-CEdG, respectively. The data represent the mean and standard deviations of results from three independent experiments.
It turned out that the bypass efficiencies for S- and R-N2-CEdG in wild-type AB1157 cells are ≈75% and 39%, respectively; deficiency in pol II or V in the isogenic AB1157 background does not appreciably affect bypass efficiencies for the two diastereomers of N2-CEdG (Fig. 4A). In stark contrast, the bypass efficiencies dropped to ≈28% and 13%, respectively, in the corresponding pol IV-deficient cells (Fig. 4A). Therefore, pol IV is the major DNA polymerase involved in the bypass of N2-CEdG in E. coli cells. In addition, regardless of the E. coli strains used, the bypass efficiency for R-N2-CEdG is approximately one-half of that for S-N2-CEdG (Fig. 4A), demonstrating that R-N2-CEdG is a stronger block to DNA replication than S-N2-CEdG. Moreover, the bypass efficiencies for N2-CEdG were similar in pol IV-deficient and triple-knockout AB1157 cells (Fig. 4A), underscoring the lack of involvement of pol II and V in bypassing the N2-CEdG in vivo. We also measured the bypass efficiencies by using the recently introduced LC-MS/MS method (32), and the results are in keeping with those measured by using the conventional CRAB assay (Fig. S4).
We then assessed the mutation frequencies of N2-CEdG in wild-type and bypass polymerase-deficient E. coli strains with the REAP assay (29, 31), and we used LC-MS/MS for interrogating the replication fragments (Fig. 3) (32). In this respect, the restriction digestion mixture was analyzed by LC-MS/MS, and we monitored the fragmentation of the [M–2H]2− ions of d(NCGACGCC), where “N” is an A, T, C, or G. It turned out that only d(GCGACGCC) and d(TCGACGCC) could be detected in the digestion mixture. We then quantified the relative amounts of different replication products with the consideration of the difference in ionization efficiencies for different ODNs [LC-MS/MS for monitoring the formation of d(GCGACGCC) and d(TCGACGCC) are shown in Fig. S5, and calibration curves are depicted in Fig. S6]. Our results revealed that the two diastereomers of N2-CEdG are weakly mutagenic in wild-type AB1157 cells, and the deficiency in pol II or V does not confer apparent increase in mutation frequency (Fig. 4B). However, deficiency in pol IV causes considerable increase in G → T mutation. Moreover, the mutation frequency induced by R-N2-CEdG is approximately one-half of that by S-N2-CEdG in the pol IV-deficient background (1.1% and 2.3%, respectively; Fig. 4B). Along this line, triple-knockout cells again exhibited significant increase in G → T mutation relative to the wild-type strain (Fig. 4B). On the grounds that low mutation frequencies were found for N2-CEdG, we may conclude that DinB plays an important role in avoiding the cytotoxic, rather than the mutagenic, effects of N2-CEdG and its structurely related derivatives.
The above results unveiled that approximately two-thirds of the bypass of N2-CEdG in AB1157 E. coli cells requires pol IV, and the deficiency in pol IV leads to a marked increase in mutation frequency. Therefore, the pol IV-mediated bypass of N2-CEdG in E. coli cells is error-free. To further substantiate this conclusion, we assessed quantitatively the efficiency and fidelity of pol IV-mediated nucleotide insertion opposite both S- and R-N2-CEdG by using the steady-state kinetic measurements (Figs. S7 and S8 and Table 1) (33, 34). It turned out that the nucleotide insertion opposite both diastereomers of N2-CEdG is remarkably accurate, with misinsertion frequencies similar to those found for nucleotide incorporation opposite an unmodified dG. Furthermore, the replacements of dG with an S- or R-N2-CEdG caused decreases in the efficiency (Vmax/Km, Table 1) for nucleotide insertion by 2.5- and 4.6-fold, respectively, which is consistent with the observation that R-N2-CEdG is a stronger block to DNA replication than S-N2-CEdG in E. coli cells (see above).
Table 1.
Steady-state kinetic parameters for nucleotide incorporation opposite the two diastereomers of N2-CEdG and unmodified dG by E. coli DNA polymerase IV (Km and Vmax are average values based on three independent measurements)
| dNTP | Vmax, nM min−1 | Km, nM | Vmax/Km, min−1 | finc |
|---|---|---|---|---|
| S-N2-CEdG-containing substrate | ||||
| dATP | 0.62 ± 0.07 | (8.41 ± 0.74) × 105 | 7.37 × 10−7 | 2.91 × 10−4 |
| dGTP | 0.23 ± 0.02 | (6.83 ± 0.63) × 105 | 3.36 × 10−7 | 1.33 × 10−4 |
| dCTP | 0.19 ± 0.02 | 75 ± 8.4 | 2.53 × 10−3 | 1.00 |
| dTTP | 0.46 ± 0.06 | (5.83 ± 0.68) × 105 | 7.89 × 10−7 | 3.12 × 10−4 |
| R-N2-CEdG-containing substrate | ||||
| dATP | 0.16 ± 0.01 | (7.90 ± 0.89) × 105 | 2.02 × 10−7 | 1.44 × 10−4 |
| dGTP | 0.49 ± 0.08 | (1.49 ± 0.26) × 106 | 3.28 × 10−7 | 2.34 × 10−4 |
| dCTP | 0.16 ± 0.03 | (1.14 ± 0.28) × 102 | 1.40 × 10−3 | 1.00 |
| dTTP | 0.48 ± 0.05 | (1.23 ± 0.05) × 106 | 3.89 × 10−7 | 2.78 × 10−4 |
| dG-containing substrate* | ||||
| dCTP | 0.34 ± 0.07 | 53 ± 8.4 | 6.42 × 10−3 | 1.00 |
| dTTP | 0.073 ± 0.001 | (4.61 ± 0.56) × 104 | 1.58 × 10−6 | 2.46 × 10−4 |
*The incorporation of dATP and dGTP opposite undamaged dG was barely detectable even at extraordinarily high dNTP concentrations (Fig. S8).
We also measured the steady-state kinetic parameters for nucleotide incorporation opposite the N2-CEdG lesion by human pol κ (Figs. S9–S11). The nucleotide insertion by human pol κ is again highly accurate, and the polymerase inserts preferentially dCMP opposite the lesion (Table S1). More strikingly, the efficiencies for human pol κ to incorporate the correct nucleotide, dCMP, opposite S- and R-N2-CEdG, were increased by ≈6- and 3.5-fold, respectively, relative to the unmodified substrate (Table S1). Thus, N2-CEdG is a better substrate for human pol κ than an unmodified dG.
Discussion
MG is induced endogenously as a byproduct of glycolysis (16, 18), a metabolic process conserved in all organisms (17). The concentration of MG in human cells can be elevated under various pathological conditions (e.g., diabetes) (19–21). It was found recently that MG induces modifications in calf thymus DNA mainly on dG to give N2-CEdG (22). N2-CEdG could also be detected in urine samples of healthy human subjects (23) and in kidney and aorta cells of diabetic and uremic patients (24). LC-MS/MS with the isotope dilution method revealed that N2-CEdG can be formed in untreated WM-266-4 cells at a level of approximately one lesion per 107 nucleosides; treatment of cells with MG or glucose can further enhance the formation of the lesion, supporting that N2-CEdG is an endogenous DNA lesion, and the amount of the lesion can be increased by byproducts of glycolysis.
Replication studies using single-stranded M13 shuttle vectors harboring a site-specifically incorporated N2-CEdG revealed that the two diastereomers of the lesion exhibited significantly different bypass efficiencies in E. coli cells. The R-N2-CEdG is twice as effective as S-N2-CEdG in blocking DNA replication in all strains of E. coli cells that we examined. Although the absence of pol II or V does not give rise to apparent alteration in bypass efficiency, deficiency in pol IV results in a significant drop in bypass efficiency by 63% and 66% for the S and R diastereomers, respectively (Fig. 4A).
Both diastereomers are weakly mutagenic in wild-type AB1157 cells and isogenic E. coli cells deficient in pol II or V. However, the frequency of G → T mutation in the pol IV-deficient background was increased significantly for both diastereomers, supporting that the pol IV-mediated lesion bypass is largely error-free. Along this line, it was found that G → T transversion accounts for 70% of benzo[a]pyrene-induced mutations in pol κ-defective cells, whereas G → T and G → A mutations occur at an equal frequency of ≈30% of total mutations in parental cells (35).
These results are also in accordance with in vitro replication data showing that nucleotide insertion opposite N2-CEdG is both accurate and efficient. In this respect, the efficiencies (Vmax/Km) for E. coli pol IV to incorporate the correct nucleotide, dCMP, opposite dG, S-, and R-N2-CEdG were 6.42 × 10−3, 2.53 × 10−3, and 1.40 × 10−3 min−1, respectively (Table 1). In addition, the corresponding efficiencies for human pol κ to insert dCMP were 5.40 × 10−5, 3.10 × 10−4, and 1.91 × 10−4 min−1, respectively (Table S1). In keeping with our observations, E. coli pol IV and human pol κ insert dCMP opposite N2-furfuryl-dG with 10- to 15-fold greater catalytic efficiency than opposite an undamaged dG (8). It is of note that N2-furfuryl-dG is a structure analog of the principal N2-dG adduct induced by nitrofurazone (8), and there is no evidence showing that N2-furfuryl-dG is an endogenously induced DNA adduct. Furthermore, E. coli pol IV and human pol κ can bypass accurately the bulky N2-dG-BPDE adduct (11–14), and the M13 genome bearing an N2-dG-BPDE gave 4-fold fewer plaques when transfected into SOS-induced pol IV-deficient E. coli cells relative to the isogenic wild-type cells (13). Different from what we found for the bypass of N2-CEdG, the efficiency for human pol κ to insert dCMP across N2-dG-BPDE was at least 70 times less than opposite an undamaged dG (11).
Recently, the x-ray crystal structure for the catalytic core of human pol κ in ternary complex with DNA and an incoming nucleotide has been solved (36). The structure reveals the absence of steric hindrance in the minor groove at the primer-template junction (36), which may explain the tolerance of the polymerase toward the minor-groove adduct, N2-CEdG.
Taken together, the results from the present study offer solid evidence supporting that N2-CEdG, a DNA adduct arising from MG, is an endogenous substrate for DinB DNA polymerase.
Materials and Methods
A full description of Materials and Methods can be found in the SI Text.
[2,2,2-D3]-N2-CEdG (D3-N2-CEdG) was synthesized from 2-fluoro-2′-deoxyinosine and D3-DL-alanine by using reported procedures (28). The resulting two diastereomers of D3-N2-CEdG were separated by HPLC and used as internal standards for the LC-MS/MS quantification of the lesion formed in human melanoma cells. ODNs containing a site-specifically inserted and stereochemically defined N2-CEdG were synthesized according to recently described procedures (ESI-MS and MS/MS shown in Fig. S12) (28).
Quantification of N2-CEdG Formed in WM-266-4 Human Melanoma Cells.
WM-266-4 human melanoma cells [American Type Culture Collection (ATCC)] were cultured under conditions recommended by ATCC. At 80% confluence, cells were detached and harvested by centrifugation. The cell pellets were washed twice with PBS and resuspended in 20 ml of PBS (106 cells per ml) containing the desired concentrations of MG. The cells were incubated with MG at room temperature for 3.0 h with occasional shaking. For d-glucose treatment, the cells were cultured in the media containing 5 or 25 mM d-glucose. The media were discarded, and the cells were cultured in fresh media containing the same concentration of d-glucose on day 3. The cells were then harvested at the end of day 5.
Nuclear DNA was isolated from cell lysates with phenol extraction and desalted by ethanol precipitation. The resulting DNA was digested to mononucleosides with four enzymes (details shown in SI Text), and to the digestion mixture were added the two diastereomers of D3-N2-CEdG. Quantitative analysis of N2-CEdG in the above DNA hydrolysates was performed by online HPLC-ESI-MS/MS on an LTQ linear ion-trap mass spectrometer (Thermo Fisher Scientific), which was set up for monitoring the fragmentation of protonated ions of N2-CEdG and D3-N2-CEdG. To eliminate the isobaric impurities present in MS/MS, we quantified the N2-CEdG by using MS3, which monitored the further fragmentation of the ions of m/z 224 and 227, corresponding to the protonated nucleobase portions of N2-CEdG and D3-N2-CEdG, respectively.
Construction of N2-CEdG-Carrying Single-Stranded M13 Genome and in Vivo Replication Studies.
The lesion-carrying genomes were constructed by inserting a 5′-phosphorylated d(GAAGACCAXCGACGCC), where “X” designates S- or R-N2-CEdG, into the EcoRI-linearized M13mp7(L2) genome via enzymatic ligation (31, 32). To determine bypass efficiency, we also prepared a competitor genome by inserting a 19-mer unmodified ODN, d(GAAGACCAGCTAGCTGCGG), into the linearized M13 genome. The amount of the lesion-containing genome was normalized against that of the competitor genome (31). The lesion-containing genome was then mixed with the competitor genome at a molar ratio of 6/1, transformed into wild-type and DNA polymerase-deficient AB1157 E. coli cells [Δpol B1::spec (pol II-deficient), ΔdinB (pol IV-deficient), ΔumuC::kan (pol V-deficient), and ΔumuC::kan ΔdinB Δpol B1::spec (triple knockout)] (37) by electroporation, and allowed for replication in the host E. coli cells for 6 h. In this regard, the AB1157 strains are proficient in nucleotide excision repair. The AB1157 cells were then pelleted and the supernatant, which contained progeny phages, was collected. To minimize the effect of residual lesion-containing genomes in subsequent analysis, the progeny/lesion-genome ratio was increased by several orders of magnitude via infecting SCS110 E. coli with the viable progeny phage (31). After culturing the cells at 37°C for 5 h, single-stranded DNA was extracted from the progeny phage.
PCR amplification of the region of interest in the resulting progeny genome was performed by using primers 5′-YCAGGGTTTTCCCAGTCACGACGTTGTAA-3′ and 5′-YCAGCTATGACCATGATTCAGTGGAAGAC-3′ (Y is an amino group) (31). PCR products were treated sequentially with BbsI, shrimp alkaline phosphatase, and Tsp509I, which gave rise to d(NCGACGCC) (“N” designates the nucleobase present at the original lesion site after DNA replication in vivo) and d(GCTAGCTGCGG) for the lesion-carrying and competitor genomes, respectively.
Restriction fragments were subjected to LC-MS/MS analysis, and a gradient of 5 min of 0−20% methanol followed by 35 min of 20–50% methanol in 400 mM hexafluoro-2-propanol buffer (pH was adjusted to 7.0 by the addition of triethylamine) was used for the separation (32, 38, 39). The mass spectrometer was set up for monitoring the fragmentation of the [M-2H]2− ions of the 8- and 11-mer ODNs.
For the CRAB assay, the ODN fragments were 5′ 32P-labeled before digestion with Tsp509I, and the resulting ODN fragments were resolved by denaturing PAGE analysis and quantified by using a phosphorimager.
Primer Extension Assays and Steady-State Kinetic Measurements.
A 20-mer ODN, d(ATGGCXCACTATGATCCTAG) (X = S/R-N2-CEdG) and a 5′-[32P]-labeled 15-mer primer, d(GCTAGGATCATAGTG), were used for primer extension assays and steady-state kinetic measurements.
Supplementary Material
Acknowledgments.
We thank Profs. John M. Essigmann and Graham Walker (Massachusetts Institute of Technology, Boston) for providing the E. coli strains and M13 shuttle vector and Prof. Myron F. Goodman (University of Southern California, Los Angeles) for offering the recombinant E. coli DNA pol IV. This work was supported by the National Institutes of Health (Grants R01 CA96906 and R01 CA101864).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711546105/DCSupplemental.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg EC, et al. DNA Repair and Mutagenesis. Washington, DC: Am Soc Microbiol Press; 2006. [Google Scholar]
- 3.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs RP, Fujii S, Wagner J. Properties and functions of Escherichia coli: Pol IV and Pol V. Adv Protein Chem. 2004;69:229–264. doi: 10.1016/S0065-3233(04)69008-5. [DOI] [PubMed] [Google Scholar]
- 5.Tang M, et al. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 6.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 7.Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc Natl Acad Sci USA. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 9.Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J Biol Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, et al. Error-free and error-prone lesion bypass by human DNA polymerase κ in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Wu X, Guo D, Rechkoblit O, Wang Z. Activities of human DNA polymerase κ in response to the major benzo[a]pyrene DNA adduct: error-free lesion bypass and extension synthesis from opposite the lesion. DNA Repair. 2002;1:559–569. doi: 10.1016/s1568-7864(02)00055-1. [DOI] [PubMed] [Google Scholar]
- 12.Rechkoblit O, et al. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J Biol Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 13.Shen X, et al. Efficiency and accuracy of SOS-induced DNA polymerases replicating benzo[a]pyrene-7,8-diol 9,10-epoxide A and G adducts. J Biol Chem. 2002;277:5265–5274. doi: 10.1074/jbc.M109575200. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki N, et al. Translesion synthesis by human DNA polymerase κ on a DNA template containing a single stereoisomer of dG-(+)- or dG-(-)-anti-N2-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 2002;41:6100–6106. doi: 10.1021/bi020049c. [DOI] [PubMed] [Google Scholar]
- 15.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 16.Thornalley PJ. The glyoxalase system: New developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem J. 1990;269:1–11. doi: 10.1042/bj2690001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fothergill-Gilmore LA, Michels PA. Evolution of glycolysis. Prog Biophys Mol Biol. 1993;59:105–235. doi: 10.1016/0079-6107(93)90001-z. [DOI] [PubMed] [Google Scholar]
- 18.Thornalley PJ. Pharmacology of MG: formation, modification of proteins and nucleic acids, and enzymatic detoxification−a role in pathogenesis and antiproliferative chemotherapy. Gen Pharmacol. 1996;27:565–573. doi: 10.1016/0306-3623(95)02054-3. [DOI] [PubMed] [Google Scholar]
- 19.Thornalley PJ. Modification of the glyoxalase system in human red blood cells by glucose in vitro. Biochem J. 1988;254:751–755. doi: 10.1042/bj2540751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips SA, Mirrlees D, Thornalley PJ. Modification of the glyoxalase system in streptozotocin-induced diabetic rats. Effect of the aldose reductase inhibitor Statil. Biochem Pharmacol. 1993;46:805–811. doi: 10.1016/0006-2952(93)90488-i. [DOI] [PubMed] [Google Scholar]
- 21.McLellan AC, Thornalley PJ, Benn J, Sonksen PH. Glyoxalase system in clinical diabetes mellitus and correlation with diabetic complications. Clin Sci. 1994;87:21–29. doi: 10.1042/cs0870021. [DOI] [PubMed] [Google Scholar]
- 22.Frischmann M, Bidmon C, Angerer J, Pischetsrieder M. Identification of DNA adducts of methylglyoxal. Chem Res Toxicol. 2005;18:1586–1592. doi: 10.1021/tx0501278. [DOI] [PubMed] [Google Scholar]
- 23.Schneider M, et al. Determination of glycated nucleobases in human urine by a new monoclonal antibody specific for N2-carboxyethyl-2′-deoxyguanosine. Chem Res Toxicol. 2004;17:1385–1390. doi: 10.1021/tx049929d. [DOI] [PubMed] [Google Scholar]
- 24.Li H, et al. N2-carboxyethyl-2′-deoxyguanosine, a DNA glycation marker, in kidneys and aortas of diabetic and uremic patients. Kidney Int. 2006;69:388–392. doi: 10.1038/sj.ki.5000064. [DOI] [PubMed] [Google Scholar]
- 25.Pischetsrieder M, Seidel W, Munch G, Schinzel R. N2-(1-Carboxyethyl)deoxyguanosine, a nonenzymatic glycation adduct of DNA, induces single-strand breaks and increases mutation frequencies. Biochem Biophys Res Commun. 1999;264:544–549. doi: 10.1006/bbrc.1999.1528. [DOI] [PubMed] [Google Scholar]
- 26.Murata-Kamiya N, Kamiya H, Kaji H, Kasai H. Methylglyoxal induces G:C to C:G and G:C to T:A transversions in the supF gene on a shuttle vector plasmid replicated in mammalian cells. Mutat Res. 2000;468:173–182. doi: 10.1016/s1383-5718(00)00044-9. [DOI] [PubMed] [Google Scholar]
- 27.Kaiser N, et al. Differential regulation of glucose transport and transporters by glucose in vascular endothelial and smooth muscle cells. Diabetes. 1993;42:80–89. doi: 10.2337/diab.42.1.80. [DOI] [PubMed] [Google Scholar]
- 28.Cao H, Jiang Y, Wang Y. Stereospecific synthesis and characterization of oligodeoxyribonucleotides containing an N2-(1-carboxyethyl)-2′-deoxyguanosine. J Am Chem Soc. 2007;129:12123–12130. doi: 10.1021/ja072130e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaney JC, Essigmann JM. Context-dependent mutagenesis by DNA lesions. Chem Biol. 1999;6:743–753. doi: 10.1016/s1074-5521(00)80021-6. [DOI] [PubMed] [Google Scholar]
- 30.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci USA. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaney JC, Essigmann JM. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- 32.Hong H, Cao H, Wang Y. Formation and genotoxicity of a guanine cytosine intrastrand cross-link lesion in vivo. Nucleic Acids Res. 2007;35:7118–7127. doi: 10.1093/nar/gkm851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi S, Valentine MR, Pham P, O'Donnell M, Goodman MF. Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J Biol Chem. 2002;277:34198–34207. doi: 10.1074/jbc.M204826200. [DOI] [PubMed] [Google Scholar]
- 35.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polκ protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci USA. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lone S, et al. Human DNA polymerase κ encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 37.Jarosz DF, Beuning PJ, Cohen SE, Walker GC. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 2007;15:70–77. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Apffel A, Chakel J, Fischer S, Lichtenwalter K, Hancock W. Analysis of oligonucleotides by HPLC-electrospray ionization mass spectrometry. Anal Chem. 1997;69:1320–1325. doi: 10.1021/ac960916h. [DOI] [PubMed] [Google Scholar]
- 39.Gu C, Wang Y. In vitro replication and thermodynamic studies of methylation and oxidation modifications of 6-thioguanine. Nucleic Acids Res. 2007;35:3693–3704. doi: 10.1093/nar/gkm247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.