Abstract
The p57kip2 gene encodes a member of the cyclin-dependent kinase inhibitor family, proteins known to block G1/S transition during the mammalian cell cycle. We observed that expression of p57kip2 in Schwann cells of the developing and injured adult peripheral nervous system is dynamically regulated. Using gene knockdown by means of vector-based RNA interference in cultured primary Schwann cells we found that reduced levels of p57kip2 lead to cell cycle exit, actin filament stabilization, altered cell morphology and growth, and down-regulation of promyelinating markers as well as induction of myelin genes and proteins. In addition, we could demonstrate that in vitro myelination is enhanced by p57kip2-suppressed Schwann cells. Using microarray technology we found that these cellular reactions are specific to lowered p57kip2 expression levels and detected a shift of the transcriptional expression program toward the pattern known from Schwann cells in developing peripheral nerves. Because in the absence of axons primary Schwann cells normally do not display differentiation-associated reactions, we conclude that we have identified a mechanism and an important intrinsic negative regulator of myelinating glia differentiation.
Keywords: glia, immune neuropathies, myelin, peripheral nerve
Axons are tightly associated with myelinating glial cells in the vertebrate nervous system, Schwann cells in the peripheral nervous system, and oligodendrocytes in the CNS. These cells wrap around axons in a radial growth and migration process and establish multiple lipid- and protein-rich layers to support and electrically insulate them, thus enabling saltatory propagation of electrical signals. As a result of axonal segregation, myelinating Schwann cells are associated with large-caliber axons in a 1:1 relationship and represent highly specialized and differentiated cells. Schwann cells derive from neural crest, and their differentiation during peripheral nerve development was shown to depend on a variety of genes, such as integrins, the transcription factors Sox10, Oct-6, and Krox20, or neuregulin-, p75-LNGFR, and Lgi4 signaling cascades (1–7).
Cell cycle exit is a prerequisite for myelinating glial cell differentiation followed by morphological changes as well as the expression and deposition of myelin proteins within the layers of lipid sheaths. The establishment of such a complex morphology depends on regulated cytoskeletal dynamics, e.g., actin filament turnover. This was demonstrated by means of application of cytochalasin D, a mycotoxin that interferes with actin filament assembly. Differentiation and myelination of Schwann cells cocultured with dorsal root ganglion (DRG) neurons were found to be blocked when this drug was added to the culture medium, indicating that actin filament dynamics is imperative for Schwann cell growth and morphological adaptation during the myelination process (8). Nevertheless, the question as to whether naturally occurring regulators of cytoskeletal dynamics, acting during nerve development and/or after injury, exist is so far unsolved.
In contrast to oligodendrocytes, fully differentiated Schwann cells of the adult peripheral nerve can adopt to injury, dedifferentiate, redifferentiate, and promote nerve repair (9). Peripheral myelinating glial cells are thus supposed to posses a plastic differentiation potential. On the other hand, cultured Schwann cells do not differentiate in the absence of axons, whereas cultured oligodendrocyte precursor cells readily change morphology and express myelin proteins upon withdrawal of growth factors or after induction by thyroid hormones. Apart from differential needs for axonal signals to execute differentiation programs it is also conceivable that this depends on the presence or appropriate regulation of intrinsic differentiation inhibitors, such as the recently described effect of LINGO-1 toward differentiating oligodendroglial cells (10).
In an attempt to identify Schwann cell-specific factors that might account for their adaptive behavior, we have performed screening experiments revealing a group of genes the differential expression of which correlated with redifferentiation activities within the regenerating sciatic nerve (11). Among these genes were the mammalian homolog of achaete scute 2 (Mash2), a basic helix–loop–helix transcription factor expressed in myelinating Schwann cells, the Krox24 transcription factor, the CXCR4 chemokine receptor, and the CXCL10 chemokine, as well as the cyclin-dependent kinase inhibitor (CKI) p57kip2. This gene belongs to the cip/kip family of cell cycle regulators and is known to interfere with the G1/S transition. Mice lacking p57kip2 suffer from a number of tissue defects and die around birth (12). It appeared that these are p57kip2-specific effects because they cannot be compensated by other CKIs. Such an early lethality, however, precludes functional investigations beyond embryonic development including the establishment of peripheral nerves.
To determine the function of this gene regarding Schwann cell differentiation, we have used RNA-interference-based gene suppression, which offers the advantage to address functional roles under physiological conditions as compared with gene overexpression. This experimental approach revealed that p57kip2 is an intrinsic negative regulator of Schwann cell differentiation affecting a number of important processes such as cell cycle control, morphology, and myelin expression. Our data provide evidence that these cellular effects are not associated with p57kip2's described function as cell cycle inhibitor and indicate that we have identified a molecular mechanism that is important for myelinating glial cell differentiation and the establishment of myelin sheaths.
Results
p57kip2 Is Down-Regulated During Peripheral Nerve Development.
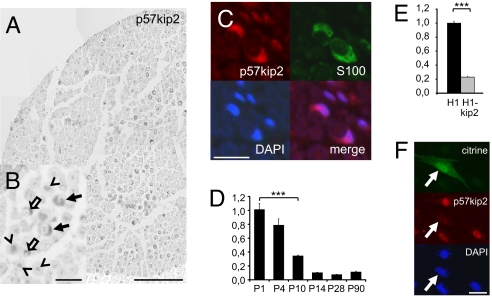
We have previously observed that p57kip2 transcripts can be detected in healthy adult and injured rat sciatic nerves, the expression of which was shown to be transiently down-regulated during the repair process (11). To investigate p57kip2's function regarding Schwann cell differentiation, we determined the expression profile during postnatal nerve development. Our measurements demonstrated high transcript levels around birth and steadily decreased expression during postnatal sciatic nerve development (Fig. 1D; significant down-regulation from postnatal day 10 onwards). This indicated that glial p57kip2 expression is most likely not involved in cell cycle control given that proliferation can still be observed in the first postnatal days (13). Immunohistochemistry using anti-p57kip2 antibodies on young adult rat sciatic nerve sections revealed the p57kip2 protein in Schwann cells as well as in axons (Fig. 1 A and B), which is in accordance with earlier results (11, 14). No specific staining was observed when primary antibodies were omitted (data not shown). Double labeling with anti-S100 antibodies and nuclear DAPI staining demonstrated that this protein is present in Schwann cell nuclei and cytoplasm (Fig. 1C). Whether the axonal expression is also regulated and exerts a particular function will be explored elsewhere. However, this observation precluded p57kip2 protein quantification in developing Schwann cells.
Fig. 1.
Expression of p57kip2 in Schwann cells of the peripheral nerve. (A and B) Anti-p57kip2 immunostainings of young adult rat sciatic nerve cross sections. Shown is p57kip2 expression in axons and Schwann cells (arrows), in axons only (arrowheads), and in Schwann cells only (open arrows). (C) Anti-p57kip2 and anti-S100 double immunostaining demonstrated that p57kip2 is present in Schwann cell nuclei and cytoplasm. (D and E) Quantitative RT-PCR analysis revealed down-regulation of p57kip2 expression during postnatal nerve development (D) and confirmed p57kip2 down-regulation in suppressed and sorted Schwann cells (E). One representative measurement of four different experiments is shown; GAPDH expression was used as reference, and data are mean values ± SEM. t test analysis demonstrated significant regulation (***, P < 0.001). (F) Anti-p57kip2 immunostaining of citrine-labeled and p57kip2-suppressed Schwann cells confirmed lowered p57kip2 protein expression. [Scale bars: 100 μm (A) and 20 μm (B, C, and F).]
RNA Interference-Mediated Down-Regulation of p57kip2 Leads to Cell Cycle Exit.
The observed down-regulation of p57kip2 expression during postnatal nerve development prompted us to use a gene suppression approach to investigate its role in cultured primary rat Schwann cells. To this end, pSUPER (15)-based suppression vectors for rat p57kip2 (H1-kip2) and p27kip1 (H1-kip1) were generated for short hairpin RNA-mediated gene knockdown. To prove the functionality of these constructs, transfected Schwann cells were isolated after 24 h in culture by using magnetic sorting as described previously (11), and expression levels were determined by means of quantitative RT-PCR analysis. This demonstrated that transcript levels of p57kip2 (Fig. 1E) and p27kip1 (data not shown) were strongly decreased. Anti-p57kip2 immunofluorescent staining of p57kip2-suppressed Schwann cells that have been labeled by means of a cotransfected citrine expression vector confirmed that this led to lowered p57kip2 protein expression (Fig. 1F).
To assess whether the down-regulation of p57kip2 affects the Schwann cell cycle, we performed BrdU labeling of transfected proliferating cells. To this end Schwann cells were transfected with either empty (H1) or p57kip2-suppression (H1-kip2) vectors and incubated in BrdU-containing medium. This revealed that the suppression of p57kip2 did not enhance but led to a >4-fold decreased proliferation rate (data not shown). Because we were mainly interested in exploring long-term cellular effects, we modified the suppression strategy and cotransfected an expression vector delivering hygromycin B resistance and citrine expression. Antibiotic-dependent selection allowed observing the functional consequences of p57kip2 gene suppression up to 14 days in culture. Quantitative RT-PCR confirmed that gene suppression was maintained in suppressed cells but also revealed that in control-transfected cells p57kip2 transcript levels steadily increased over time (Fig. 2D). The fact that these cells keep on proliferating is yet another indication that the role of p57kip2 in Schwann cells is not cell cycle-associated.
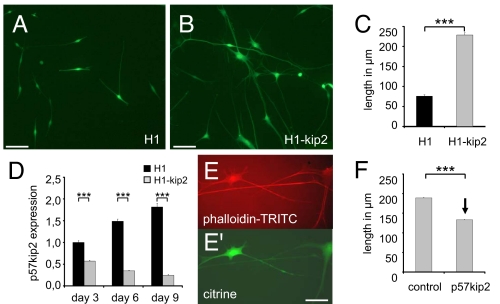
Fig. 2.
Suppression of p57kip2 affects Schwann cell morphology. (A and B) Morphology and size are changed in p57kip2-suppressed Schwann cells 9 days after transfection. (C) Quantification of average protrusion lengths. (D) Quantitative RT-PCR analysis revealed a strong and significant down-regulation of p57kip2 expression in knockdown Schwann cells and a continuous up-regulation in control-transfected cells at days 3, 6, and 9. One representative measurement of three experiments is shown; GAPDH expression was used as reference, and data are mean values ± SEM. Black bars, control-transfected (H1) cells; gray bars, p57kip2-suppressed (H1-kip2) cells. (E) Phalloidin-TRITC labeling of p57kip2-suppressed Schwann cells revealed the accumulation of long actin filaments. (Scale bars: 100 μm.) (F) Overexpression of p57kip2 in growing (p57kip2-suppressed) Schwann cells led to significantly reduced average protrusion length (control, empty expression vector; p57kip2, p57kip2 expression vector). ***, P < 0.001 (t test).
Long-Term Suppression of p57kip2 Leads to Striking Morphological Changes and Myelin Induction.
Regulated cell cycle exit is imperative for Schwann cell differentiation from precursors to myelinating cells. We then investigated whether the p57kip2 suppression also affects further differentiation-associated processes, such as cell morphology or marker gene expression. We observed that ≈3 days after transfection p57kip2-suppressed but not control-transfected Schwann cells started to change their morphology. These differences were most prominent around day 9 (Fig. 2 A and B) and included increased somata and cellular extensions. Interestingly, the morphologies of p57kip2-suppressed cells closely resembled those of premyelinating Schwann cells in cocultures (see arrows in Fig. 4 for comparison). The distribution of cellular extension lengths was shifted toward longer protrusions, and the average extension length after 9 days was significantly increased by a factor of three (Fig. 2C). On the other hand, p57kip2 overexpression did not affect cell morphology or gene expression (data not shown). However, when growing p57kip2-suppressed Schwann cells were retransfected with a p57kip2-expressing vector, the average extension length was significantly reduced (shown in Fig. 2F for suppressed Schwann cells at day 8, which were retransfected at day 5). Nevertheless, it is currently not known whether p57kip2-dependent differentiation is fully reversible. This, together with the fact that morphological transitions can be induced by using two different p57kip2 suppression constructs, encoding different RNA interfering sequences that are unique to the rat p57kip2 transcript sequence, provided strong evidence that alterations in cellular p57kip2 are solely responsible for this effect. These morphological alterations have to be seen in the context of protrusion extension during the normal myelination process, which features Schwann cell growth along and around axons. They depend on cytoskeletal rearrangements, which have already been shown to be imperative for Schwann cell maturation and myelination (8). Because cytoskeletal dynamics depends on the actin filament turnover and stabilization, we visualized actin filaments by means of phalloidin-TRITC staining. This revealed long actin filaments within the protrusions of p57kip2-suppressed Schwann cells (Fig. 2 E and E′), indicating that actin filament turnover was shifted toward polymerization.
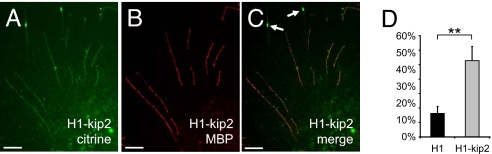
Fig. 4.
In vitro myelination of DRG axons by p57kip2-suppressed Schwann cells. (A–C) Immunostaining revealed that p57kip2 knockdown Schwann cells (labeled by means of citrine expression) formed MBP-positive internodes when seeded on DRG cocultures. (Scale bar: 100 μm.) Arrows mark elongated, premyelinating (MBP-negative) cells. (D) Quantification of the number of MBP-positive control-transfected (H1) versus p57kip2-suppressed (H1-kip2) Schwann cells. One representative experiment of two is shown. **, P < 0.01 (t test).
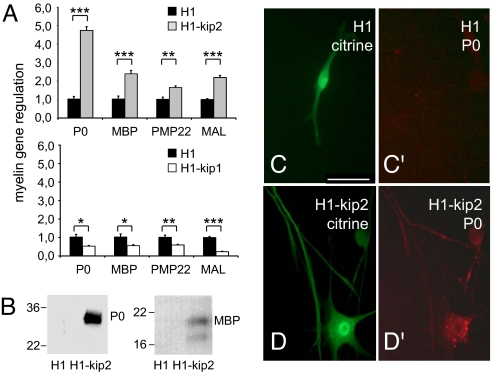
As Schwann cells wrap around axons, they express myelin genes and present myelin proteins within the layers of wraps. Determination of myelin transcript levels revealed that genes coding for P0, MBP, PMP22, and MAL were significantly induced in p57kip2-suppressed cells. This could already be observed after 3 days (data not shown) and was most prominent in cells 9 days after transfection (Fig. 3A). In contrast to this, suppression of p27kip1 led to decreased myelin expression levels (Fig. 3A), indicating that myelin gene induction is restricted to the down-regulation of p57kip2. P0 and MBP Western blot analysis (Fig. 3B) as well as immunofluorescent staining for P0 on nonpermeabilized Schwann cells (Fig. 3 C–D′) demonstrated that control-transfected cells were devoid of myelin proteins and that upon p57kip2 suppression myelin proteins were induced and presented at the Schwann cell surface.
Fig. 3.
Suppressed levels of p57kip2 expression lead to myelin induction. (A) Quantitative RT-PCR analysis revealed induced myelin gene expression in p57kip2 knockdown Schwann cells and down-regulated levels upon p27kip1 suppression at day 9 after transfection. One representative measurement of five experiments is shown; GAPDH expression was used as reference, and data are mean values ± SEM. Black bars, control-transfected cells; gray bars, p57kip2-suppressed cells; white bars, p27kip1-suppressed cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (t test). (B) Western blot analysis demonstrated that P0 and MBP proteins were induced in p57kip2-suppressed cells. (C–D′) Immunostaining revealed P0 protein expression on p57kip2-suppressed Schwann cells but not on control-transfected cells. H1, control-transfected cells; H1-kip2, p57kip2-suppressed cells. (Scale bar: 50 μm.)
Suppression of p57kip2 Leads to Accelerated in Vitro Myelination of DRG Axons.
Because p57kip2 suppression was shown to promote Schwann cell maturation, we then investigated whether the myelination process was also affected. To test this, we suppressed p57kip2 in Schwann cell cultures, harvested transfected cells after 3 days, and seeded them onto dissociated DRG cocultures before endogenous in vitro myelination could be observed. Transplanted cells were labeled by means of citrine expression vector cotransfection. Seven days after control-transfected and p57kip2-suppressed cells were added, cocultures were fixed and processed for anti-MBP immunofluorescent staining. This revealed that within this period p57kip2-suppressed Schwann cells formed significantly more MBP-positive internodes compared with control-transfected cells (Fig. 4), indicating that p57kip2 is also an intrinsic inhibitor of peripheral myelination.
GeneChip Analysis Revealed a Complete Shift of the Schwann Cell Transcriptome Without Induction of the IFN Response.
To assess to what degree the p57kip2-dependent Schwann cell maturation process is reflected by changes in the transcriptome and to rule out the possibility that our gene suppression strategy induces a nonspecific defense reaction, the so-called IFN response (16–18), we conducted a GeneChip analysis. Total RNA from nondifferentiating (control-transfected) and differentiating (p57kip2-suppressed) Schwann cells was prepared at 9 days after transfection. cRNA probes were generated and used to hybridize Affymetrix Rat Genome 230 2.0 arrays. This analysis revealed that, of 55 genes known to be affected by this defense reaction, 36 genes were detected but not regulated, five genes were found to be slightly up-regulated, and six genes were down-regulated [supporting information (SI) Table S1]. Of note, the IFN-inducible double-stranded RNA-dependent protein kinase (Prkr), one of the major IFN response regulator genes (17), was also repressed. This supported our conclusion that the observed cellular reactions of Schwann cells were a specific consequence of the p57kip2 knockdown.
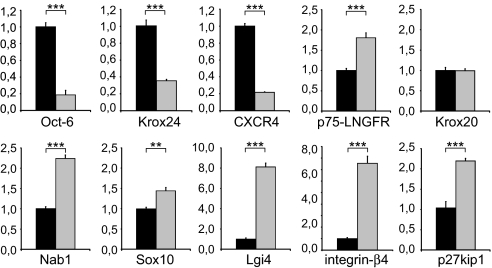
On the other hand, significant regulation of many Schwann cell-related genes was observed. This analysis is likely to shed light on the underlying molecular mechanisms. We have validated the regulation of several genes of interest by means of quantitative RT-PCR (Fig. 5). Down-regulation of the promyelinating transcription factor gene Oct-6 (5), the precursor and nonmyelinating Schwann cell marker Krox24 (19), and the chemokine receptor CXCR4 (20) is in accordance with their down-regulation during postnatal nerve development. We also observed clear inductions of the p75-LNGFR and Lgi4 genes. These proteins have been shown to be imperative for peripheral myelination (1, 3). Krox20 expression (7) was not changed, but expression levels of Krox20 coactivators Nab1 (21) and Nab2 (data not shown) and of the transcription factor Sox10 (6) were induced upon p57kip2 suppression. We also detected significant inductions of integrin-β4 (4), but not of integrin-β1 (data not shown), and of erbB2 and erbB3 receptors (2) (data not shown). Whether cell cycle exit is a consequence of elevated p21cip1 (data not shown) and p27kip1 (Fig. 5) expression levels or whether this reflects compensatory mechanisms regarding cytoskeletal modifications via the Rho pathway (22) remains to be shown by future studies. Induction of both CKI genes could already be observed when first morphological changes became apparent. Because all of these regulations are in accordance with the developmental expression patterns, this provides additional evidence that upon p57kip2 suppression Schwann cells differentiate as they would do in their natural context.
Fig. 5.
GeneChip gene expression analysis at day 9 revealed a number of differentially regulated genes. Quantitative RT-PCR analysis was used to validate the regulation of Schwann cell differentiation-related genes in p57kip2-suppressed versus control-transfected Schwann cells. For each gene one representative measurement of three experiments is shown; GAPDH expression was used as reference, and data are mean values ± SEM. Black bars, control-transfected cells; gray bars, p57kip2-suppressed Schwann cells. **, P < 0.01; ***, P < 0.001 (t test).
Discussion
In contrast to oligodendroglial precursor cells, cultured primary Schwann cells have so far been regarded as blocked in their differentiation (23, 24). Here we demonstrate that silencing of p57kip2 expression results in Schwann cell cycle exit, cellular growth, myelin induction, shifted gene expression pattern, and enhanced in vitro myelination. We show that this process is specific for p57kip2, and, considering its down-regulation during postnatal peripheral nerve development, we conclude that p57kip2 is an intrinsic inhibitor of myelinating Schwann cell differentiation, the down-regulation of which is a prerequisite to allow differentiation to progress.
As a member of the cip/kip family of CKIs, p57kip2 is primarily considered to negatively interfere with the G1/S transition of the cell cycle. However, we found that suppression of p57kip2 leads to cell cycle exit. This can be explained by the observed induction of the related CKIs p27kip1 and p21cip1 and is in accordance with previously published results (25, 26). Currently we have no evidence that the members of the INK4 family, namely p15, p16, p18, and p19, are involved in regulating Schwann cell cycle exit in response to p57kip2 suppression. Interestingly, it has already been shown that CKIs can act as regulators of neural differentiation independent of their cell cycle inhibitory function (27–29). These effects are probably mediated by p57kip2 interacting with additional binding partners. Such a model is further supported by the observation that the severe developmental defects seen in p57kip2-deficient mice cannot be compensated by the other CKIs (12). Thus, they are probably specific to p57kip2's additional protein domains (30). It is conceivable that the observed Schwann cell differentiation effect is also based on such additional interaction partners as Nurr1 (28) or LIM domain-containing proteins (31), a hypothesis that remains to be explored in future experiments.
Apart from stimulated cellular growth and morphology, widespread alterations in Schwann cell gene expression were observed that were found to be highly similar to regulatory events during peripheral nerve development. Whether these are primary changes in cell morphology inducing secondary gene regulatory events or whether these are parallel processes based on different p57kip2 binding partners remains to be elucidated. In this regard it is important to note that cytochalasin D-dependent disruption of actin filament formation was shown to prevent myelin gene expression in a dose-dependent manner (8). This implies that functional links between these two processes exist. Whether p57kip2 represents such a missing link or, alternatively, whether soluble or filamentous actin can exert this function via association with signal transduction molecules (32) remains to be shown. Interestingly, our analysis also indicated that the degree of myelin gene induction occurring in p57kip2-suppressed Schwann cells is not mediated by the Krox20 transcription factor. This appears rather to be a consequence of Nab coactivator and Sox10 transcription factor induction. It will be interesting to see whether Krox20 can be further induced once p57kip2-suppressed cells myelinate axons.
Because the central and peripheral nervous systems differ fundamentally in their potential to regenerate spontaneously, our findings are of biomedical relevance. Successful nerve regeneration depends on the generation of a growth-permissive environment by dedifferentiating and redifferentiating Schwann cells within the degenerating distal nerve segment. This adaptive behavior facilitates and actively promotes axonal regeneration (9). However, such an adaptation to pathological situations is not observed with oligodendrocytes, which tend to degenerate as a consequence of trauma or disease (33, 34). That this fundamental difference in nerve repair is indeed (among other reasons) dependent on the plasticity of myelinating glial cells is supported by observations of Schwann cell-dependent CNS regeneration (35, 36). As to whether differential p57kip2 regulation represents the main determinant for a glial cell's redifferentiation capacity remains yet to be proven. Nevertheless, Dugas et al. (37) observed that increasing p57kip2 levels constitute the differentiation-associated timer preceding oligodendrocyte differentiation. This suggests that p57kip2 does not exert the same function in CNS and peripheral nervous system myelinating glial cells. It also remains to be shown whether p57kip2 dysregulation or aberrant subcellular localization accounts for delayed or remyelination failure as it can be observed in inherited or inflammatory demyelinating diseases. Because unspecific defense reactions could be excluded, silencing the p57kip2 gene might provide a therapeutic tool to treat peripheral demyelinating diseases or to promote myelin restoration after traumatic injury.
In our experiments, we mimicked the postnatal down-regulation of p57kip2 by means of RNA interference; however, it is currently unsolved what signals mediate down-regulation in vivo in both developing and regenerating peripheral nerves. Furthermore, it has yet to be demonstrated to what degree p57kip2 influences peripheral myelination in vivo. Because the existing knockout mouse mutants die perinatally (12) this question awaits the generation and analysis of specific mouse mutants.
Materials and Methods
Cloning Procedures.
pSUPER-based vectors for p57kip2 and p27kip1 were designed according to the protocol of the manufacturer (OligoEngine). Briefly, complementary 64-nt oligonucleotides containing 19-nt inverted repeats of the target mRNA and separated by a spacer element were annealed, phosphorylated, and ligated with the pSUPER vector. After integration, the insert sequence was verified by using BigDye terminator sequencing on an ABI 310 Genetic Analyzer (Applied Biosystems). For details see SI Materials and Methods.
Schwann Cell Culture and Transfection.
Primary rat Schwann cells were cultured, and transfection was carried out as described in ref. 11. For short-term experiments, cells were cotransfected with suppression/expression vectors together with the CD14 expression vector pMACS-CD14.1 (Miltenyi Biotec) at a ratio of 5:1 before magnetic sorting (11). For long-term experiments, cells were cotransfected with the pcDNA3-hyg-citrine vector (ratio 5:1) allowing identification and selection of transfected cells. Selection of transfected cells was done by application of 50 μg/ml hygromycin B (Invitrogen Life Technologies) to the culture medium. Morphological measurements were done on a Nikon Eclipse TE 200 microscope using Lucia software (Nikon). Cocultures of dissociated DRG were prepared from E17 rats according to ref. 38. For details see SI Materials and Methods.
Staining Procedures.
Immunostainings of either paraformaldehyde-fixed rat Schwann cells and dissociated DRG cocultures or paraffin sections from sciatic nerves were performed with rabbit anti-p57kip2 (Sigma–Aldrich; diluted 1/100) as described in ref. 30, mouse anti-S100 (Abcam; 1/500), mouse anti-MBP (Sternberger Monoclonals; 1/500), and mouse anti-P0 (39) (diluted 1/1,000) antibodies; as well as phalloidin-TRITC (Sigma–Aldrich; 1/1,000). Alexa Fluor 488-, Alexa Fluor 594-, or horseradish peroxidase-conjugated antibodies (all Molecular Probes; 1/500) were used for signal visualization. Nuclei were stained with DAPI (Roche). Determination of proliferation rates was done by using the BrdU-Kit I (Roche) according to the manual of the supplier.
RNA Preparation, cDNA Synthesis, and Quantitative RT-PCR Analysis.
Total RNA derived from cultured cells and sciatic nerves was prepared by using the RNeasy procedure following the protocols of the supplier (Qiagen). Reverse transcription of total RNA was performed by using the TaqMan Reverse Transcription kit (Applied Biosystems) and dT16(A/C/G) oligonucleotide primers. Quantitative determination of gene expression levels was performed on an ABI 7000 sequence detection system using TaqMan and SybrGreen universal master mixes (Applied Biosystems). Primer sequences were determined by means of PrimerExpress 2.0 software (Applied Biosystems) and subsequently tested for the generation of specific amplicons (see SI Materials and Methods for sequences). GAPDH and ornithine decarboxylase were used as reference genes, and relative gene expression levels were determined according to the manufacturer's ΔΔCt method (Applied Biosystems). Each sample was measured in quadruplicate; data are shown as mean values ± SEM.
Western Blot Analysis.
Western blotting was performed as described in refs. 40 and 41 using mouse anti-P0 (39) and mouse anti-MBP (Chemicon) antibodies at a dilution of 1/10,000 and 1/1,000, respectively. Visualization was conducted by using horseradish peroxidase-conjugated goat anti-mouse antibodies at a dilution of 1/10,000 (Southern Biotechnology) and an ECL Western blot detection system (Amersham Pharmacia).
GeneChip Analysis.
Two micrograms of total RNA from control-transfected and p57kip2-suppressed Schwann cells was used to generate cRNA probes using the GeneChip one-cycle target labeling and Control Reagents kits (Affymetrix). Labeled cRNAs were hybridized to GeneChip Rat Genome 230 2.0 Arrays (Affymetrix) according to the manufacturer's protocol before chip readout and signal digitalization. Data quality verification and analysis were performed by means of ArrayAssist software (Stratagene) using five different algorithms (MAS5.0, MBEI, PLIER, RMA, and GC-RMA) for low-level analysis. After variance stabilization (+16) and log-transformation (base 2), statistical analysis was carried out via t test. The thresholds to consider genes to be regulated were >1.5 (fold change) and <0.02 (P values). Genes were considered as being significantly regulated if they met the above threshold criteria after PLIER or GC-RMA analysis and at least according to one additional preprocessing algorithm.
Supplementary Material
Acknowledgments.
We thank Sabine Hamm and Kristin Zimmermann for assistance with histology and D. Abankwa (University of Queensland, Brisbane, Australia) for providing the citrine vector. This work was supported by Deutsche Forschungsgemeinschaft Grant SFB590 and by the Research Promotion Fund of the Heinrich Heine University of Düsseldorf.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802659105/DCSupplemental.
References
- 1.Bermingham JR, Jr, et al. The claw paw mutation reveals a role for Lgi4 in peripheral nerve development. Nat Neurosci. 2006;9:76–84. doi: 10.1038/nn1598. [DOI] [PubMed] [Google Scholar]
- 2.Britsch S, et al. The ErbB2 and ErbB3 receptors and their ligand, neuregulin-1, are essential for development of the sympathetic nervous system. Genes Dev. 1998;12:1825–1836. doi: 10.1101/gad.12.12.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosgaya JM, Chan JR, Shooter EM. The neurotrophin receptor p75NTR as a positive modulator of myelination. Science. 2002;298:1245–1248. doi: 10.1126/science.1076595. [DOI] [PubMed] [Google Scholar]
- 4.Feltri ML, et al. Beta 4 integrin expression in myelinating Schwann cells is polarized, developmentally regulated and axonally dependent. Development. 1994;120:1287–1301. doi: 10.1242/dev.120.5.1287. [DOI] [PubMed] [Google Scholar]
- 5.Jaegle M, et al. The POU factor Oct-6 and Schwann cell differentiation. Science. 1996;273:507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- 6.Peirano RI, Goerich DE, Riethmacher D, Wegner M. Protein zero gene expression is regulated by the glial transcription factor Sox10 4. Mol Cell Biol. 2000;20:3198–3209. doi: 10.1128/mcb.20.9.3198-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topilko P, et al. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Valle C, Gorman D, Gomez AM, Bunge MB. Actin plays a role in both changes in cell shape and gene-expression associated with Schwann cell myelination. J Neurosci. 1997;17:241–250. doi: 10.1523/JNEUROSCI.17-01-00241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Son YJ, Thompson WJ. Schwann cell processes guide regeneration of peripheral axons. Neuron. 1995;14:125–132. doi: 10.1016/0896-6273(95)90246-5. [DOI] [PubMed] [Google Scholar]
- 10.Mi S, et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8:745–751. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- 11.Küry P, Greiner-Petter R, Cornely C, Jürgens T, Müller HW. Mammalian achaete scute homolog 2 is expressed in the adult sciatic nerve and regulates the expression of Krox24, Mob-1, CXCR4, and p57kip2 in Schwann cells. J Neurosci. 2002;22:7586–7595. doi: 10.1523/JNEUROSCI.22-17-07586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang P, et al. Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role in Beckwith-Wiedemann syndrome. Nature. 1997;387:151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- 13.Stewart HJ, Morgan L, Jessen KR, Mirsky R. Changes in DNA synthesis rate in the Schwann cell lineage in vivo are correlated with the precursor–Schwann cell transition and myelination. Eur J Neurosci. 1993;5:1136–1144. doi: 10.1111/j.1460-9568.1993.tb00968.x. [DOI] [PubMed] [Google Scholar]
- 14.Schmetsdorf S, Gartner U, Arendt T. Constitutive expression of functionally active cyclin-dependent kinases and their binding partners suggests noncanonical functions of cell cycle regulators in differentiated neurons. Cereb Cortex. 2007;17:1821–1829. doi: 10.1093/cercor/bhl091. [DOI] [PubMed] [Google Scholar]
- 15.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 16.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 17.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 18.Sledz CA, Williams BR. RNA interference and double-stranded-RNA-activated pathways. Biochem Soc Trans. 2004;32:952–956. doi: 10.1042/BST0320952. [DOI] [PubMed] [Google Scholar]
- 19.Topilko P, et al. Differential regulation of the zinc finger genes Krox-20 and Krox-24 (Egr-1) suggests antagonistic roles in Schwann cells. J Neurosci Res. 1997;50:702–712. doi: 10.1002/(SICI)1097-4547(19971201)50:5<702::AID-JNR7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 20.Küry P, et al. Cyclic AMP and tumor necrosis factor-alpha regulate CXCR4 gene expression in Schwann cells. Mol Cell Neurosci. 2003;24:1–9. doi: 10.1016/s1044-7431(03)00132-5. [DOI] [PubMed] [Google Scholar]
- 21.Le N, et al. Nab proteins are essential for peripheral nervous system myelination. Nat Neurosci. 2005;8:932–940. doi: 10.1038/nn1490. [DOI] [PubMed] [Google Scholar]
- 22.Denicourt C, Dowdy SF. Cip/Kip proteins: More than just CDKs inhibitors. Genes Dev. 2004;18:851–855. doi: 10.1101/gad.1205304. [DOI] [PubMed] [Google Scholar]
- 23.Abney ER, Bartlett PP, Raff MC. Astrocytes, ependymal cells, and oligodendrocytes develop on schedule in dissociated cell cultures of embryonic rat brain. Dev Biol. 1981;83:301–310. doi: 10.1016/0012-1606(81)90476-0. [DOI] [PubMed] [Google Scholar]
- 24.Morrison S, et al. P0 gene expression in cultured Schwann cells. J Neurocytol. 1991;20:769–780. doi: 10.1007/BF01187850. [DOI] [PubMed] [Google Scholar]
- 25.Atanasoski S, et al. Cell cycle inhibitors p21 and p16 are required for the regulation of Schwann cell proliferation. Glia. 2006;53:147–157. doi: 10.1002/glia.20263. [DOI] [PubMed] [Google Scholar]
- 26.Shen AG, et al. Dynamic changes of p27(kip1) and Skp2 expression in injured rat sciatic nerve. Cell Mol Neurobiol. 2007 Jul 24; doi: 10.1007/s10571-007-9167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dyer MA, Cepko CL. p57(Kip2) regulates progenitor cell proliferation and amacrine interneuron development in the mouse retina. Development. 2000;127:3593–3605. doi: 10.1242/dev.127.16.3593. [DOI] [PubMed] [Google Scholar]
- 28.Joseph B, et al. p57(Kip2) cooperates with Nurr1 in developing dopamine cells. Proc Natl Acad Sci USA. 2003;100:15619–15624. doi: 10.1073/pnas.2635658100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohnuma S, Philpott A, Wang K, Holt CE, Harris WA. p27Xic1, a Cdk inhibitor, promotes the determination of glial cells in Xenopus retina. Cell. 1999;99:499–510. doi: 10.1016/s0092-8674(00)81538-x. [DOI] [PubMed] [Google Scholar]
- 30.Potikha T, Kassem S, Haber EP, Ariel I, Glaser B. p57Kip2 (cdkn1c): Sequence, splice variants and unique temporal and spatial expression pattern in the rat pancreas. Lab Invest. 2005;85:364–375. doi: 10.1038/labinvest.3700229. [DOI] [PubMed] [Google Scholar]
- 31.Yokoo T, et al. p57Kip2 regulates actin dynamics by binding and translocating LIM-kinase 1 to the nucleus. J Biol Chem. 2003;278:52919–52923. doi: 10.1074/jbc.M309334200. [DOI] [PubMed] [Google Scholar]
- 32.Carraway KL, Carraway CA. Signaling, mitogenesis and the cytoskeleton: Where the action is. BioEssays. 1995;17:171–175. doi: 10.1002/bies.950170212. [DOI] [PubMed] [Google Scholar]
- 33.Crowe MJ, Bresnahan JC, Shuman SL, Masters JN, Beattie MS. Apoptosis and delayed degeneration after spinal cord injury in rats and monkeys. Nat Med. 1997;3:73–76. doi: 10.1038/nm0197-73. [DOI] [PubMed] [Google Scholar]
- 34.Hisahara S, Okano H, Miura M. Caspase-mediated oligodendrocyte cell death in the pathogenesis of autoimmune demyelination. Neurosci Res. 2003;46:387–397. doi: 10.1016/s0168-0102(03)00127-5. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Raisman G. Integration of transplanted cultured Schwann cells into the long myelinated fiber tracts of the adult spinal cord. Exp Neurol. 1997;145:397–411. doi: 10.1006/exnr.1997.6502. [DOI] [PubMed] [Google Scholar]
- 36.Woodhoo A, et al. Schwann cell precursors: A favourable cell for myelin repair in the central nervous system. Brain. 2007;130:2175–2185. doi: 10.1093/brain/awm125. [DOI] [PubMed] [Google Scholar]
- 37.Dugas JC, Ibrahim A, Barres BA. A crucial role for p57(Kip2) in the intracellular timer that controls oligodendrocyte differentiation. J Neurosci. 2007;27:6185–6196. doi: 10.1523/JNEUROSCI.0628-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Podratz JL, Rodriguez E, Windebank AJ. Role of the extracellular matrix in myelination of peripheral nerve. Glia. 2001;35:35–40. doi: 10.1002/glia.1068. [DOI] [PubMed] [Google Scholar]
- 39.Archelos JJ, et al. Production and characterization of monoclonal antibodies to the extracellular domain of P0. J Neurosci Res. 1993;35:46–53. doi: 10.1002/jnr.490350107. [DOI] [PubMed] [Google Scholar]
- 40.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 41.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.