Abstract
Recently, it has been reported that there is a differential subcellular distribution of components of the minor U12-dependent and major U2-dependent spliceosome, and further that the minor spliceosome functions in the cytoplasm. To study the subcellular localization of the snRNA components of both the major and minor spliceosomes, we performed in situ hybridizations with mouse tissues and human cells. In both cases, all spliceosomal snRNAs were nearly exclusively detected in the nucleus, and the minor U11 and U12 snRNAs were further shown to colocalize with U4 and U2, respectively, in human cells. Additionally, we examined the distribution of several spliceosomal snRNAs and proteins in nuclear and cytoplasmic fractions isolated from human cells. These studies revealed an identical subcellular distribution of components of both the U12- and U2-dependent spliceosomes. Thus, our data, combined with several earlier publications, establish that, like the major spliceosome, components of the U12-dependent spliceosome are localized predominantly in the nucleus.
Keywords: localization, pre-mRNA splicing, snRNA, U12-dependent spliceosome
Pre-mRNA splicing is an essential step in the co/posttranscriptional processing of eukaryotic transcripts before their export from the nucleus. The low-abundance U12-dependent “minor” spliceosome exists in parallel with the U2-dependent “major” spliceosome in most multicellular eukaryotes (1). It catalyzes the removal of a rare class of introns (U12-type) that represent <1% of introns in mammals (2, 3). The U12-dependent spliceosome contains four unique snRNAs: U11, U12, U4atac, and U6atac, which are paralogs of U1, U2, U4, and U6 snRNAs of the U2-dependent spliceosome, respectively (4, 5). U5 snRNA, in contrast, is shared between the two spliceosomes. In addition to the snRNAs, both spliceosomes contain numerous snRNP and non-snRNP proteins (6, 7).
The five snRNP components of the major spliceosome are predominantly localized in the nucleus, but the biogenesis of four of them involves a cytoplasmic step (8). That is, after transcription by RNA Pol II, the snRNAs U1, U2, U4, and U5, which all contain an Sm protein-binding site, are first exported into the cytoplasm. The Sm proteins subsequently bind, and the snRNA's cap is hypermethylated to 2,2,7-tri-methyl-guanosine (m3G) (9, 10). These processing steps are a prerequisite for their reimport into the nucleus (11). In contrast to these so-called Sm-class snRNAs, U6 snRNA is transcribed by RNA pol III, acquires Sm-like proteins (Lsm2–8) (12) and a γ-monomethyl phosphate cap, and does not leave the nucleus. The minor snRNAs U11, U12, and U4atac are also Sm-class snRNAs, containing an Sm-binding site that is also bound by Sm proteins and an m3G cap (5, 13), whereas U6atac is transcribed by RNA pol III, possesses a γ-monomethyl phosphate cap (5), and is assembled with Lsm proteins (14). After nuclear import, spliceosomal snRNPs first pass through Cajal bodies, where they undergo further modifications and assembly (15, 16) and then subsequently localize in the nucleoplasm. The presence of an m3G cap and Sm proteins in U11, U12, and U4atac snRNPs, and the monomethyl cap and Lsm proteins in U6atac suggest that these minor snRNPs share the same biogenesis pathways and localization with their major counterparts. Indeed, previous studies have localized U11 and U12 snRNAs in the nucleus in human cells (17) and U11 in the nucleus of Drosophila cells (18).
The vast majority of snRNP and spliceosomal proteins appear to be shared by both spliceosomes, with the notable exception of seven proteins that are specifically associated with the U11 and/or U11/U12 snRNPs (14, 19). Significantly, GFP-tagged versions of proteins specific to the U12-dependent spliceosome, namely the U11-35K, U11/U12-31K (ZCRB1, alias MADP-1), and U11/U12-65K (RNPC3) proteins, are also detected predominantly in the nucleus of Arabidopsis (20) and/or human cells (21, 22).
Recently, it has been reported that, in contrast to the major spliceosomal snRNAs, the minor snRNAs are enriched in the cytoplasm of both zebrafish and mammalian cells (23). Based on this observation, and the apparent enrichment of pre-mRNAs with unspliced U12-type introns in the cytoplasm, König et al. (23) postulated that the splicing of U12-dependent introns takes place in the cytoplasm.
Here, we have examined the intracellular localization of the snRNP components of the U12-dependent spliceosome in mouse tissues and also in human and mouse cell lines. In all mouse tissues examined, either embryonic or adult, we detected nearly exclusive nuclear localization of both major and minor spliceosomal snRNAs. FISH experiments further revealed that major and minor snRNAs colocalize within the nucleus of HeLa cells, and subsequent HeLa cell fractionation studies confirmed an identical subcellular distribution for components of both the U12- and U2-dependent spliceosomes. Thus, our results clearly demonstrate that minor spliceosome components are predominantly localized in the nucleus.
Results
Major and Minor snRNAs Are Predominantly Nuclear in Mouse Tissues.
The similarities in the function and structural modifications of the major and minor spliceosomal snRNPs suggest that the minor snRNPs also localize in the nucleus. To learn more about the cellular localization of components of the major and minor spliceosomes, we performed in situ hybridizations (ISH) with sections of embryonic and adult mouse tissues, using two different detection methods.
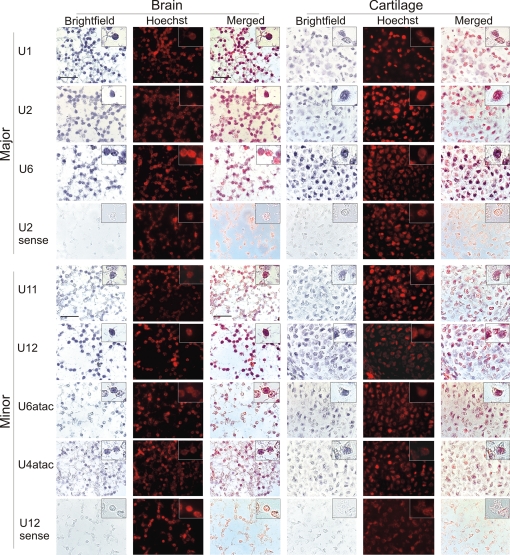
First, we performed nonradioactive ISH with full-length cRNA probes labeled with digoxigenin (DIG) to detect U1, U2, U6, U11, U12, U6atac, and U4atac snRNAs in sections of embryonic day 15 (E15) mouse embryos [Fig. 1, supporting information (SI) Fig. S1]. Sense-strand probes for several snRNAs were used as controls and gave no significant signals (control hybridizations for U12 and U2 in Fig. 1 and Fig. S1). Because of the ≈100-fold difference in abundance between major and minor snRNAs (5, 13), staining reactions were incubated for 40 min for the major snRNAs and 4 h for minor snRNAs. The staining signals obtained with probes for both the major and minor snRNAs clearly overlap with the nuclear counterstain (Hoechst) in all tissues, confirming that both classes of snRNAs are found predominantly in the nucleus. The nuclear signals appear slightly granular, which may be due to the enrichment of snRNPs in subnuclear structures such as nuclear speckles and Cajal bodies. Sections from three representative types of tissue hybridized with different snRNA probes are shown. Well separated nuclei can be clearly seen in brain, where axons and dendrites fill the space between individual cells, and in cartilage, which contains considerable amounts of extracellular matrix (Fig. 1). In skin, the nuclei close to the surface of the epithelium are flat, and the staining appears more diffuse than in the other tissue types shown, but the signals from the probes are nevertheless clearly nuclear (Fig. S1).
Fig. 1.
Localization of spliceosomal snRNAs in embryonic mouse tissues. ISH was performed on sagittal sections of E15 mouse embryos using DIG-labeled full-length RNA probes complementary to the snRNAs indicated. NBT/BCIP staining was performed for 40 min (major snRNAs) or 4 h (minor snRNAs). Blue color in the brightfield images represents the signal from the snRNA probes. Hoechst staining of nuclei in the darkfield images was converted to red by image processing. In the merged images, colocalization of the snRNA and Hoechst signals is indicated by purple. (Inset) An enlarged Inset of individual cells is included at the top right corner of each image. (Scale bar is 20 μm.)
The specificity of the probes used in nonradioactive ISH was tested in Northern hybridizations using corresponding 32P-labeled probes (which allows quantification of the specificity of each probe) and total RNA preparations derived from mouse E13 embryos, adult brain, or adult liver. The hybridization and washing conditions were identical to those used in nonradioactive ISH (SI Text). Significantly, U11, U12, U6atac, and U2-specific probes hybridized only to their cognate snRNAs (Fig. S2 A, B, D, and E), whereas with U4atac a weak cross-reacting band, with an intensity of ≈1% of the authentic signal, was detected (Fig. S2C). Consistent with the ISH results, no detectable signal was observed with the U12 or U2 sense probes (Fig. S2 F and G). We conclude that probes used for the ISH experiments are specific for their cognate snRNAs, exhibiting very little or no cross-reactivity.
To provide independent evidence for the nuclear localization of minor snRNAs, we also performed radioactive ISH to localize U2, U12, U6atac, and U4atac snRNAs in sections of adult mouse brain (Fig. S3, SI Text). Consistent with the above results, signals from all snRNAs tested mainly overlap with the nuclear counterstain, although the scattering of the radioactive signal impedes precise localization. Apart from the expected differences in signal intensity, we detected no differences in localization between major and minor snRNAs. Thus, our results, obtained with two different detection methods, are in perfect accordance with earlier studies reporting nuclear localization for components of the minor spliceosome (see above).
Major and Minor snRNAs Colocalize in the Nucleoplasm of HeLa Cells.
To learn more about the nuclear distribution of components of the major and minor spliceosomes, we also performed FISH with fluorescently labeled DNA oligonucleotides against U2 or U4, or RNAs complementary to the U11 or U12 snRNAs (Fig. 2) using human HeLa SS6 cells. The specificity of the U11 and U12 cRNA probes was tested by Northern analysis, using the identical hybridization and wash conditions used for FISH (SI Text). Similar to the nonradioactive ISH probes, the FISH probes hybridized specifically with their cognate snRNAs when RNA from nuclear or cytoplasmic extract was analyzed; the U11 cRNA also cross-reacted slightly with an RNA solely present in cytoplasmic extract (Fig. S4).
Fig. 2.
Subcellular localization of U2, U4, U11, and U12 snRNAs in HeLa cells. Fluorescence microscopy was performed with HeLa cells stained with fluorescently labeled DNA oligonucleotides (U2 and U4) or cRNAs (U12 and U11) complementary to the snRNA indicated in each image. Left and Center show the staining patterns of the various snRNAs alone, and Right shows the confocal overlay of Center and Left beside them.
Consistent with previous observations (17), nucleoplasmic staining with several intensely stained foci (likely Cajal bodies) amid diffuse less intense speckles was observed with the anti-U2 and -U12 probes (Fig. 2 Lower). Most of the U2 and U12 colocalized with one another, as evidenced by the predominantly yellow color in the confocal overlay (Fig. 2 Lower Right). FISH with probes against U11 and U4 yielded a similar nucleoplasmic staining pattern (Fig. 2 Upper), albeit with less intense staining of specific foci, and the confocal overlay (Fig. 2 Upper Right) indicated that the majority of these snRNAs colocalize in the nucleus. Thus, these data also indicate that snRNA components of the major and minor spliceosome are found, for the most part, in the same subcellular compartment.
Major and Minor snRNPs Are Found Predominantly in the Nucleoplasm of HeLa Cells.
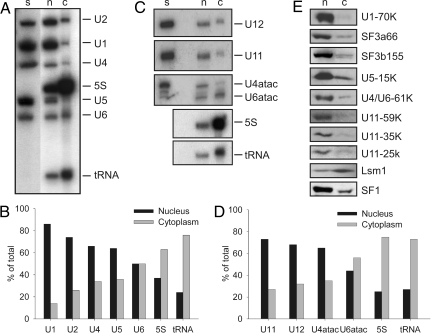
To confirm our ISH/FISH data, we assayed for spliceosomal components in cytoplasmic and nuclear fractions prepared from HeLa cells. Nuclear and cytoplasmic extracts were prepared from the same batch of cells. RNA and protein were subsequently isolated from an identical cell equivalent amount of each extract and then analyzed by Northern or Western blotting. As shown in Fig. 3A and quantitated by PhosphorImager analysis in Fig. 3B, the majority of the human U1, U2, U4, and U5 snRNAs were found in nuclear, as opposed to cytoplasmic, extract. U6 snRNA was found in approximately equal amounts in both extracts. This result seems surprising considering the nuclear biogenesis of U6 but is consistent with previous studies (24) and most probably reflects leakage of monomeric U6 during cell fractionation (see Discussion). Indeed, the distribution of snRNPs after cell fractionation reflects not only their original localization but also how well separated the nuclear and cytoplasmic fractions are and the degree to which they leak from the nucleus during fractionation (24). In contrast to the U1, U2, U4, and U5 snRNAs, 5S RNA and tRNA, which are known cytoplasmic RNAs, were found predominantly in HeLa cytoplasmic extract, confirming that a good separation of the cytoplasm and nucleus was achieved during cell fractionation. Like their major snRNA counterparts, the majority of the U11, U12, and U4atac snRNAs was also found in nuclear vs. cytoplasmic extract (Fig. 3 C and D). Similar to the U6 snRNA, approximately half of U6atac was found in cytoplasmic extract. This suggests that the subcellular distribution of both U6 and U6atac differs somewhat from the other spliceosomal snRNAs, or alternatively that they more readily leak out of the nucleus during the cell fractionation process.
Fig. 3.
Major and minor snRNPs are found predominantly in the nucleus of HeLa cells. (A, C) RNA was recovered from HeLa nuclear (n) or cytoplasmic (c) extract (in both cases corresponding to 3 × 106 cell equivalents) or from purified snRNPs (s) and fractionated by denaturing PAGE. Northern blot analysis was performed with a mixture of 32P-labeled probes against U1, U2, U4, U5, and U6, followed by 5S RNA and tRNA (A), or against U11, U12, or U4atac and U6atac (C). (B, D) The amount of a given RNA in nuclear and cytoplasmic extract was quantitated by PhosphorImager analysis and the percent of the total (nucleus plus cytoplasm) is shown graphically. (E) Proteins were recovered from 3 × 106 cell equivalents of nuclear (n) or cytoplasmic extract (c) and separated by SDS/PAGE. Immunoblotting was performed with the indicated antibodies.
Consistent with our Northern blot results, Western blot analysis with antibodies directed against the U1-70K and the U2-associated SF3a66 revealed that these major spliceosomal proteins are present predominantly in the nucleus (Fig. 3E). A similar distribution was also observed for SF1, a splicing factor that is not directly associated with snRNPs and is thought to display entirely nuclear localization (Fig. 3E). Likewise, the vast majority of SF3b155, which is present in both U2 and U11/U12 di-snRNPs (25), as well as U5-15K and U4/U6-61K, which are found both in U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs (14), were more abundant in nuclear extract. Significantly, the majority of the U11- and U11/U12-associated 59K, 35K, and 25K proteins (19) were also detected in HeLa nuclear extract. In contrast, Lsm1, a component of the cytoplasmic Lsm1-7 complex (26), was found predominantly in HeLa cytoplasmic extract. Taken together, our results demonstrate that both major and minor spliceosomal snRNPs are predominantly localized in the nucleus.
Discussion
Here, we have investigated the localization of several snRNA and protein components of the minor spliceosome. By performing ISH with different detection methods, we show that the snRNAs unique to the U12-dependent spliceosome are all nearly exclusively localized in the nucleus in embryonic (Fig. 1, Fig. S1) and adult mouse tissues (Fig. S3), and in human cells (Fig. 2). We also show that the U11 and U12 snRNAs largely colocalize with major snRNAs within the nucleus of HeLa cells, consistent with previous observations. Furthermore, by performing cell fractionation studies, we demonstrate that the U11-59K, -35K, and -25K proteins, which are specific to the minor spliceosome (19), are also enriched in the nucleus (Fig. 3). These results complement previous studies in human cells where GFP-tagged versions of the U11/U12-specific 31K and 65K proteins were detected in the nucleus (21, 22) and studies in plants, where GFP-tagged U11-35K also was located in the nucleus (20). Taken together, our data strongly support a predominantly nuclear localization for both major and minor snRNPs.
A similar subcellular distribution of major and minor snRNPs is consistent with the well characterized biogenesis pathway of m3G-capped snRNPs (reviewed in ref. 8). Spliceosomal snRNAs are transcribed in the nucleus and then, with the exception of U6 and U6atac, are exported to the cytoplasm, where they bind the common Sm proteins. After cap hypermethylation, they are reimported into the nucleus. The minor snRNAs contain all of the signals required for nuclear import, including an m3G cap and an Sm site that is bound by an apparently identical set of Sm proteins. It is thus difficult to imagine a mechanism that would specifically prevent their reimport into the nucleus. Cytoplasmic retention could theoretically be achieved via one or more proteins that specifically associate with the minor snRNPs. However, five of the seven proteins specifically associated with the human U11/U12 snRNP (65K, 59K, 35K, 31K, and 25K) have been shown to be located predominantly in the nucleus (this study; refs. 21 and 22). The localization of the remaining two (48K and 20K) remains to be determined, but the 48K protein contains a putative nuclear localization signal (data not shown), indicating that it also most likely accumulates in the nucleus. More significantly, the minor U4atac and U6atac snRNPs appear to possess protein compositions identical to the major U4 and U6 snRNPs (14), arguing against the idea that the former could be selectively retained in the cytoplasm due to differences in their protein complement. Likewise, despite significant differences in their sequences, the secondary structures of the minor snRNAs are very similar to those of their major snRNA counterparts (5, 13), suggesting that unique RNA structural elements are not potentially responsible for a selective enrichment of minor snRNPs in the cytoplasm.
Upon reimport into the nucleus, the major snRNPs are known to transiently localize to Cajal bodies where the final steps of snRNP maturation, such as snRNA base modification, the assembly of snRNP-specific proteins, and the formation of the U4/U6.U5 tri-snRNP, occur (reviewed in ref. 27). Significantly, the minor snRNAs also contain modified bases (28), and they are associated with a number of proteins (e.g., SF3b subunits) that are thought to first associate with snRNPs in Cajal bodies (29), once again arguing that the minor snRNAs are also reimported into the nucleus. Likewise, in vitro data indicate that U4atac/U6atac and U5 also form a tri-snRNP before their association with the minor spliceosome.
Recently, it has been reported by König et al. (23) that minor snRNAs are enriched in the cytoplasm. What could be the explanation for the discrepancy between our results and those described by König et al. (23)? In their study, U12 and U6atac snRNAs were detected in the cytoplasm of zebrafish tissues and mouse fibroblasts by ISH using DIG-labeled, locked nucleic acid (LNA) oligonucleotide probes. It is conceivable that the single antigen molecule in the oligonucleotide probe used in their experiments was in this case not sufficient for the accurate detection of minor snRNAs, considering the low abundance of U12-dependent spliceosome components in cells (5, 13). König et al. (23) also detected approximately half of U6atac snRNA in the cytoplasm of NIH 3T3 cells by nuclear-cytoplasmic fractionation followed by RT-PCR; unfortunately, U6 snRNA was not used as control in their study. Our results show that approximately half of both U6 and U6atac snRNAs are present in cytoplasmic extract from HeLa cells, which in the case of U6 is consistent with previous cell fractionation studies (24). Given that it has been well documented that the U6 snRNA is localized in the nucleus of intact cells (refs. 30 and 31; reviewed in ref. 32), its enrichment in the cytoplasmic fraction relative to the other major snRNPs is most likely due to enhanced leakage from the nucleus during extract preparation. Significantly, the relative proportion of U4atac snRNA (or U4) in the cytoplasmic fraction is much less compared to that of U6atac (or U6) snRNA (Fig. 3), suggesting that only monomeric U6atac (and U6) is prone to leak into the cytoplasm during cell fractionation. Indeed, there is a considerable excess of monomeric U6 snRNP over U4 in vertebrate cells (33, 34), and thus a selective leakage of U6 is not at all surprising. The similar distribution of U6 and U6atac snRNAs upon cellular fractionation indicates that their localization in intact cells is also similar, and is thus consistent with our ISH results (Fig. 1).
Based on their observations that some minor snRNAs localize in the cytoplasm of vertebrate cells, and also RT-PCR studies indicating that some pre-mRNAs with unspliced U12-type introns are found in the cytoplasm, König et al. (23) conclude that, in contrast to the major spliceosome, U12-dependent splicing is a cytoplasmic, and not a nuclear, event. A key characteristic of U12-type introns is that they are removed more slowly than U2-type introns, suggesting they may serve as rate-limiting controls for the expression of their host genes (35, 36). The slow processing rate of the U12-dependent spliceosome was suggested by König et al. (23) to lead to the export of unspliced pre-mRNAs from the nucleus and entail the need for splicing activity in the cytoplasm. Although the presence of a small proportion of Sm-class snRNPs in the cytoplasm is expected considering the cytoplasmic steps in their biogenesis, our data, together with previous studies by others, strongly argue against any significant level of U12-type splicing occurring outside the nucleus. Indeed, with the exception of some highly specialized cells (e.g., anucleate platelets) (37) or in the case of neuronal dendrites (38), there is little evidence that pre-mRNA splicing in general takes place outside of the nucleus. Furthermore, the splicing of both U2- and U12-type introns is routinely performed in vitro in nuclear extract, and, in the absence of added factors (i.e., SR proteins), neither spliceosome is active in cytoplasmic extracts (39). Taken together, the bulk of the data currently available supports the idea that the splicing of both U2- and U12-type introns is exclusively a nuclear event in the vast majority of cells.
Materials and Methods
Nonradioactive ISH.
The subcellular localization of U1, U2, U6, U11, U12, U4atac, and U6atac snRNAs in several embryonic mouse tissues was analyzed by nonradioactive cRNA ISH on tissue sections. Sagittal sections of E15 NMRI mouse embryos were fixed in 4% paraformaldehyde, embedded in paraffin, and serially sectioned at 5 μm. cRNA probes were labeled with DIG by performing in vitro transcription of snRNA clones (5, 36) in the presence of DIG RNA labeling mix (Roche). Sense probes were synthesized from the complementary strand of the probe templates. ISH was carried out with Ventana Discovery automatic staining instrument using commercial buffers (Ventana Medical Systems). Hybridization programs were as follows: proteinase K treatment at 37°C for 4 min, denaturation at 75°C (except for U4atac and U6atac at 80°C) for 6 min, hybridization in Ribohybe buffer (Ventana Medical Systems) at 65°C (except for U4atac and U6atac at 60°C) for 6 h, and three washes with 0.1× SSC at 75°C for 6 min. Detection was performed by incubation with anti-DIG antibody fused to alkaline phosphatase for 20 min, followed by NBT/BCIP staining for 40 min (U1, U2, U6) or 4 h (U11, U12, U4atac, U6atac). Afterward, the sections were counterstained with Hoechst (3 μg/ml in PBS) (Sigma–Aldrich) for 30 min at room temperature and then mounted with 20% Mowiol 4–88 (Calbiochem). The samples were photographed with a Zeiss Axioplan II epifluorescence microscope.
FISH.
HeLa SS6 cells were grown under standard conditions on coverslips. After fixation and permeabilization of the cell membrane, FISH was performed with 10 ng of Cy3- or Cy5-labeled oligonucleotide complementary to nucleotides 4–44 of the human U2 snRNA or to nucleotides 42–90 of human U4 snRNA, respectively, or 10 ng of in vitro transcribed, internally Cy3 (U11) or Cy5 (U12) -labeled, full-length RNA complementary to human U11 or U12 snRNA, essentially as described (40). Briefly, cells were incubated in buffer containing denatured probe, 0.5 μg/μl tRNA, 0.5 μg/μl SSS-DNA, 15% formamide, 10 mM NaPO4 (pH 7.0), 10% dextran sulfate, 2× SSC (300 mM sodium chloride, 30 mM sodium citrate, pH 7.0) at 37°C overnight. The cells were then washed twice for 30 min at 37°C with buffer containing 10 mM NaPO4 (pH 7.0), 15% formamide, and 2× SSC, twice for 15 min at room temperature (RT) with 2× SSC containing 0.1% Triton, and finally twice for 15 min at RT with 1× SSC containing 0.1% Triton. Fluorescence microscopy was carried out with a Leica SP2 confocal laser scanning microscope.
HeLa Cell Fractionation and Northern and Western Blots.
Cytoplasmic and nuclear extracts were prepared from HeLa SS6 cells as described (41), with minor modification. After washing with PBS and centrifugation (1,000 × g for 10 min), pelleted cells were resuspended in 5 cell volumes of Roeder A buffer (20 mM Hepes·KOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithioerythritol) and incubated on ice for 10 min. Cells were pelleted by centrifugation (1,000 × g for 10 min), resuspended in 2 cell volumes of Roeder A buffer, and the cell membrane was disrupted by 15 strokes with a dounce homogenizer (B pestle). Nuclei were pelleted first by centrifuging for 10 min at 1,000 × g. The resultant supernatant containing the cytoplasm was used directly for the isolation of cytoplasmic RNA and protein (41). After additional centrifugation, (20 min at 25,000 × g), 1.4 cell volumes (starting cell volume) of Roeder C (Hepes·KOH, pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% vol/vol glycerol) were added to the nuclear pellet, and nuclei were homogenized by 10 strokes with a Dounce homogenizer (B pestle). The homogenate was incubated for 30 min on ice with constant mixing and centrifuged for 30 min at 25,000 × g. The resultant supernatant (nuclear extract) was used directly for the isolation of nuclear RNA and protein.
RNA and protein were recovered from cytoplasmic or nuclear extract isolated from an identical number of cells, by extraction with phenol:chloroform followed by ethanol (RNA) or acetone (protein) precipitation. RNA was then fractionated on a 7 M urea–10% polyacrylamide gel and blotted to a nylon membrane. Northern blot analysis and the generation of 32P-DNA probes complementary to the spliceosomal snRNAs, 5S RNA or arginine/threonine tRNA, were performed as described (42). The percent of RNA in the nucleus or cytoplasm was quantitated by PhosphorImager analysis (Molecular Dynamics).
Proteins were fractionated on a 10/13% SDS-polyacrylamide gel, transferred to nitrocellulose, and immunostained using an ECL detection kit (Pierce). Antibodies against the following human proteins were used: U1-70K (43); SF3b155 (44); SF3a66 (29); U5-15K (45); U4/U6-61K (46); U11-59K, U11-35K, and U11-25K (19); Lsm1 (47); or SF1 (rabbit antibodies raised against the peptide SRWNQDTMEQKTVIPC comprising amino acids 20–34, plus a cysteine residue).
Supplementary Material
Acknowledgments.
We thank Gabi Heyne, Thomas Conrad, Hossein Kohansel, and Marja-Leena Peltonen for excellent technical assistance; Ira Lemm (Max Planck Institute, Göttingen) for providing genes encoding arginine and threonine tRNA; Marjo Salminen for advice with in situs, and Jouni Kvist, Elina Niemelä, and Jens Verbeeren for comments on the manuscript. This work was supported by grants from the Academy of Finland (to M.J.F. and X.M.) and from the Deutsche Forschergruppe (Lu294/12-1), Fonds der Chemischen Industrie, and the Ernst Jung Stiftung (R.L.). H.K.J.P. was supported by the Viikki Graduate School in Biosciences and N.P. and J.J.T. by the Helsinki Graduate School in Biotechnology and Molecular Biology.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 8485.
‡Deceased December 27, 2005.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803646105/DCSupplemental.
References
- 1.Patel AA, Steitz JA. Splicing double: Insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 2.Levine A, Durbin R. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 2001;29:4006–4013. doi: 10.1093/nar/29.19.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheth N, et al. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tarn W-Y, Steitz JA. A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 5.Tarn W-Y, Steitz JA. Highly divergent U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996;273:1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- 6.Jurica MS, Moore MJ. Pre-mRNA splicing: Awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 7.Will CL, Lührmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol Chem. 2005;386:713–724. doi: 10.1515/BC.2005.084. [DOI] [PubMed] [Google Scholar]
- 8.Will CL, Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 9.Mattaj IW. Cap hypermethylation of U snRNA is cytoplasmic and dependent on snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- 10.Yong J, Golembe TJ, Battle DJ, Pellizzoni L, Dreyfuss G. snRNAs Contain Specific SMN-Binding Domains That Are Essential for snRNP Assembly. Mol Cell Biol. 2004;24:2747–2756. doi: 10.1128/MCB.24.7.2747-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer U, Sumpter V, Sekine M, Satoh T, Lührmann R. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 1993;12:573–583. doi: 10.1002/j.1460-2075.1993.tb05689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Achsel T, et al. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montzka KA, Steitz JA. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12 etc. Proc Natl Acad Sci USA. 1988;85:8885–8889. doi: 10.1073/pnas.85.23.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider C, Will CL, Makarova OV, Makarov EM, Lührmann R. Human U4/U6. U5 and U4atac/U6atac.U5 Tri-snRNPs Exhibit Similar Protein Compositions. Mol Cell Biol. 2002;22:3219–3229. doi: 10.1128/MCB.22.10.3219-3229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanek D, Neugebauer KM. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol. 2004;166:1015–1025. doi: 10.1083/jcb.200405160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matera AG, Shpargel KB. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr Opin Cell Biol. 2006;18:317–324. doi: 10.1016/j.ceb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Matera AG, Ward DC. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J Cell Biol. 1993;121:715–727. doi: 10.1083/jcb.121.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider C, Will CL, Brosius J, Frilander MJ, Lührmann R. Identification of an evolutionarily divergent U11 small nuclear ribonucleoprotein particle in Drosophila. Proc Natl Acad Sci USA. 2004;101:9584–9589. doi: 10.1073/pnas.0403400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Will CL, et al. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA. 2004;10:929–941. doi: 10.1261/rna.7320604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorković ZJ, Lehner R, Forstner R, Barta A. Evolutionary conservation of minor U12-type spliceosome between plants and humans. RNA. 2005;11:1095–1107. doi: 10.1261/rna.2440305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, et al. Isolation, expression, and characterization of the human ZCRB1 gene mapped to 12q12. Genomics. 2007;89:59–69. doi: 10.1016/j.ygeno.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Zhao E, et al. Cloning and identification of a novel human RNPC3 gene that encodes a protein with two RRM domains and is expressed in the cell nucleus. Biochem Genet. 2003;41:315–323. doi: 10.1023/b:bigi.0000006032.04031.d0. [DOI] [PubMed] [Google Scholar]
- 23.König H, Matter N, Bader R, Thiele W, Müller F. Splicing Segregation: The Minor Spliceosome Acts outside the Nucleus and Controls Cell Proliferation. Cell. 2007;131:718–729. doi: 10.1016/j.cell.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 24.Fury MG, Zieve GW. U6 snRNA maturation and stability. Exp Cell Res. 1996;228:160–163. doi: 10.1006/excr.1996.0311. [DOI] [PubMed] [Google Scholar]
- 25.Will CL, Schneider C, Reed R, Lührmann R. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science. 1999;284:2003–2005. doi: 10.1126/science.284.5422.2003. [DOI] [PubMed] [Google Scholar]
- 26.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanek D, Neugebauer KM. The Cajal body: A meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 2006;115:343–354. doi: 10.1007/s00412-006-0056-6. [DOI] [PubMed] [Google Scholar]
- 28.Massenet S, Branlant C. A limited number of pseudouridine residues in the human atac spliceosomal UsnRNAs as compared to human major spliceosomal UsnRNAs. RNA. 1999;5:1495–1503. doi: 10.1017/s1355838299991537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Will CL, et al. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spiller MP, Boon KL, Reijns MA, Beggs JD. The Lsm2–8 complex determines nuclear localization of the spliceosomal U6 snRNA. Nucleic Acids Res. 2007;35:923–929. doi: 10.1093/nar/gkl1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vankan P, McGuigan C, Mattaj IW. Domains of U4 and U6 snRNAs required for snRNP assembly and splicing complementation in Xenopus oocytes. EMBO J. 1990;9:3397–3404. doi: 10.1002/j.1460-2075.1990.tb07541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiss T. Biogenesis of small nuclear RNPs. J Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 33.Black DL, Pinto AL. U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol Cell Biol. 1989;9:3350–3359. doi: 10.1128/mcb.9.8.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamm J, Mattaj IW. An abundant U6 snRNP found in germ cells and embryos of Xenopus laevis. EMBO J. 1989;8:4179–4187. doi: 10.1002/j.1460-2075.1989.tb08603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel AA, McCarthy M, Steitz JA. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 2002;21:3804–3815. doi: 10.1093/emboj/cdf297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pessa HKJ, Ruokolainen A, Frilander MJ. The abundance of the spliceosomal snRNPs is not limiting the splicing of U12-type introns. RNA. 2006;12:1883–1892. doi: 10.1261/rna.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denis MM, et al. Escaping the nuclear confines: Signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122:379–391. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glanzer J, et al. RNA splicing capability of live neuronal dendrites. Proc Natl Acad Sci USA. 2005;102:16859–16864. doi: 10.1073/pnas.0503783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hastings ML, Krainer AR. Functions of SR proteins in the U12-dependent AT-AC pre-mRNA splicing pathway. RNA. 2001;7:471–482. doi: 10.1017/s1355838201002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taneja KL, Lifshitz LM, Fay FS, Singer RH. Poly(A) RNA codistribution with microfilaments: evaluation by in situ hybridization and quantitative digital imaging microscopy. J Cell Biol. 1992;119:1245–1260. doi: 10.1083/jcb.119.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartmuth K, et al. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc Natl Acad Sci USA. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kastner B, Kornstadt U, Bach M, Lührmann R. Structure of the small nuclear RNP particle U1: Identification of the two structural protuberances with RNP-antigens A and 70K. J Cell Biol. 1992;116:839–849. doi: 10.1083/jcb.116.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Will CL, et al. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 2001;20:4536–4546. doi: 10.1093/emboj/20.16.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reuter K, Nottrott S, Fabrizio P, Lührmann R, Ficner R. Identification, characterization and crystal structure analysis of the human spliceosomal U5 snRNP-specific 15 kD protein. J Mol Biol. 1999;294:515–525. doi: 10.1006/jmbi.1999.3258. [DOI] [PubMed] [Google Scholar]
- 46.Makarova OV, Makarov EM, Liu S, Vornlocher H-P, Lührmann R. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6·U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 2002;21:1148–1157. doi: 10.1093/emboj/21.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ingelfinger D, Arndt-Jovin DJ, Lührmann R, Achsel T. The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.