Abstract
Many developmentally regulated genes contain a poised RNA polymerase II (Pol II) at their promoters under conditions where full-length transcripts are undetectable. It has been proposed that the transcriptional activity of such promoters is regulated at the elongation stage of Pol II transcription. In Drosophila, the heat-shock loci expressing the Hsp70 genes have been used as a model for the regulation of the transcriptional activity of poised Pol II. Drosophila ELL (dELL) is a Pol II elongation factor capable of stimulating the rate of transcription both in vivo and in vitro. Although ELL and the elongation factor Elongin A have indistinguishable effects on RNA polymerase in vitro, the loss-of-function studies indicate that these proteins are not redundant in vivo. In this article, we use RNAi to investigate the physiological properties of dELL and a dELL-associated factor (dEaf) in a living organism. Both ELL and Eaf are essential for fly development. dELL is recruited to heat shock loci upon induction, and its presence with Pol II at such loci is required for proper heat-shock gene expression. Consistent with a role in elongation, dELL knockdown reduces the levels of phosphorylated Pol II at heat-shock loci. This study implicates dELL in the expression of loci regulated by Pol II elongation.
Keywords: gene expression, heat-shock response, transcription elongation
Efficient transcription by RNA polymerase II (Pol II) is an intricate process that requires multiple contacts with the DNA template and nascent RNA that inevitably leads to frequent stalling during the transcription of a gene (1). The average rate of transcription by Pol II in vivo is an order of magnitude higher than that obtained in vitro despite additional impediments, such as traversing through nucleosomes. Using biochemical approaches, two Pol II elongation factors, Eleven nineteen lysine-rich leukemia (ELL) and Elongin A, were isolated from cell extracts as factors capable of stimulating Pol II activity by suppressing transient pausing. Despite similar in vitro activities, the Drosophila orthologs of ELL and Elongin A are each essential for development (2, 3). This observation indicates that their in vivo activity is not redundant.
Recent genome-wide studies have found a large number of developmentally regulated genes that contain a paused Pol II at their promoters (4, 5). Therefore, it has been proposed that the transcriptional activity of such poised Pol IIs is regulated at the level of transcription elongation. The classic model for studying genes regulated by promoter-proximal paused polymerase is Hsp70 gene induction in Drosophila (6). Previous studies have shown that several Pol II elongation factors are rapidly recruited to the Hsp70 genes after heat shock (7–13). Although much work has been done on the role of these factors in gene regulation in cultured cells, less is known about the role of these factors in the regulation of heat-shock gene expression in the whole organism. Although there are several mutants in the gene encoding Drosophila ELL (dELL), all of these alleles are embryonic lethal (2). Therefore, we have not been able to use these alleles to further characterize the role of the elongation factor ELL in the regulation of the transcriptional activity of poised Pol II and Hsp70 loci. To test the role for dELL in gene expression, we have used RNAi to reduce expression levels of both dELL and dELL-associated factor (dEaf) expression levels during development, and we have examined the in vivo effect of their reduction on transcription and development. We find that knockdown of dELL and dEaf results in lethality. Furthermore, knockdown of these elongation factors results in reduced Hsp70 transcript accumulation after heat shock. Immunolocalization of phosphorylated Pol II in heat-shocked dELL knockdown salivary glands demonstrates reduced levels of the elongating form of Pol II at the Hsp70 loci in the absence of dELL. Our studies demonstrate that dELL is essential for full induction of heat-shock gene expression and are consistent with a role for dELL in Pol II elongation. These findings provide a role for an RNA Pol II elongation factor in the transcriptional regulation of poised Pol II.
Results and Discussion
dELL Knockdown by RNAi Leads to Loss of Viability.
dELL was previously shown to be essential; homozygous mutant clones do not survive in the eye (14) and homozygotes for loss-of-function alleles die at the end of embryogenesis or in early first instar (2). To investigate the role of dELL in transcription in flies, we chose to knock down dELL by RNAi, which typically reduces, but does not eliminate, the targeted gene products. A 600-bp portion of the dELL coding region was inserted into a P-element vector that drives the expression of dsRNA through two convergent Gal4 UAS promoters that flank the insert (see Materials and Methods). Several transgenic lines were generated and tested for effects on viability by crossing to an Actin5C-Gal4 driver line that expresses yeast Gal4 under the cytoplasmic actin promoter. All eight dELL RNAi lines show significant loss of viability when expressed under this driver (Table 1). When adult escapers were obtained, very few males were observed, indicating that males are more susceptible to loss of dELL. We also observed greater numbers of females than males at the third instar larval stage, indicating that males are dying earlier than females (data not shown). A significant genome-wide reduction of dELL protein is observed by immunofluorescence analysis of dELL RNAi larval polytene chromosomes (Fig. 1).
Table 1.
Effect of dELL RNAi on viability
| RNAi line | Act5c-Gal4 |
CyO, y+ |
||||
|---|---|---|---|---|---|---|
| Female | Male | Total | Female | Male | Total | |
| dELL 1 | 0 | 0 | 0 | 58 | 55 | 113 |
| dELL 3 | 2 | 0 | 2 | 46 | 37 | 83 |
| dELL 4 | 0 | 0 | 0 | 43 | 58 | 101 |
| dELL 5 | 0 | 0 | 0 | 78 | 82 | 160 |
| dELL 6 | 0 | 0 | 0 | 54 | 46 | 100 |
| dELL 7 | 0 | 0 | 0 | 62 | 54 | 116 |
| dELL 8 | 45 | 1 | 46 | 62 | 80 | 142 |
| dELL 9 | 14 | 0 | 14 | 67 | 57 | 124 |
Fig. 1.
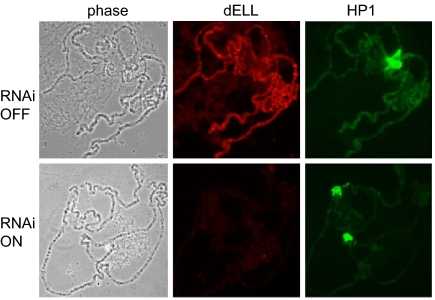
Reduction of dELL levels on polytene chromosomes after RNAi. Flies carrying a Gal4-responsive dsRNA construct were crossed to flies expressing one copy of Gal4 driven by the Actin5C promoter. (Upper) Salivary gland polytene chromosomes from control larvae (RNAi OFF) carrying the second chromosome balancer CyO were seen to express dELL at numerous interband sites of polytene chromosomes. (Lower) In contrast, RNAi larvae (RNAi ON), expressing Gal4, have greatly reduced levels of dELL throughout the salivary gland nucleus. HP1 levels on control and dELL knockdown larvae were comparable.
Through two-hybrid analysis, two interacting partners of ELL have been characterized in humans, Eaf1 and Eaf2 (15, 16). Eaf1 and Eaf2 are highly related and can stimulate the elongation activity of ELL in vitro (17). Recently, the association of Eaf with ELL was shown to be evolutionarily conserved, with the finding that Schizosaccharomyces pombe homologs SpEaf and SpELL directly interact with each other (18). Additionally, SpEaf enhances the stimulation by SpELL of Pol II transcription in vitro (18). Because Drosophila also has a single Eaf homolog, we used RNAi to knock down dEaf levels and assessed the viability of dEaf-knockdown flies in six different transgenic RNAi lines (Table 2). In all lines, we observed significant reductions in the number of adult progeny of RNAi-expressing flies compared with control siblings. In addition, a consistent reduction in the male–female sex ratio was observed for dEaf RNAi, suggesting that the male-enhanced lethal phenotype (not observed by us for other elongation factors) is due to loss of a dELL–dEaf complex.
Table 2.
Effect of dEaf RNAi on viability
| RNAi line | Act5c-Gal4 |
CyO, y+ |
||||
|---|---|---|---|---|---|---|
| Female | Male | Total | Female | Male | Total | |
| dEaf 1 | 143 | 60 | 203 | 228 | 240 | 468 |
| dEaf 2 | 204 | 73 | 277 | 241 | 221 | 462 |
| dEaf 3 | 195 | 64 | 259 | 189 | 152 | 341 |
| dEaf 4 | 60 | 5 | 65 | 223 | 228 | 451 |
| dEaf 6 | 235 | 97 | 332 | 288 | 324 | 612 |
| dEaf 8 | 268 | 139 | 397 | 240 | 310 | 547 |
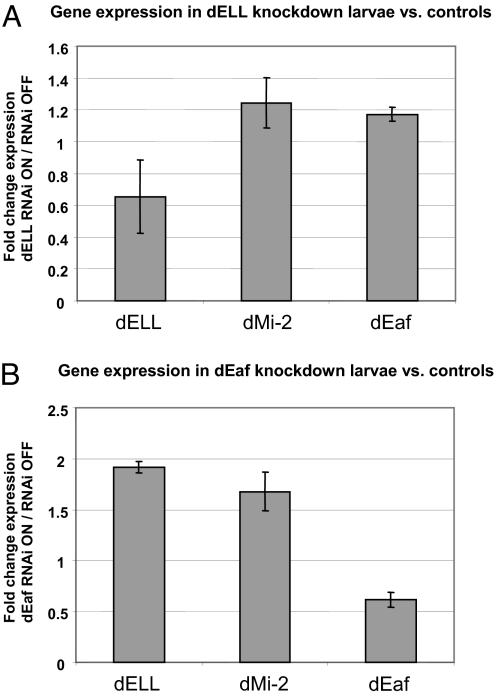
To test for the effectiveness of the RNAi knockdowns, we measured dELL and dEaf mRNA levels in knockdown larvae and their control siblings. Significant reductions in dELL transcripts are observed in the dELL RNAi larvae (Fig. 2A). dELL transcripts, as measured by RT-PCR, are not reduced by RNAi to the same level as dELL protein, as assessed by immunofluorescence on polytene chromosomes. Previously, we observed that knockdown of dRTF1 by RNAi was more effective at the protein than the RNA levels presumably because the long dsRNAs produced are processed as miRNAs and interfere with translation (13). Because dELL is nested in an intron of the gene encoding the chromatin remodeling enzyme dMi-2, we also measured transcript levels for this gene and found no reduction of dMi-2 RNA in dELL RNAi larvae (Fig. 2A). Additionally, we find that dEaf RNA levels are reduced in dEaf RNAi larvae (Fig. 2B). Interestingly, a significant increase in dELL levels is observed in dEaf RNAi larvae, possibly compensating for the lower dEaf levels (Fig. 2B).
Fig. 2.
dELL and dEaf mRNA levels are reduced by RNAi in transgenic flies. (A) dELL mRNA levels are reduced in dELL knockdown larvae relative to control larvae. RNA was extracted from dELL knockdown larvae or their control siblings and used for real-time RT-PCR using primers to dELL. For comparison, expression levels of dMi-2, the gene whose large intron contains the dELL gene, and expression levels of dEaf, an interacting partner of dELL, were measured. Values are normalized to rp49 and represent the average of three collections of control and knockdown larvae. Error bars indicate the standard error. (B) dEaf mRNA levels are reduced in dEaf knockdown larvae. RT-PCR was performed as in A, but with dEaf knockdown larvae and their control siblings.
dELL Is Required for Heat-Shock Gene Expression.
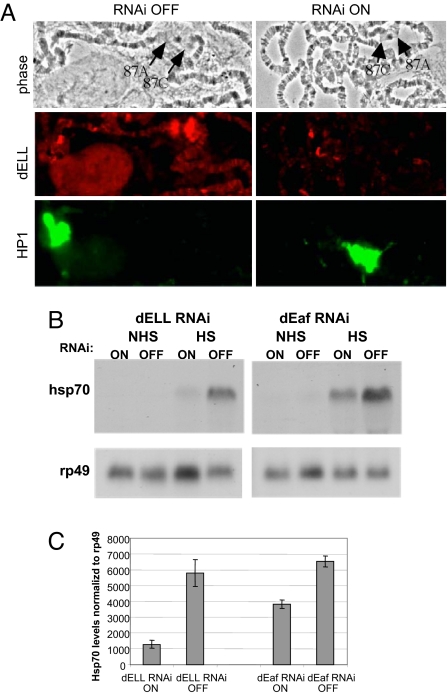
dELL was previously shown to be recruited to heat-shock genes upon heat shock (7). To determine whether dELL is required for heat-shock gene expression, we compared the levels of Hsp70 transcripts after heat shock in dELL knockdown larvae and their control siblings. By immunofluorescence analysis, little or no dELL is seen at the Hsp70 gene after heat shock in dELL knockdown larvae, whereas the control siblings without the Gal4 driver showed the expected recruitment of dELL to the Hsp70 gene (Fig. 3A). Northern blot analysis showed reduced levels of Hsp70 mRNA levels in the dELL RNAi larvae (Fig. 3B). A similar analysis was done with dEaf RNAi larvae, and reduced Hsp70 mRNA also occurs after heat shock, although the deficit was less than observed for the dELL RNAi larvae (Fig. 3B). Similar results were observed when Hsp70 levels were measured by RT-PCR, showing greater reductions in Hsp70 RNA levels in dELL RNAi than dEaf RNAi larvae (Fig. 3C).
Fig. 3.
Hsp70 induction is reduced in dELL and dEaf RNAi knockdown larvae. (A) Knockdown dELL RNAi larvae (RNAi ON) or control siblings (RNAi OFF) were heat-shocked and stained with antibodies to dELL. Although dELL normally goes to the major heat-shock loci, it cannot be detected at the 87A/C loci in the knockdown larvae. HP1 levels are comparable in the knockdown and control larvae. Phase contrast images are shown. (B) Heat-shock response is lower in dELL and dEaf knockdown larvae. (Upper) Hsp70 mRNA levels were measured by Northern blotting in dELL and dEaf knockdown and control larvae, before heat shock [no heat shock (NHS)], and after heat shock (HS). (Lower) rp49 was probed as a loading control. (C) Similar reductions in Hsp70 mRNA levels after heat shock induction are seen by RT-PCR. Error bars indicate the standard error.
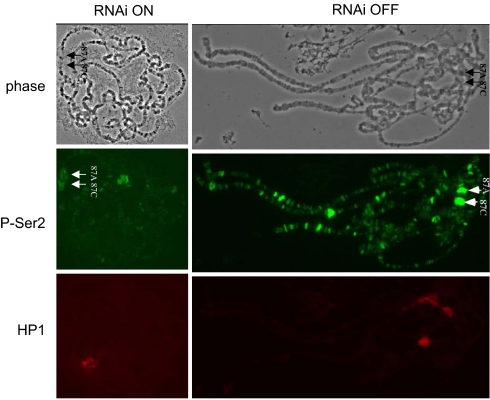
Chromosomal levels of dELL are markedly reduced in the absence of Cdk9, the catalytic subunit of the Pol II C-terminal domain (CTD) kinase PTEF-B (19). To determine whether dELL knockdown affects the recruitment of Pol II to the Hsp70 genes, dELL knockdown and control polytene chromosomes were probed with antibodies to the Ser-2-phosphorylated, elongating form of Pol II. We consistently observed lower levels of Ser-2-phosphorylated Pol II at the Hsp70 heat-shock loci in dELL-knockdown larvae (Fig. 4), suggesting a close link between dELL function and phosphorylation of the Pol II CTD.
Fig. 4.
dELL knockdown results in the reduction of the Ser-2-phosphorylated form of Pol II. Polytene chromosome preparations from dELL knockdown (RNAi ON) and control larvae (RNAi OFF) were probed with the H5 monoclonal antibody to the Ser-2-phosphorylated, elongating form of Pol II. Representative images of the average staining level of Ser-2-phosphorylation levels are shown for the dELL knockdown and control. HP1 levels were unchanged.
dELL and dEloA Are Not Redundant in Vivo.
ELL belongs to a class of transcription elongation factors that have been shown to stimulate the Km and/or Vmax of RNA Pol II in vitro by alleviating pausing on a purified DNA template. Another member of this class is Elongin A and its Drosophila ortholog dEloA (1, 3). From the present and previous studies, it is clear that both dELL and dEloA localize to the Hsp70 gene upon heat shock, and each is required for full levels of heat-shock gene expression, suggesting that the in vivo roles of these elongation factors in Hsp70 gene transcription are not redundant (8). Similarly, we have observed that the knockdown phenotypes of these two proteins can be unique, such as the enhanced male lethality in dELL RNAi larvae (Table 1). How could both elongation factors be redundant in vitro, yet nonredundant in vivo? The in vitro studies were performed on naked DNA templates, whereas the chromatin environment of RNA Pol II-transcribed genes can provide additional challenges to the polymerase. Each of these elongation factors has its own interaction partners and may be recruited to distinct states of the polymerase, such as initiating, elongating, or stalled polymerase. Consistent with this view, knockdown of dELL, but not dEloA, results in decreased levels of Ser-2-phosphorylated Pol II at the Hsp70 and other loci (Fig. 4) (8). Interestingly, the chromosomal targeting of dELL, but not dEloA, is dramatically reduced by the knockdown of CDK9, the Pol II CTD kinase (19), suggesting that dELL and dEloA are recruited to genes by distinct mechanisms. Fine mapping of dELL and dEloA on the well characterized Hsp70 gene at different time points after activation could clarify the distinct roles for these enzymes.
The lesser effect of dEaf knockdown on Hsp70 gene induction could be indicative of a requirement of dEaf for optimal function of dELL, whereas dELL can partially function without dEaf. Indeed, in vitro transcription studies have demonstrated that human Eaf proteins, in combination with ELL, stimulate transcription elongation by Pol II above the levels obtained with ELL alone (17). In dEaf RNAi larvae, we observed that dELL levels are increased, conceivably as a cellular response to increased pausing resulting from lower dEaf levels.
Males Are More Sensitive to dELL Knockdown.
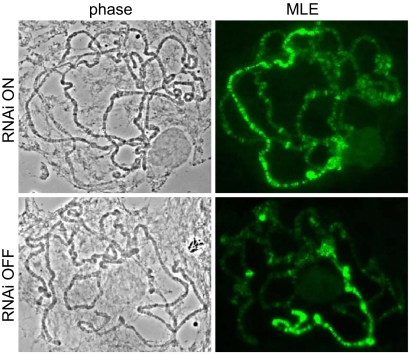
Previous work on the function of dELL made use of alleles of the Su(Tpl) locus, which encodes dELL (2). All known Su(Tpl) alleles are embryonic lethal. In contrast, RNAi of dELL allows survival to the larval or adult stages depending on the insertion line of the dsRNA construct. Interestingly, the few “escaper” dELL RNAi adults are overwhelmingly female. As seen with the heat-shock defect, the difference in male and female viability is less in dEaf RNAi flies than in dELL RNAi flies, consistent with dEaf enhancing, but not being absolutely required for, dELL function. A previous study showed that males express much higher levels of a dELL transcript than females, although the functional significance of this difference has not been investigated (7). One hypothesis is that dELL is needed in males as part of the process of X chromosome dosage compensation; Drosophila dosage compensation factors are thought to enhance transcription elongation of X-linked genes in males, and loss of any of these factors leads to male-specific lethality (20). In addition, reduced levels of several global chromatin regulators, including the supercoiling factor, Jil-1 H3 kinase, heterochromatin protein HP1, and the chromatin remodeler ISWI, have been reported to differentially affect the survival of males and/or the morphology of the X chromosome (21–24). However, in dELL knockdowns, MSL localization and the male polytene X chromosome morphology appears similar in dELL knockdown male larvae and their control brothers (Fig. 5). Whether there are specific defects in dosage compensation of X-linked genes may be an interesting avenue for future investigations. Alternative explanations for a male-enhanced lethality also should be considered. For example, Drosophila males differ from females not just in having one less X chromosome, but also in carrying a Y chromosome, which comprises ≈12% of the male genome. A number of genes are male-lethal due to the presence of the mostly heterochromatic Y chromosome, including modulators of position effect variegation, such as the Su(var)3-3 gene that encodes the histone demethylase LSD1, the uncharacterized Su(var)2-1, as well as the HP1-interacting protein Bonus (dTIF1), an enhancer and suppressor of position-effect variegation (reviewed in ref. 25). For Su(var)2-1 and Bonus, the Y-lethal effect is not Y-specific but can be phenocopied by other sources of heterochromatin (25). A role for dELL in the regulation of heterochromatin is unknown but could conceivably be required for the expression of heterochromatin components.
Fig. 5.
X chromosome morphology and male-specific lethal protein localization are not apparently affected by reduction of dELL. Polytene chromosome preparations from dELL knockdown males (RNAi ON) or their control brothers (RNAi OFF) were probed with antibodies to MLE, an RNA helicase that is required for dosage compensation in Drosophila and spreading of the complex along the X chromosome (20). MLE staining patterns appear similar in knockdown and control brothers.
The finding that a large number of developmentally regulated genes have Pol II poised at their promoters in the absence of detectable full-length transcripts suggests that regulated Pol II elongation is an important transcription regulatory mechanism in eukaryotes. Consistent with that view, the Drosophila ELL is an essential protein that, as shown here, is required for the full induction of Hsp70, one of the earliest examples of a gene regulated by Pol II elongation. Our current model is that dELL is a Pol II elongation factor that controls the expression levels of diverse genes. Importantly, our studies in Drosophila are beginning to define distinct regulatory roles for different Pol II elongation factors that otherwise behave similarly in vitro. A biochemical description of how the binding of dELL (and its associated factor dEaf) enhances Pol II elongation is essential to understanding gene regulation during development.
Materials and Methods
Fly Stocks and Crosses.
A 600-bp region (1,200–1,800 bp) of the dELL ORF or a 600-bp region (200–800 bp) of the dEaf ORF was cloned into Sym-pUAST-w P element vector (26) vector and injected into yw embryos to generate transgenic flies. To induce dsRNA, yw; P(Sym-pUAST-dELL) or yw; P(Sym-pUAST-dEaf), males were crossed to yw; Act5C-Gal4/CyO, y+ females.
Antibodies.
dELL rabbit polyclonal serum was raised against full-length recombinant His6-tagged dELL and was described previously (27). Anti-HP1 monoclonal antibody C1A9 was a gift from Sarah Elgin (Washington University, St. Louis). Anti-Ser-2-phosphorylated CTD of Pol II H5 monoclonal was purchased from Covance. MLE rabbit polyclonal serum was a gift from John Lucchesi (Emory University, Atlanta). Polytene chromosome staining was performed as described in ref. 28. Briefly, salivary glands were fixed for 30 s in 2% paraformaldehyde and then in 45% acetic acid/2% formaldehyde for 3 min. RNAi knockdown and control larvae were chosen based on the presence of yellow or black mouth hooks, respectively.
Northern Blot Analysis and Real-Time PCR.
Northern blotting was done as described previously (8). Briefly, third-instar larvae, sorted by the color of the mouthooks as described above, were untreated or heat shocked for 30 min. Then 15 μg of total RNA was separated on 1% formaldehyde-agarose gel, blotted to Biotrans nylon membrane (ICN), and hybridized with a riboprobe in 50% formamide at 65°C.
For real-time PCR, total RNA from larvae was treated with DNase and repurified by RNeasy columns (Qiagen). Finally, 50 ng of total RNA was used in a 25-μl reaction with the iScript one-step RT-PCR kit with SYBR Green in a Bio-Rad MyCycler.
Primers used were: rp49, AGAGTTCTTGTAACGTGGTCGGAATA and CAATGGTGCTGCTATCCCAATC; Hsp70, GGGTGTGCCCCAGATAGAAG and TGTCGTTCTTGATCGTGATGTTC (29); dELL, TTCCAGGACTTATCCGAACG and CAATGGTGCGATCGTTATTG; dEaf, CAGGGAGCATGAACTGACCT and TCTGTGCTGTCATCCGAGTC; and dMi-2, TTCAGCGACACAAGGACAAG and TGCCGTACGATGGTTGATTA.
Acknowledgments.
We thank Tyler Marquart for technical assistance and Sarah Elgin (Washington University, St. Louis, MO) and John Lucchesi (Emory University, Atlanta, GA) for providing reagents. This work was supported by National Science Foundation Grants MCB 0131414 and MCB 0615831 (to J.C.E.) and National Institutes of Health Grant CA089455 (to A.S.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu Rev Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- 2.Eissenberg JC, et al. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc Natl Acad Sci USA. 2002;99:9894–9899. doi: 10.1073/pnas.152193699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerber M, et al. In vivo requirement of the RNA polymerase II elongation factor elongin A for proper gene expression and development. Mol Cell Biol. 2004;24:9911–9919. doi: 10.1128/MCB.24.22.9911-9919.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rougvie AE, Lis JT. Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990;10:6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerber M, Ma J, Dean K, Eissenberg JC, Shilatifard A. Drosophila ELL is associated with actively elongating RNA polymerase II on transcriptionally active sites in vivo. EMBO J. 2001;20:6104–6114. doi: 10.1093/emboj/20.21.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerber M, et al. Regulation of heat shock gene expression by RNA polymerase II elongation factor, Elongin A. J Biol Chem. 2005;280:4017–4020. doi: 10.1074/jbc.C400487200. [DOI] [PubMed] [Google Scholar]
- 9.Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 12.Saunders A, et al. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 13.Tenney K, et al. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc Natl Acad Sci USA. 2006;103:11970–11974. doi: 10.1073/pnas.0603620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neufeld TP, Tang AH, Rubin GM. A genetic screen to identify components of the sina signaling pathway in Drosophila eye development. Genetics. 1998;148:277–286. doi: 10.1093/genetics/148.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simone F, et al. EAF1, a novel ELL-associated factor that is delocalized by expression of the MLL-ELL fusion protein. Blood. 2001;98:201–209. doi: 10.1182/blood.v98.1.201. [DOI] [PubMed] [Google Scholar]
- 16.Simone F, Luo RT, Polak PE, Kaberlein JJ, Thirman MJ. ELL-associated factor 2 (EAF2), a functional homolog of EAF1 with alternative ELL binding properties. Blood. 2003;101:2355–2362. doi: 10.1182/blood-2002-06-1664. [DOI] [PubMed] [Google Scholar]
- 17.Kong SE, Banks CA, Shilatifard A, Conaway JW, Conaway RC. ELL-associated factors 1 and 2 are positive regulators of RNA polymerase II elongation factor ELL. Proc Natl Acad Sci USA. 2005;102:10094–10098. doi: 10.1073/pnas.0503017102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banks CA, et al. Identification and Characterization of a Schizosaccharomyces pombe RNA polymerase II elongation factor with similarity to the metazoan transcription factor ELL. J Biol Chem. 2007;282:5761–5769. doi: 10.1074/jbc.M610393200. [DOI] [PubMed] [Google Scholar]
- 19.Eissenberg JC, Shilatifard A, Dorokhov N, Michener DE. Cdk9 is an essential kinase in Drosophila that is required for heat shock gene expression, histone methylation and elongation factor recruitment. Mol Genet Genomics. 2007;277:101–114. doi: 10.1007/s00438-006-0164-2. [DOI] [PubMed] [Google Scholar]
- 20.Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 21.Furuhashi H, Nakajima M, Hirose S. DNA supercoiling factor contributes to dosage compensation in Drosophila. Development (Cambridge, UK) 2006;133:4475–4483. doi: 10.1242/dev.02620. [DOI] [PubMed] [Google Scholar]
- 22.Deuring R, et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Zhang W, Jin Y, Johansen J, Johansen KM. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell. 2001;105:433–443. doi: 10.1016/s0092-8674(01)00325-7. [DOI] [PubMed] [Google Scholar]
- 24.Liu LP, Ni JQ, Shi YD, Oakeley EJ, Sun FL. Sex-specific role of Drosophila melanogaster HP1 in regulating chromatin structure and gene transcription. Nat Genet. 2005;37:1361–1366. doi: 10.1038/ng1662. [DOI] [PubMed] [Google Scholar]
- 25.Beckstead RB, et al. Bonus, a Drosophila TIF1 homolog, is a chromatin-associated protein that acts as a modifier of position-effect variegation. Genetics. 2005;169:783–794. doi: 10.1534/genetics.104.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giordano E, Rendina R, Peluso I, Furia M. RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics. 2002;160:637–648. doi: 10.1093/genetics/160.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerber MA, Shilatifard A, Eissenberg JC. Mutational analysis of an RNA polymerase II elongation factor in Drosophila melanogaster. Mol Cell Biol. 2005;25:7803–7811. doi: 10.1128/MCB.25.17.7803-7811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eissenberg JC. Functional genomics of histone modification and non-histone chromosomal proteins using the polytene chromosomes of Drosophila. Methods. 2006;40:360–364. doi: 10.1016/j.ymeth.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;21:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]