Abstract
Analysis of marine cyanobacteria and proteobacteria genomes has provided a profound understanding of the life strategies of these organisms and their ecotype differentiation and metabolisms. However, a comparable analysis of the Bacteroidetes, the third major bacterioplankton group, is still lacking. In the present paper, we report on the genome of Polaribacter sp. strain MED152. On the one hand, MED152 contains a substantial number of genes for attachment to surfaces or particles, gliding motility, and polymer degradation. This agrees with the currently assumed life strategy of marine Bacteroidetes. On the other hand, it contains the proteorhodopsin gene, together with a remarkable suite of genes to sense and respond to light, which may provide a survival advantage in the nutrient-poor sun-lit ocean surface when in search of fresh particles to colonize. Furthermore, an increase in CO2 fixation in the light suggests that the limited central metabolism is complemented by anaplerotic inorganic carbon fixation. This is mediated by a unique combination of membrane transporters and carboxylases. This suggests a dual life strategy that, if confirmed experimentally, would be notably different from what is known of the two other main bacterial groups (the autotrophic cyanobacteria and the heterotrophic proteobacteria) in the surface oceans. The Polaribacter genome provides insights into the physiological capabilities of proteorhodopsin-containing bacteria. The genome will serve as a model to study the cellular and molecular processes in bacteria that express proteorhodopsin, their adaptation to the oceanic environment, and their role in carbon-cycling.
Keywords: Bacteroidetes, marine bacteria, whole-genome analysis, heterotrophic CO2 fixation
Bacteroidetes are successful in the degradation of particulate organic matter in the ocean (1, 2), and at least one molecular study showed them to be more abundant on particles than free-living in the water column (3). Many representatives have gliding motility and the capacity to degrade polymers, possibly allowing them to grow on detritus particles or algal cells using the polymeric substances as carbon and energy sources. Using microautoradiography combined with FISH, for example, Cottrell and Kirchman (4) showed that these bacteria are better adapted to the consumption of proteins over that of amino acids. Functional analysis of the genome of Gramella forsetii indicated that this marine Bacteroidetes has a substantial number of hydrolytic enzymes and a predicted preference for polymeric carbon sources (5). Although the diversity of Bacteroidetes is large, the adaptation to the degradation of polymeric substances seems a common theme. This trait contrasts that of another major group of marine bacteria, the proteobacteria: both alpha- and gammaproteobacteria seem better adapted to use monomers rather than polymers (4) and to a free-living existence in the water column. Therefore, the study of Bacteroidetes promises to reveal novel life strategies for successfully populating the surface ocean different from those of the proteobacteria whose complete genomes have been analyzed thus far (6, 7).
Here, we present the genome of Polaribacter sp. MED152. This genome was chosen for manual annotation and analysis for two reasons. First, it is representative of marine Bacteroidetes. Direct counts by FISH repeatedly show Bacteroidetes to account for ≈10–20% of the prokaryotes in seawater (8, 9), most belonging to the flavobacteria (10, 11). In 2004, there were a total of 864 16S rRNA gene sequences from marine Bacteroidetes in GenBank, of which 76 (9%) belonged to the genus Polaribacter (12). Of the Polaribacter sequences, 27 (36%) were most closely related to Polaribacter dokdonensis. Thus, Polaribacter is one of the major genera of Bacteroidetes found in the marine environment. Second, screening of the draft genome revealed the proteorhodopsin gene. The gene for this membrane protein was first found in DNA fragments directly obtained from seawater and functions as a light-driven H+ pump in the ocean (13). Subsequent work has demonstrated a wide diversity and distribution of proteorhodopsin in the surface ocean bacterioplankton. Escherichia coli transformed with the proteorhodopsin gene can in fact use light energy for photophosphorylation (14) and cellular activities such as flagellar motion (15). Recently, proteorhodopsin genes have been found in some cultured isolates (7, 16), a few belonging to the Bacteroidetes phylum (17). The presence of the proteorhodopsin gene in cultured bacteria opens the possibility to study the function of proteorhodopsin in vivo.
We have recently shown (17) that Dokdonia sp. MED134 (a relative of Polaribacter) can use light energy gathered through proteorhodopsin to grow better in the light than in the dark. This is different from the light response of the alphaproteobacterium Pelagibacter ubique and gammaproteobacterium strain HTCC2207, neither of which has been shown to grow better in the light despite the presence of a functional proteorhodopsin gene (7, 16). Accordingly, genome analysis of proteorhodopsin-containing flavobacteria opens a unique window to understand evolutionary adaptations to grow in a sunlit environment. Our present genome analysis indicates that the strategy of Polaribacter sp. MED152 to grow in seawater is different from that of other groups of abundant marine bacteria.
Results and Discussion
Genome Properties.
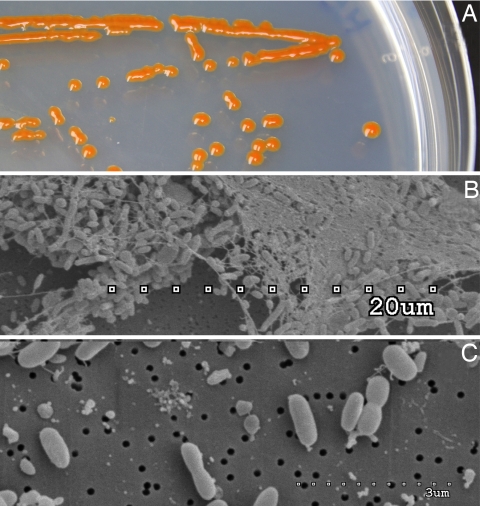
MED152 forms bright-orange colonies on agar plates and tends to aggregate into large flocks in liquid culture (Fig. 1). The genome contains 2,967,150 bp with 2,692 predicted genes. This is a relatively small genome size for a marine bacterium. For example, 75% of the genomes in the Gordon and Betty Moore Foundation Marine Microbiology Initiative (total of 116 sequenced prokaryotes so far) have genomes larger than MED152 (with primarily SAR11 and Prochlorococcus genomes being smaller). Moreover, this is among the smallest genomes of Bacteroidetes isolates sequenced until now. The reduced genome size of MED152 is a consequence of a reduced number of protein-coding genes and gene families compared with most other marine bacteria, in combination with a low number of paralogs in each family. General genome features are presented in supporting information (SI) Table S1. Although not completely closed, the genome sequence is on a single contig. G+C skew analysis indicates that the chromosome is circular (data not shown). The largest protein families are peptidases (93 ORFs), glycosyl hydrolases (30 ORFs), TonB-dependent outer membrane channels (27 ORFs), response regulators (25 ORFs), glycosyl transferases (25 ORFs), ABC transporters (22 ORFs), and His kinases (21 ORFs).
Fig. 1.
Images of MED152. (A) Colonies on marine agar showing the characteristic orange color. (B) SEM image of a typical aggregate showing abundant extracellular material. (C) SEM image showing individual cells.
Most peptides annotated as conserved hypothetical or as functional proteins are most similar to peptides in the closely related Polaribacter irgensii 23-P (1,506 genes; based on BLASTP). Remaining peptides had homologues in other members of the Bacteroidetes phylum. It does not contain any insertion sequence element or peptides related to lysogenic phages, but five genes encode phage integrase family proteins that could allow for site-specific recombination.
Central Metabolism.
The metabolic pathways identified in the genome of MED152 are shown in Fig. S1–S3 and Table S2. As expected in a bacterium living in surface waters, the genome of MED152 has adaptations for protection against stress from reactive oxygen species and repair of DNA damage (Tables S3 and S4). Consistent with its life in the ocean, MED152 has a Na+- rather than a H+-translocating NADH:quinone oxidoreductase as a part of the respiratory chain to establish a Na+ gradient for energy coupling (18).
There are two main characteristics of the central metabolism of MED152: one is logical, and the other intriguing. The first one is logical, considering the relatively small size of the genome: MED152 has a modest number of metabolic capabilities. It can grow only by aerobic respiration, although it does have a cbb3-type cytochrome oxidase mainly found in microaerophiles; it is unable to use fermentation or anaerobic respiration for energy conservation. Its genome also lacks the Entner–Doudouroff pathway; ammonia and sulfate are the single inorganic sources of nitrogen and sulfur, respectively (Table S2). MED152 cannot use dimethylsulfoniopropionate or common products in algal exudates such as glycolate, taurine or polyamines, and it cannot oxidize inorganic sulfur compounds or carbon monoxide. We could identify only the metabolic pathways for the main cellular components (Table S2) but none of the rich variety of pathways found for example in Silicibacter pomeroyi (6).
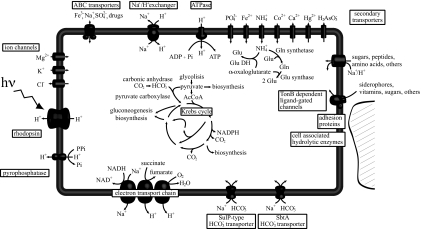
The second characteristic is the remarkable number of genes potentially involved in anaplerotic metabolism (Table S2). First, MED152 has two types of putative bicarbonate importer genes similar to those found in cyanobacteria. The first, bicA (MED152_09030, a SulP-type Na+-dependent bicarbonate transporter), has a relatively low affinity for the substrate but a high flux rate (19), whereas the second, sbtA (MED152_03855, also a Na+-dependent transporter), has high affinity for bicarbonate (20). These bicarbonate importer genes are complemented by a carbonic anhydrase gene that interconverts CO2 and bicarbonate (Fig. 2).
Fig. 2.
Diagram of a MED152 cell. Processes that generate a H+ or Na+ gradient are shown Lower and Left. Central metabolism pathways, as well as anaplerotic reactions, are indicated. Bicarbonate transporters are shown Lower.
Further, MED152 has two putative anaplerotic enzymes that use bicarbonate. These are PEP carboxylase (MED152_09950) and pyruvate carboxylase (MED152_04060), which generate oxaloacetate from bicarbonate and PEP and from bicarbonate and pyruvate, respectively. Both enzymes are found in two other proteorhodopsin-containing Bacteroidetes, P. irgensii 23-P and Dokdonia sp. MED134. None of the remaining genome-sequenced marine proteobacteria with proteorhodopsin contain both pyruvate and PEP carboxylase genes (Table S5). Pyruvate carboxylase activity requires ATP, but PEP carboxylase does not. Because ATP generation in proteorhodopsin-containing bacteria could be independent of organic matter respiration in the presence of light, it may be speculated that some of this ATP could be used in biosynthesis through pyruvate carboxylase. In any case, the response of different proteorhodopsin-containing bacteria to light may differ depending on the genome context (14), in this case, presence of specific transporters or carboxylases.
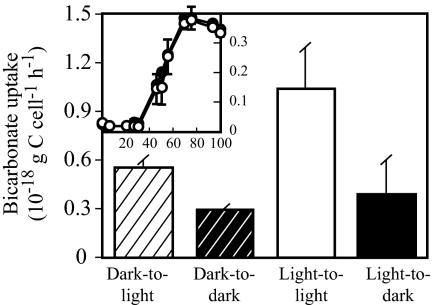
Carbonate concentrating mechanisms similar to those mentioned above play a critical role in CO2-fixation pathways in autotrophic organisms but could also be essential for anaplerotic CO2 fixation in heterotrophs. MED152 does not have any of the conventional bicarbonate fixation pathways of autotrophs, and it does not grow in a medium without organic matter. It is, therefore, a heterotroph. Growth of MED152 was measured in diluted marine broth cultures exposed to light or darkness, to examine a potential benefit of harboring proteorhodopsin. In this relatively rich medium, growth was similar in the light and in the dark (Fig. 3Inset). In the related Dokdonia sp. MED134, growth in the light was better than in the dark only in medium with very low organic matter (17). We have not been successful in designing a simple poor medium where a light effect on growth could be shown at its best if MED152 behaved exactly as Dokdonia sp. MED134. Nevertheless, experiments with radioactively labeled bicarbonate showed that, even though no differences were found in cell numbers, MED152 fixed more bicarbonate in the light than in the dark (Fig. 3). Notably, bicarbonate uptake rates were higher in the light irrespective of light conditions before the uptake experiment. These results suggest that light-stimulated anaplerotic fixation of bicarbonate could play a significant role in the life strategy of MED152.
Fig. 3.
Bicarbonate uptake. Striped columns denote subsamples from cultures grown in the dark, incubated with H14CO3− in the light (dark-to-light) or dark (dark-to-dark). Solid columns indicate subsamples from cultures grown in the light, incubated with H14CO3− in the light (light-to-light) or dark (light-to-dark). (Inset) Growth of cultures in the dark (filled circles) and in the light (open circles) from where subsamples for bicarbonate uptake assays were collected at 50 h; the x axis shows time (hours) and the y axis, optical density. Error bars denote SD for duplicates.
Proteorhodopsin phototrophy has been suggested to meet all energy requirements of the cell in the presence of light (15). Proteorhodopsin phototrophy allows the bacterium to invest reduced carbon substrates in biosynthesis instead of respiring them for ATP generation. This is advantageous in oligotrophic environments where labile organic matter is frequently in short supply. However, replacing respiration with biosynthesis represents a challenge to the cell, because it will result in an imbalance of TCA cycle intermediates necessary as precursors. Anaplerotic CO2 fixation plays an important role in compensating for such imbalances (Fig. S4). Thus, when intermediates of the TCA cycle are used for biosynthesis, the TCA cycle needs to be replenished through the activity of bicarbonate fixation pathways. This could explain the presence of this type of bicarbonate-concentrating mechanism in MED152. Taken together, our findings suggest that light-stimulated anaplerotic inorganic carbon fixation could be a means allowing proteorhodopsin-containing flavobacteria to efficiently use organic matter for biosynthesis.
Light Utilization and Sensing.
Despite its small genome, MED152 harbors an ample suite of genes involved in sensing and using light (Table S6) when compared with most other heterotrophic bacteria analyzed to date (21). The genome harbors the gene encoding proteorhodopsin and the genes for synthesis of the proteorhodopsin chromophore retinal (crtEBIY, blh). Moreover, the proteorhodopsin gene encodes a protein with conserved key amino acid positions required for light-driven H+ pumping.
The synthesis of proteorhodopsin is expected to be regulated in response to light conditions. This has been shown in Dokdonia sp. MED134 (17). We therefore searched for genes potentially involved in such regulation. MED152 has a complex array of genes allowing it to respond to changes in the environment. The genome contains 21 two-component system sensor His kinases and 25 response regulators. In addition, it contains five hybrid His kinase/response regulators. Such sensor- and signal-transducing proteins likely provide means for controlling gene expression in response to environmental stimuli such as light and nutrients.
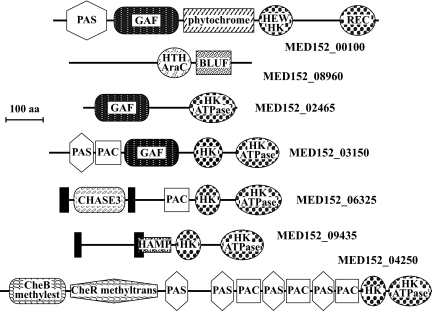
A number of sensor membrane molecules contain domains that respond to light. Fig. 4 shows some of the remarkable architectures found in peptides with signaling domains. The PAS and GAF domains were each found in four of the two-component system sensor His kinases. PAS domains can respond to cellular energy levels, oxygen levels, redox potential, and light (22), whereas GAF domains act as phototransducers. Both PAS and GAF domains are known to bind ligands and to interact with other proteins, and they are common components of phytochromes. MED152, like proteorhodopsin-containing Dokdonia sp. MED134, contains a putative phytochrome known to detect red and far-red light (MED152_00100; Table S7) (23). This protein has a modular structure with PAS, GAF, phytochrome, HWE-type His kinase, and response regulator receiver domains. In cyanobacteria, similar phytochrome photoreceptors control gene expression through signal transduction pathways (24). This suggests that proteins with PAS and GAF domains, such as phytochromes, contribute to the regulation of the synthesis of proteins that respond to light in MED152.
Fig. 4.
Domain architectures of selected peptides involved in signal transduction in MED152. MED152_00100 is a hybrid two-component His kinase sensor with both a HEW type His kinase (HEW HK) and a response regulator (REC). It contains a phytochrome domain that detects red and far-red light in a number of organisms. PAC, PAS, and GAF domains are common components of phytochromes. HK is a His kinase domain with its cognate ATPase domain (HK ATPase). In MED152_08960, a BLUF domain (shown to sense blue light) is next to a DNA-interacting AraC-type helix-turn-helix domain (HTC AraC). CHASE3 is an extracellular sensory domain, and HAMP has been suggested to regulate signal transduction. CheB methylesterase and CheR methyltransferase are part of chemiotaxis signaling in bacteria. Solid bars represent transmembrane regions.
Another remarkable set of genes in MED152 is that of the photolyases. Most of these are DNA repair flavoproteins that, in response to blue light, repair UV-induced DNA damage. The cryptochromes are a particular set of photolyase homologs, originally described in multicellular eukaryotes, where they function as blue light photoreceptors or regulators of circadian rhythm. Later, they have been described in prokaryotes as well (25). MED152 contained a genome region with three different DNA photolyase/cryptochrome genes (Fig. S5), whereas most marine bacteria had one or two genes only (Table S7). This region also contained a photolyase-related protein and folE, the latter possibly involved in the synthesis of the folate derived chromophore attached to the DNA photolyase/cryptochromes (26). Phylogenetic analysis showed that each of the three DNA photolyase/cryptochrome genes in MED152 belonged to a different gene family (Fig. S6). The “class I CPD photolyase” and “cryptochrome DASH” families are found in several Bacteroidetes genomes (Fig. S6, Table S7). Strikingly, however, proteorhodopsin-containing flavobacteria also have an additional DNA photolyase/cryptochrome belonging to the “animal cryptochrome and (6–4) photolyase” family, and they form a previously undescribed cluster within this family. Except for P. ubique, most marine bacteria with the proteorhodopsin gene have at least two types of DNA photolyase/cryptochrome (Table S7). If this correlation holds when more genomes are analyzed, some of these peptides are likely part of a light response mechanism that controls gene expression in marine bacteria.
Further, MED152_08960 contains one BLUF domain, which has been shown to sense blue light (Fig. 4). Similar domains are found in marine heterotrophic proteobacteria, aerobic anoxygenic photoheterotrophs, and cyanobacteria (27). Interestingly the two proteorhodopsin-containing flavobacteria that contain a BLUF domain also contain a phytochrome (Table S7). None of these genes are found in other proteorhodopsin-containing bacteria.
MED152 is orange in color because of the presence of carotenoids (Fig. 1A). HPLC analysis showed that MED152 readily synthesizes three main carotenoids: β-carotene, zeaxanthin, and a myxoxanthophyll-like carotenoid, and seven crt genes were identified. In addition to being precursors of retinal, carotenoids can function as accessory pigments in photosystems or as protective antioxidants. Interestingly, most carotenoid synthesis genes in MED152 are localized in two gene clusters next to two different types of regulators, suggesting they respond to different environmental stimuli. Thus, crtZDA are next to a two-component system response regulator of the LytR/AlgR type, whereas crtYZBI (responsible for the synthesis of β-carotene, the precursor of both retinal and the other carotenoids) is part of a different gene cluster next to a transcriptional regulator of the MerR family. crtZ is responsible for the synthesis of zeaxanthin from β-carotene and is present in both clusters. Similar gene content and organizations are found in other marine Bacteroidetes (e.g., Dokdonia sp. MED134, Leeuwenhoekiella blandensis MED217, and P. irgensii 23-P), regardless of the presence or absence of the proteorhodopsin gene. In summary, MED152 has a large range of genes that allow it to mediate, and possibly optimize, its reaction/response to light, a major environmental variable in the surface waters from which it was isolated.
Adhesion and Degradation of Polymers.
It has been proposed that marine flavobacteria are specialized in using proteins and other polymers for growth, whereas proteobacteria would be specialized in using monomeric compounds (3, 4, 28). Genome analysis of G. forsetii revealed an abundance of genes involved in attachment and a wide range of hydrolytic enzymes (5). These include 40 TonB dependent/ligand-gated channel genes (susC). In the related Bacteroides thetaiotaomicron, for example, the starch utilization system (Sus) operates such that the bacterium attaches to starch before starting its degradation (29, 30). The TonB-dependent/ligand-gated channel is an important component of the Sus complex. In these complexes, hydrolytic enzymes are located on the surface of the bacterium, but also outer membrane proteins that bind the polysaccharides to the bacterial surface. One such outer membrane protein is SusD (31). This lifestyle requires these and other proteins to bind the polymer of interest and degrade it, and to transport the monomers to the interior of the cell.
MED152 seems to have a similar system where the polymer degradation complex is associated to the cell surface. Twenty-seven genes were annotated as susC and five of these where next to their respective susD (Fig. S7). Other homologs of the eight-component Sus system in B. thetaiotaomicron were not found in the vicinity. However, the susCD were next to membrane protein genes with or without adhesion domains next to genes that encode enzymes that degrade polymers and transporters (Fig. S7). Although the TonB-dependent/ligand-gated channels are better known for the iron uptake in Gram-negative bacteria, they may be involved in the degradation and immediate uptake of the products of hydrolytic enzymes (32).
MED152 has a number of predicted periplasmic or extracellular peptidases and glycosyl hydrolases similar to Bacteroidetes that are well known for their capability to degrade polymeric compounds: Flavobacterium johnsoniae, Flavobacterium psychrophilum, Cytophaga hutchinsonii, and Bacteroides spp. This suggests that proteins and complex carbohydrates are important carbon and nitrogen sources also for MED152. Indeed, in the laboratory, MED152 grows well on both peptone and starch. However, the number of transporters specific for free amino acids or oligopeptides was low, and very few dedicated sugar transporters were found (Table S8). Most notably, MED152 lacked sugar-specific ABC-transporters, whereas Roseobacter clade bacteria and marine gammaproteobacteria, in contrast, had up to 20 such transporters (SI Methods). In accordance with previous studies of substrate utilization in marine bacteria (4, 28), our data suggest that proteins and polysaccharides, rather than monomers, are important carbon and nitrogen sources for at least certain marine flavobacteria.
Moreover, MED152 has a complete set of genes involved in gliding motility (15 genes), which could be beneficial in the exploration of solid surfaces. Suspended particles are particularly abundant during and after algal blooms, providing a potentially abundant source of food in the form of particulate organic matter. Flavobacteria are typically associated with the decay phase of phytoplankton blooms (1, 2, 10). Further, up to 26 ORFs contained domains that are involved in attachment to surfaces (Table S9). Several of these ORFs were located within hydrolytic clusters that included susCD (Fig. S7).
MED152 has a large array of genes involved in the synthesis and export of extracellular polysaccharide material (e.g., 25 glycosyl transferases). Indeed, MED152 cells grown in liquid medium have a strong tendency to aggregate and form large visible flocks (Fig. 1B). Flavobacteria might also benefit from biofilm formation on particles, which can give protection against grazing or against compounds that inhibit their growth and that are synthesized by competitors (33, 34).
Sigma Factors.
As seen thus far, MED152 has several sensors for light, possibly allowing regulation of gene expression under light and dark conditions. Further, it is reasonable to assume that the genome contains regulatory mechanisms for life attached to aggregates/particles. As expected, the genome of MED152 encodes one RpoD type σ-70 transcription factor (MED152_11764) but has as many as 15 alternative σ-70 family of σ factors. Whereas P. ubique has only two and E. coli has six, the number of such transcription factors in other marine bacteria is as high as in MED152. Alternative sigma factors regulate the synthesis of particular sets of genes in response to different environmental stimuli. Thus, genes in the vicinity of the alternative σ-70 transcription factors could give clues to the type of positively regulated genes and the type of signals MED152 might respond to. For example, MED152_02685 is next to a predicted transmembrane peptide, and both are next to a transcriptional regulator of the MerR type and genes involved in the synthesis of carotenoids and retinal. Both important features of Polaribacter metabolism, light utilization and polymer degradation, appear to be regulated by this mechanism, because other σ-70 transcription factor genes are next to peptidase, glycosyl hydrolase, transporter, and lipase genes.
Transporters.
Membrane transporters play critical roles in making use of available nutrients and responding to variations in the environment. Genome analysis identified only 106 cell membrane transporters in MED152 (Table 1, Table S8). Our comparative analysis showed this is a remarkably low number (see SI Methods for analysis details). For example, marine Roseobacter or gammaproteobacteria can harbor up to 330. The number of transporters in MED152 is only marginally higher than in P. ubique, which has 88, despite the genome size of MED152 being more than double that of P. ubique (Table 1). A large number of transporters is typically found in versatile, and generalist bacteria. In contrast, a low number of transporters can be expected in specialized bacteria or bacteria living in a stable and predictable environment (35). Thus, it seems that MED152 is a relatively specialized bacterium when it comes to transporters, which is in accordance with its constrained cell metabolism and small genome size.
Table 1.
Summary of transporters detected in the MED152 and selected genomes
| Transporters | PELAGIBACTER | MED152 | ROSEOBACTER | E. coli |
|---|---|---|---|---|
| Total | 88 | 106 | 210 – 330 | ≈ 350 |
| ATP-dependent | 28 | 28 | 69 – 107 | 72 |
| ABC superfamily | 24 | 22 | 65 – 102 | 67 |
| Ion channels | 1 | 12 | 7 – 12 | 15 |
| PTS | 0 | 0 | 2 – 4 | 29 |
| Secondary | 57 | 62 | 112 – 156 | 233 |
| DMT | 20 | 6 | 17 – 37 | 16 |
| MFS | 7 | 10 | 14 – 25 | 70 |
| MOP | 3 | 7 | 4 – 7 | 8 |
PELAGIBACTER is P. ubique HTCC1062; ROSEOBACTER includes LoktanelIa sp. SKA53, Roseobacter sp. MED193, Jannaschia sp. CCS1, Roseovarius nubinhibens ISM, Sulfitobacter sp. TM1040, and S. pomeroyi DSS-3; and E. coli is strain K12-MG1655.
Conclusion
The general features of the MED152 genome are consistent with life in the surface ocean, including both utilization of and protection from light and oxygen, and a significant number of Na+-dependent proteins. We propose that the more specific features of the genome are related to the need of Polaribacter to alternate between two lifestyles. On the one hand, MED152 is very well equipped to attach to surfaces, glide in search for polymers, and degrade them for carbon, nutrients, and energy. However, once suitable polymeric substrates have been exhausted, MED152 needs to find new particles to colonize. This forces the bacterium to carry on a free-living existence in a carbon-poor environment where it cannot move and is not prepared to take up most of the simple carbon compounds available. Thus, MED152 needs to somehow survive through the potentially long “traverse of the desert.” MED152 seems to solve the problem by using proteorhodopsin to capture light energy, increase anaplerotic bicarbonate fixation, and use the few carbon compounds it can get hold of exclusively for biosynthesis. In addition, proteorhodopsin phototrophy could be useful in providing energy for the degradation of complex/recalcitrant organic matter. This strategy is completely different from that of proteobacteria and may be common to many of the other marine flavobacteria accounting for up to 30% of the prokaryotic cells in surface oceans. Analysis of the genome of MED152 has been extremely helpful in generating hypotheses about the life strategy of Polaribacter that can now be tested experimentally.
Materials and Methods
Isolation of Flavobacteria.
Bacteria were isolated from Northwestern Mediterranean Sea surface water (0.5 m depth), collected 1 km off the Catalan coast at the Blanes Bay Microbial Observatory (41° 40′ N, 2° 48′ E; Spain). Strain MED152 was isolated on Marine Agar 2216 (Difco; Fig. 1A). 16S rRNA sequence analysis indicated it belongs to the genus Polaribacter, and its sequence is 99.6% similar to that of P. dokdonensis (Fig. S8). However, there are a few phenotypic differences between the two: unlike P. dokdonensis, MED152 is β-galactosidase-positive and is able to degrade gelatin.
Genomic Sequencing and Annotation.
Whole-genome sequencing was carried out by the J. Craig Venter Institute through the Gordon and Betty Moore Foundation initiative in Marine Microbiology (https://research.venterinstitute.org/moore). A Sanger/pyrosequencing hybrid method was used (36). Large (40-kb) and small (4-kb) insert random libraries were sequenced with an average success rate of 95% (large insert) and 93% (small insert) and an average high-quality read length of 837 (large insert) and 860 (small insert) nucleotides. The completed genome sequence of MED152 contains 30,151 reads, achieving an average of 10-fold sequence coverage per base.
ORFs were predicted and autoannotated using GenDB (37). All predicted genes were used to query the TransportDB database (38), and matches were assigned to transporter families within the TransportDB database (www.membranetransport.org). Further, a comparison to 19 genomes of marine bacteria in TransportDB was done (see SI Methods). All automatic annotations were curated manually. All predicted proteins from this genome were compared with selected genomes by using the basic local alignment search tool (BLAST). When necessary, analysis was carried out by using ad hoc Perl scripts.
Bacteria used in comparisons were members of the Bacteroidetes phylum in GenBank and genomes in the Gordon and Betty Moore Foundation Marine Microbiology Initiative. Gene content was also compared with available genomes of marine proteobacteria whose members are known to be abundant in sea water, such as the SAR11 and Roseobacter clades, and proteobacteria that contain the proteorhodopsin gene.
Bicarbonate Uptake.
MED152 was grown at 16°C in Marine Broth (Difco) diluted 1:8 with artificial seawater [35 practical salinity units, prepared from Sea Salts (Sigma)] in light (180 μmol of photons m−2·s−1) or dark conditions. Bicarbonate uptake rates were determined by the radioactive carbon assimilation technique (39) after 50-h incubation (during exponential growth; Fig. 3 Inset). Subsamples from each incubation condition were then placed in four 25-ml glass bottles (duplicates in both transparent and dark bottles), and incubation continued for 2 h with 20 μl of H14CO3− (3 μCi). For details, see SI Methods.
Pigment Analysis.
Pigments were identified and quantified by using HPLC analysis as described in SI Methods.
Supplementary Material
Acknowledgments.
We thank Folker Meyer, Jomuna Veronica Choudhuri, Daniela Bartels, Andreas Wilke, and Tobias Paczian, then at the University of Bielefeld, Bielefeld, Germany, for help with GenDB and José Manuel Fortuño of Institut de Ciències del Mar for help with electron microscopy. The genome was sequenced through the Marine Microbial Initiative of the Gordon and Betty Moore Foundation. This work was supported by the European Union (contract EU-FP6-505403 NoE Marine Genomics Europe), the Spanish Ministry of Education and Science (Grant CTM2004-02586/MAR), and the Swedish Research Council (Grant 621-2003-2692).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genome sequence reported in this paper has been deposited in the GenBank database (accession no. NZ_AANA00000000).
See Commentary on page 8487.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712027105/DCSupplemental.
References
- 1.Pinhassi J, et al. Changes in bacterioplankton composition under different phytoplankton regimens. Appl Environ Microbiol. 2004;70:6753–6766. doi: 10.1128/AEM.70.11.6753-6766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riemann L, Steward GF, Azam F. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl Environ Microbiol. 2000;66:578–587. doi: 10.1128/aem.66.2.578-587.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLong E, Franks DG, Alldredge AL. Phylogenetic diversity of aggregate-attached versus free-living marine bacterial assemblages. Limnol Oceanog. 1993;38:924–934. [Google Scholar]
- 4.Cottrell MT, Kirchman DL. Natural assemblages of marine Proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–1697. doi: 10.1128/aem.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer MM, et al. Whole genome analysis of the marine Bacteroidetes ‘Gramella forsetii' reveals adaptations to degradation of polymeric organic matter. Environ Microbiol. 2006;8:2201–2213. doi: 10.1111/j.1462-2920.2006.01152.x. [DOI] [PubMed] [Google Scholar]
- 6.Moran MA, et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432:910–913. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- 7.Giovannoni SJ, et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005;438:82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- 8.Cottrell MT, Kirchman DL. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl Environ Microbiol. 2000;66:5116–5122. doi: 10.1128/aem.66.12.5116-5122.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glöckner F-O, Fuchs BM, Amann RI. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abell GCJ, Bowman JP. Ecological and biogeographic relationships of class Flavobacteria in the Southern Ocean. FEMS Microbiol Ecol. 2005;51:265–277. doi: 10.1016/j.femsec.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Alonso C, Warnecke F, Amann RI, Pernthaler J. High local and global diversity of Flavobacteria in marine plankton. Environ Microbiol. 2007;9:1253–1266. doi: 10.1111/j.1462-2920.2007.01244.x. [DOI] [PubMed] [Google Scholar]
- 12.Pommier T, Pinhassi J, Hagström Å. Biogeographic analysis of ribosomal RNA clusters from marine bacterioplankton. Aquat Microb Ecol. 2005;41:79–89. [Google Scholar]
- 13.Béjà O, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 14.Martinez A, Bradley AS, Waldbauer JR, Summons RE, DeLong EF. Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host. Proc Natl Acad Sci USA. 2007;104:5590–5595. doi: 10.1073/pnas.0611470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter JM, Greenfield D, Bustamante C, Liphardt J. Light-powering Escherichia coli with proteorhodopsin. Proc Natl Acad Sci USA. 2007;104:2408–2412. doi: 10.1073/pnas.0611035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stingl U, Desiderio RA, Cho JC, Vergin KL, Giovannoni SJ. The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl Environ Microbiol. 2007;73:2290–2296. doi: 10.1128/AEM.02559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gómez-Consarnau L, et al. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature. 2007;445:210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- 18.Unemoto T, Hayashi M. Na+-translocating NADH-quinone reductase of marine and halophilic bacteria. J Bionenerg Biomembr. 1993;25:385–391. doi: 10.1007/BF00762464. [DOI] [PubMed] [Google Scholar]
- 19.Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc Natl Acad Sci USA. 2004;101:18228–18233. doi: 10.1073/pnas.0405211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata M, et al. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria. J Biol Chem. 2002;277:18658–18664. doi: 10.1074/jbc.M112468200. [DOI] [PubMed] [Google Scholar]
- 21.Van der Horst MA, Key J, Hellingwerf KJ. Photosensing in chemotrophic, non-phototrophic bacteria: let there be light sensing too. Trends Microbiol. 2007;15:554–562. doi: 10.1016/j.tim.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fankhauser C. The phytochromes, a family of red/far-red absorbing photoreceptors. J Biol Chem. 2001;276:11453–11456. doi: 10.1074/jbc.R100006200. [DOI] [PubMed] [Google Scholar]
- 24.Ashby MK, Houmard J. Cyanobacterial two-component proteins: structure, diversity, distribution, and evolution. Microbiol Mol Biol Rev. 2006;70:472–509. doi: 10.1128/MMBR.00046-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hitomi K, et al. Bacterial cryptochrome and photolyase: characterization of two photolyase-like genes of Synechocystis sp. PCC6803. Nucleic Acids Res. 2000;28:2353–2362. doi: 10.1093/nar/28.12.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanai S, Kikuno R, Toh H, Ryo H, Todo T. Molecular evolution of the photolyase-blue-light photoreceptor family. J Mol Evol. 1997;45:535–548. doi: 10.1007/pl00006258. [DOI] [PubMed] [Google Scholar]
- 27.Gomelsky M, Klug G. BLUF: A novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem Sci. 2002;27:497–500. doi: 10.1016/s0968-0004(02)02181-3. [DOI] [PubMed] [Google Scholar]
- 28.Kirchman DL. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol. 2002;39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 29.Anderson KL, Salyers AA. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch binding sites and periplasmic starch-degrading enzymes. J Bacteriol. 1989;171:3192–3198. doi: 10.1128/jb.171.6.3192-3198.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson KL, Salyers AA. Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1989;171:3199–3204. doi: 10.1128/jb.171.6.3199-3204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reeves AR, Wang G-R, Salyers AA. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol. 1997;179:643–649. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanvillain S, et al. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS ONE. 2007;2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long RA, Azam F. Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol. 2001;67:4975–4983. doi: 10.1128/AEM.67.11.4975-4983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grossart HP, Schlingloff A, Bernhard M, Simon M, Brinkhoff T. Antagonistic activity of bacteria isolated from organic aggregates of the German Wadden Sea. FEMS Microbiol Ecol. 2004;47:387–396. doi: 10.1016/S0168-6496(03)00305-2. [DOI] [PubMed] [Google Scholar]
- 35.Ren Q, Paulsen IT. Comparative analyses of fundamental differences in membrane transport capabilities in Prokaryotes and Eukaryotes. PLoS Comput Biol. 2005;1:e27. doi: 10.1371/journal.pcbi.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldberg MD, et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc Natl Acad Sci USA. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer F, et al. GenDB – An open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren Q, Kang KH, Paulsen IT. TransportDB: A relational database of cellular membrane transport systems. Nucleic Acids Res. 2004;32:D284–D288. doi: 10.1093/nar/gkh016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parsons TR, Maita Y, Lalli CM. A Manual of Chemical and Biological Methods for Seawater Analysis. Oxford, UK: Pergamon Press; 1984. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.