Abstract
Originally recognized for their role in lipoprotein metabolism and cardiovascular disease, apolipoprotein (apo) E isoforms (apoE2, apoE3, and apoE4) have also been implicated to play a key role in several biological processes not directly related to their lipid transport function. For example, apoE4 contributes significantly to neurodegeneration in Alzheimer's disease. However, the role of apoE in infectious diseases is less well defined. Here, by examining a large cohort of HIV+ European and African American subjects, we found that the APOE ε4/ε4 genotype is associated with an accelerated disease course and especially progression to death compared with the APOE ε3/ε3 genotype. However, an association between the ε4/ε4 genotype and HIV-associated dementia (HAD), a neurological condition with clinicopathological features similar to Alzheimer's disease, was not detected. Consistent with the genotype–phenotype relationships observed, compared with recombinant apoE3, apoE4 enhanced HIV fusion/cell entry of both R5 and X4 HIV strains in vitro. These findings establish apoE as a determinant of HIV-AIDS pathogenesis and raise the possibility that current efforts to convert apoE4 to an “apoE3-like” molecule to treat Alzheimer's disease might also have clinical applicability in HIV disease.
Keywords: HIV/AIDS, fusion/cell entry, infectious diseases, apoE
There is incontrovertible evidence demonstrating that polymorphisms in some host genes have a significant impact on susceptibility to HIV-1 infection and rate of disease progression. Discovery of such polymorphisms has improved our understanding of the host factors that influence HIV pathogenesis and spurred development of new antiviral therapeutics. Among the most intensely studied of such polymorphisms is a 32-bp deletion (Δ32) mutation in the gene encoding CC chemokine receptor 5 (CCR5) (1). The in vitro and in vivo genotype–phenotype relationships linked to this mutation not only established CCR5 as the major coreceptor for the cell entry of HIV but also provided the impetus to develop CCR5 blockers for the treatment of HIV-infected patients (2). Since its discovery, the CCR5-Δ32 mutation has been extensively investigated, and associations between this polymorphism and several infectious and noninfectious diseases have been established (e.g., refs. 3 and 4).
Akin to the CCR5-Δ32 mutation, polymorphisms in the coding sequence of the gene encoding apolipoprotein (apo) E have been scrutinized intensely, because they have been found to consistently convey differential susceptibility to several noninfectious diseases (5, 6). Less well known are their contributions to the pathogenesis of infectious diseases (5, 6). Given the power of combining genetic association studies with in vitro analyses to elucidate the contribution of a gene to disease pathogenesis, we used a similar approach, with HIV infection as a model system to clarify further the role of apoE in infectious diseases.
ApoE was discovered as a plasma protein involved in lipoprotein metabolism (5, 6). The three major alleles of the APOE locus, designated as ε2, ε3, and ε4, encode the three major isoforms of apoE, designated as apoE2, apoE3, and apoE4, respectively (5). Allele frequencies of ε2, ε3, and ε4 vary significantly between different ethnicities (5, 7). The ε3 allele is the most prevalent allele in all human populations, with frequencies of 50–90%, whereas the frequencies of ε4 and ε2 are 5–35% and 1–15%, respectively. A large number of epidemiological studies have demonstrated that the ε4 allele is associated with diverse adverse outcomes, including decreased longevity, increased plasma cholesterol levels, and pathogenesis of several diseases involving the CNS (5, 8), especially Alzheimer's disease (AD) (9–11). Thus, the prevailing view is that, relative to the ε3 allele, possession of the ε4 allele confers a detrimental effect in noninfectious diseases.
Although it has been hypothesized that apoE might play a role in infectious diseases (5, 6), this premise is supported by only a few in vitro studies and genetic-epidemiologic analyses (12–19). Some examples include the observation that herpes simplex virus-1 (HSV-1) infection in the CNS results in severe encephalitis and can also be a risk factor for AD, and notably, there appears to be a higher prevalence of the ε4 allele among AD patients who are HSV-positive than in those who are HSV-negative (17). Also, Corder et al. (20) showed in a study of 44 patients that, compared with apoE4-negative patients, twice as many apoE4-positive patients developed HAD and neurological symptoms.
Although the aforementioned studies suggest a possible link between apoE genotype and host responses to pathogens, particularly viruses, these genetic associations have been conducted with a small number of patients. Additionally, the mechanisms by which apoE isoforms might differentially influence susceptibility to infection remain unknown. To investigate apoE–HIV interactions, we examined a large well-characterized cohort of HIV-infected patients and seronegative subjects (21–24) and tested the hypothesis that, compared with the ε3 allele, the ε4 allele is associated with an accelerated rate of HIV disease progression and possibly with an increased risk of acquiring HIV infection. Given the link between apoE and CNS diseases, especially AD, we also examined whether APOE alleles conveyed differential susceptibility to HAD, a condition that shares many pathogenic and clinical features with AD (25, 26). We coupled these analyses with in vitro studies and investigated whether the apoE3 and apoE4 isoforms influenced cell entry of HIV in a manner that was concordant with the effect of the corresponding APOE alleles on HIV disease progression.
Results
Distribution of APOE Alleles and Genotypes.
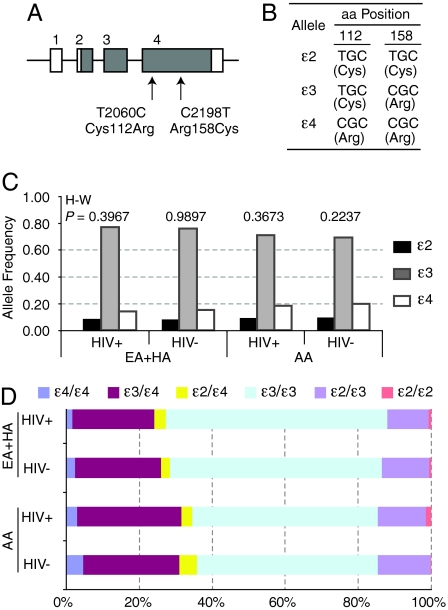
In subjects of European [European American (EA) and Hispanic American (HA)] descent and African Americans (AA) from Wilford Hall Medical Center (WHMC), the distribution of the three APOE alleles (Fig. 1 A and B) were in Hardy–Weinberg equilibrium (Fig. 1C). The APOE allele and genotype frequencies were consistent with previously published reports (5–7) and differed significantly between subjects of European and African descent (Fig. 1 C and D). The prevalence of homozygous APOE alleles was ε3/ε3 > ε4/ε4 > ε2/ε2 (Fig. 1D).
Fig. 1.
APOE gene structure, APOE polymorphisms, and frequency of the APOE alleles and genotypes in HIV+ and HIV-negative subjects from WHMC. (A) Schema of APOE gene structure. The numbers above the boxes indicate exons. Gray, coding region; white, untranslated region. Arrows, position of the two SNPs [and corresponding amino acid (aa) changes] genotyped. (B) APOE alleles. The two SNPs are in strong linkage disequilibrium and result in three APOE alleles (ε2, ε3, and ε4). The corresponding amino acids are in parentheses. (C and D) Distribution of the APOE (C) alleles and (D) genotypes in the WHMC HIV-positive (HIV+) and HIV-negative (HIV−) subjects. EA, European American; HA, Hispanic American; AA, African American. The numbers at the top of the bars in C represent significance values for the test of Hardy–Weinberg (H–W) equilibrium. χ2 = 17.37 and P = 0.0002 for differences in the APOE allele frequencies in subjects of European (EA+HA) vs. African (AA) descent.
APOE Polymorphisms and the Risk of Acquiring HIV.
In the overall cohort (i.e., without accounting for racial/ethnic background), the APOE allele and genotype frequencies did not differ between HIV-negative and -positive (HIV+) subjects (Fig. 1 C and D), suggesting that these variations in APOE do not influence the risk of acquiring HIV. Accordingly, when subjects were stratified by their racial/ethnic background, APOE alleles and genotypes were not associated with risk of acquiring HIV infection (data not shown).
APOE Polymorphisms and Rate of Disease Progression.
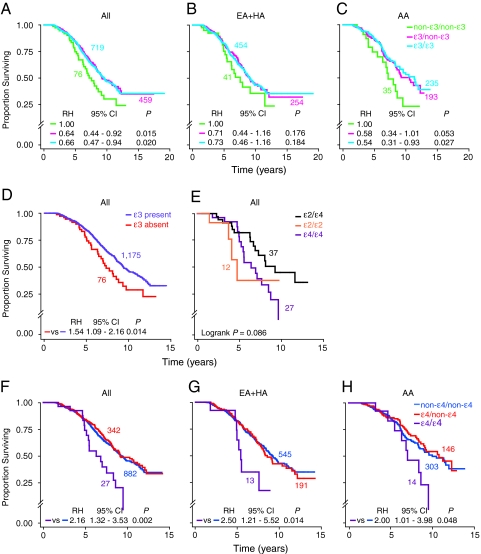
Although it remains unclear whether ε3 is the ancestral APOE allele (5–7), it is the most prevalent allele in all populations and is generally associated with beneficial effects in noninfectious diseases (5, 6, 8, 11). Accordingly, we tested the hypothesis that in the WHMC HIV+ cohort, possession of an ε3 allele might have a slower HIV disease course. Consistent with this hypothesis, compared with genotypes that do not contain an ε3 allele (non-ε3/non-ε3), heterozygosity or homozygosity for an ε3 allele was associated with a slower rate of disease progression, with a greater influence on progression to death than AIDS [Fig. 2 A–C, supporting information (SI) Fig. S1, and data not shown]. This association was evident in the overall HIV+ cohort and remained evident after stratifying the cohort according to their racial/ethnic background, although the strength of the effects appeared to be more prominent in AAs than in EAs (Fig. 2 A–C).
Fig. 2.
APOE alleles and genotypes influence the rate of disease progression in HIV-infected subjects from the WHMC cohort. Kaplan–Meier plots depict the association between the indicated APOE alleles/genotypes and rate of progression to death. (A, D–F) Kaplan-Meier plots for HIV+ subjects from the entire WHMC HIV+ cohort; (B and G) subjects of European (EA+HA) descent; (C and H) subjects of AA descent. P values in E are by log-rank test, whereas in the other plots they were obtained by Cox proportional hazard regression models. RH, relative hazard; CI, confidence interval. Color-coded numbers adjacent to each plot are the corresponding number of subjects in each genotypic group.
Because those lacking an ε3 allele had an accelerated disease course (Fig. 2D and Fig. S1), we next asked which genotypes that do not contain an ε3 allele (i.e., genotypes reflected by “ε3 absent” in Fig. 2D) conferred disease acceleration. Homozygosity for either the ε2 or the ε4 allele was associated with an accelerated disease course (Fig. 2E and Fig. S1). Given that the prevalence of the ε2/ε2 genotype was very low (Fig. 1D), subsequent analyses focused on the effects of the ε3 and ε4 alleles.
Having one or two copies of the ε4 allele increases a person's risk of developing several noninfectious diseases, although maximal effects are for the ε4/ε4 genotype (5, 27). Additionally, results from murine studies indicate that apoE4 not only is less neuroprotective than apoE3 but also acts as a dominant negative factor that interferes with the beneficial function of apoE3 (28). Although homozygosity for the ε4 allele was associated with a rapid rate of HIV disease progression (Fig. 2E), it remained unclear whether possession of one copy of ε4 was associated with disease-accelerating effects. We therefore determined the effect of homozygosity or heterozygosity for the ε4 allele on HIV disease course. We found that, compared with those lacking the ε4 allele (non-ε4/non-ε4), homozygosity but not heterozygosity for the ε4 allele was strongly associated with an accelerated rate of disease progression in the entire cohort (Fig. 2F and Fig. S1). These associations were observed among subjects of European and African descent in the WHMC HIV+ cohort (Fig. 2 G and H). These findings are consistent with a dose effect such that two but not one allele of ε4 confers disease acceleration.
APOE Polymorphisms and AIDS-Defining Illnesses.
In time-to-event analysis, the ε4/ε4 genotype was not associated with an altered rate of progression to HAD [relative hazard (RH) = 0.90, 95% C.I. = 0.12–6.56, P = 0.917]. In contrast, the ε4/ε4 genotype had a strong and significant influence on the rate of progression to Mycobacterium avium complex (Fig. S2) and toxoplasmosis (RH = 4.82, 95% C.I. = 1.10–21.1, P = 0.037; Kaplan–Meier plots not shown). Logistic regression analyses confirmed the lack of association between risk of developing HAD and possession of the ε4/ε4 genotype (SI Text).
ε4/ε4 Genotype and Viral Load.
We next conducted analyses to identify correlates of disease progression that may suggest potential mechanisms by which the ε4/ε4 genotype confers an accelerated rate of disease progression. We determined whether the effect of the ε4/ε4 genotype persisted after adjustment for the disease-influencing effects of parameters that independently affect HIV disease progression, namely: baseline CD4+ T cell count, steady-state viral load (set point), delayed type hypersensitivity (DTH) skin-test reactivity (an in vivo indicator of cell-mediated immunity), and CCL3L1-CCR5 genetic risk groups, which are genetic factors that affect HIV-AIDS pathogenesis (22, 23). These analyses showed that the influence of the ε4/ε4 genotype on rate of disease progression remained after adjustment for baseline CD4+ T cell counts, DTH responses, and CCL3L1-CCR5 genetic risk groups (Table 1), indicating that the effect of this genotype on disease progression is independent of these parameters.
Table 1.
Association of the APOE ε4/ε4 genotype with rate of progression to death in HIV-infected subjects from the WHMC cohort before (unadjusted) and after adjusting for the effects of other covariates
| Model | n | RH | 95% C.I. | P value |
|---|---|---|---|---|
| Unadjusted | 1,251 | 2.16 | 1.32–3.53 | 0.002 |
| Adjusted for | ||||
| CD4 | 1,094 | 1.97 | 1.21–2.23 | 0.006 |
| VL | 812 | 1.48 | 0.78–2.81 | 0.226 |
| DTH | 1,093 | 2.62 | 1.60–4.29 | 0.0001 |
| GRGs | 1,219 | 1.90 | 1.14–3.17 | 0.013 |
| CD4, VL | 812 | 1.55 | 0.82–2.94 | 0.176 |
| CD4, DTH | 1,092 | 2.50 | 1.53–4.09 | 0.0002 |
| CD4, GRG | 1,072 | 1.71 | 1.03–2.85 | 0.039 |
| CD4, DTH, VL | 811 | 2.16 | 1.14–4.12 | 0.019 |
| CD4, DTH, GRG | 1,070 | 2.15 | 1.29–3.59 | 0.004 |
| CD4, DTH, VL, GRG | 794 | 1.57 | 0.80–3.10 | 0.191 |
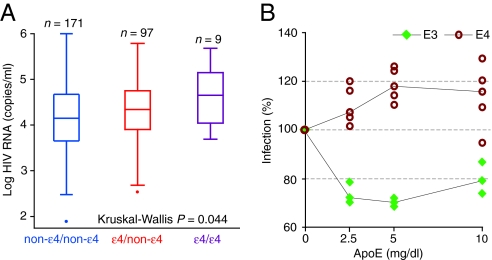
In contrast, the influence of ε4/ε4 genotype on disease was minimized after adjustment for the steady-state viral load (Table 1), suggesting that the extent of viral replication might differ according to APOE genotype. The latter possibility was supported by two observations. First, the non-ε4/non-ε4, ε4/non-ε4, and ε4/ε4 genotypes were associated with a step-wise increase in the steady-state viral load, although a significant association (P = 0.044) was observed only in AAs (Fig. 3A and data not shown). Second, each additional ε4 allele in HIV+ AA subjects was associated with an increase in the steady-state viral load, as shown by using linear regression analysis (P for trend = 0.010). Those with the ε4/non-ε4 and ε4/ε4 genotypes had 0.17 log (95% C.I.: 0–0.35, P = 0.05) and 0.48 log (95% C.I.: 0.01– 0.96, P = 0.046) higher steady-state viral loads, respectively, compared with those who did not have the ε4 allele (designated as non-ε4/non-ε4). These effects of APOE genotype on viral load were in contrast with our previous observation that heterozygosity for the ε4 allele was not associated with a major impact on disease progression rates in the overall cohort. Although this difference in viral load is statistically significant, a difference of 0.17 log between those with a ε4/non-ε4 and non-ε4/non-ε4 genotype may not be clinically meaningful. Furthermore, this weak association between ε4/non-ε4 genotype and viral load either may be related to a race-specific effect of the ε4 allele on viral load in AA subjects or may be partly explained by the lower frequency of the ε4 allele in subjects of European descent, or both.
Fig. 3.
Influence of the ε4 allele on the steady-state viral load in the WHMC HIV+ cohort, and apoE3 or apoE4 isoforms on HIV infection in vitro. (A). Box-and-whisker plot shows the distribution of the steady-state viral load according to APOE genotype. The overall difference among the genotypes was assessed by using the Kruskal–Wallis test (n) number of subjects. (B) Influence of apoE isoforms on HIV infection in vitro. MAGI-R5 cells (2 × 105) were infected with luciferase-pseudotyped HIV bearing NL4–3 gp120 after incubation for 2 h with apoE3 or apoE4 at the indicated concentrations. Luciferase activity (reflecting gene expression from integrated provirus) was measured 3 days later. Each analysis was conducted in replicates of three or five, and the results were calculated as a percentage of the untreated control (the median value of the mock-treated samples). Significance was determined by Student's t test, and P values for significance between the effects of apoE3 and apoE4 isoforms were <0.001 at each concentration tested.
ApoE Isoforms Influence HIV Infection in Vitro.
The aforementioned results indicated that the ε3 allele had beneficial effects and the ε4 allele had detrimental effects on HIV disease course. We therefore next determined whether purified apoE3 or apoE4 protein conferred in vitro resistance and susceptibility, respectively, to HIV infection. Using a luciferase pseudotyped virus assay, we found that MAGI-R5 cells infected in the presence of apoE4 were more susceptible to infection than those infected in the presence of apoE3 (Fig. 3B). At multiple concentrations (2.5, 5, 10 mg/dl), treatment with apoE4 resulted in significantly higher infection-associated luciferase activity than did treatment with an equivalent concentration of apoE3 (Fig. 3B). The difference in luciferase activity between apoE3- and apoE4-treated cells ranged from 35% to 50% (P < 0.001).
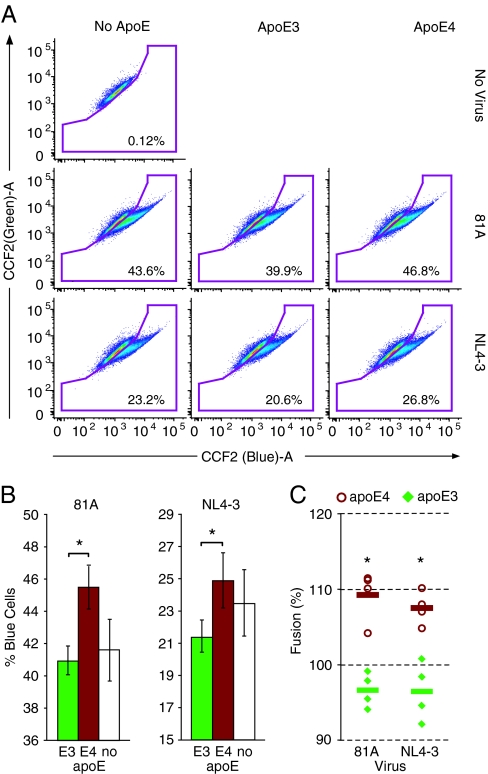
To understand the mechanism by which apoE4 protein predisposes cells to increased susceptibility to infection, we used a flow cytometry-based assay to investigate the effect of apoE isoforms on viral attachment to and fusion with the target-cell membrane (29). The fluorescence resonance energy transfer-based HIV virion fusion assay exploits the incorporation of β-lactamase (BlaM)–Vpr chimeric proteins into HIV-1 virions and their subsequent delivery into the cytoplasm of target cells as a marker of fusion. BlaM-positive cells can be revealed by loading the cells with CCF2, a fluorescent substrate of BlaM, which upon cleavage changes its fluorescence from green to blue. Fusion mediated by X4- or R5-tropic receptors was tested by using NL4–3 or 81A viruses containing BlaM-Vpr, respectively.
Infection of SupT1-CCR5 target cells in the presence of apoE4 (10 μg/ml) consistently resulted in a higher frequency of BlaM+/blue fluorescent cells compared with infection in the presence of an equal concentration of apoE3, indicating a higher frequency of HIV-target cell fusion in the presence of apoE4 than apoE3 (P < 0.05) (Fig. 4 A and B). Consistent effects were observed when using either CXCR4- or CCR5-using HIV strains. On average, the percentage of cells undergoing fusion with 81A-pseudotyped virus after apoE4 treatment was 12.6 ± 3.6% higher than after apoE3 treatment (P < 0.01) (Fig. 4C). Similarly, fusion of NL4–3-pseudotyped virus was 11.1 ± 4.9% higher when treated with apoE4 than apoE3 (P < 0.01). Given the synchronous nature of infection in this system, these results suggest a more rapid rate of infection in the presence of apoE4 compared with apoE3.
Fig. 4.
Influence of apoE isoforms on fusion of HIV to the cell membrane. (A) Flow cytograms from a representative experiment. Shown are examples of results from uninfected (No virus, Top) and untreated (No apoE, Left) controls and infection with 81A (Middle) or NL4–3 (Bottom) HIV strains following pretreatment with apoE3 (Middle) or apoE4 (Right). SupT1-CCR5 cells that have undergone fusion show increased blue fluorescence, and the percentage of cells included in the “fusion” gate are noted in the lower right-hand corner. Each plot represents a single well from each treatment. (B) Histograms representing the results for 81A and NL4-3 infection demonstrate the mean percent of blue cells per well and standard deviation for the experiment shown in A. Samples were run in triplicate, and significance values were determined by Student's t test; *, P < 0.05. (C) Comparison of data from four individual experiments. Results were calculated as a percentage of the untreated control, using the mean value of the mock-treated samples as the untreated control [(mean of experimental group/mean of untreated control group) × 100%]. These results demonstrate that 81A and NL4-3 infection in the presence of apoE4 (open circles) reproducibly resulted in significantly higher levels of fusion/infection than in the presence of apoE3 (closed diamonds). The horizontal bars represent the mean value for data points. Significance was determined by Student's t test; *, P < 0.01.
Discussion
The results of this large-scale genetic-epidemiologic investigation provide a link between APOE genotype and HIV disease course, whereas the biochemical-virologic analyses reported herein provide a link between the corresponding apoE isoforms and virus–host cell interactions. These links have important implications for the improved understanding of the pathogenesis of HIV-AIDS, risk stratification of HIV-infected subjects, and possibly development of novel antiviral therapeutic interventions.
Although we did not detect an association between APOE genotype and risk of acquiring HIV, the APOE ε4/ε4 genotype was associated with a faster rate of disease progression in HIV+ subjects. The robustness of this association is bolstered by the large number of subjects studied and consistency of the genotype–phenotype relationships detected in subjects of both European and African descent. Concordant with these associations, the in vitro studies showed that apoE4 promotes HIV infection by facilitating a greater frequency of fusion events than apoE3. These results provide a possible mechanism underlying the in vivo (genetic–epidemiologic) associations detected for the ε4/ε4 genotype. Of note, although the enhancing of HIV infection in vitro by apoE4 was relatively small, it was reproducible in different experimental systems. It is also important to consider that these in vitro studies reflect a single round of infection. However, over the course of HIV disease in a host, the cycle of infection occurs many millions of times. It is very plausible that even modest differences in effect size, such as the one we demonstrate in vitro, may be greatly amplified at the organismal level, resulting in the gradual emergence of clinical differences, such as those we observed for the ε4/ε4 and ε3/ε3 genotypes.
Also underscoring the robustness of our findings, the differential impact of the ε4/ε4 and ε3/ε3 genotypes and their corresponding apoE isoforms observed in HIV disease is similar to their contrasting effects observed in a variety of noninfectious diseases (5, 6, 8, 11). However, we noted one exception to this general rule. We could not affirm our hypothesis that, akin to its strong effect on AD, the APOE genotype would also influence risk of HAD. Why this might be the case is not clear but may suggest that, although the phenotypes of AD and HAD appear similar (25, 26), the pathogenesis of each follows a separate pathway. Nevertheless, this possibility warrants confirmation in other cohorts of HIV-infected subjects, because it is conceivable that the ε4/ε4 genotype might be associated with milder forms of HIV-associated neurological impairment but not HAD, the phenotypic endpoint we studied herein.
How might apoE isoforms influence HIV-AIDS pathogenesis? There are several possibilities. There is a large body of evidence suggesting that the amphipathic helical domains of apolipoproteins may act as viral fusion inhibitors, because of their homology with the fusogenic domains of viral fusion proteins (12, 30–33). Invoking this model, we suggest that the amphipathic helix domains of apoE inhibit HIV infection in a manner analogous to the clinical HIV fusion inhibitor, Enfurvitide (T20), that is, by binding to gp41 and blocking either the formation or the function of its N-terminal fusogenic hairpin domain (34, 35). It has been clearly established that the single amino acid difference that distinguishes the apoE3 and apoE4 isoforms has a profound impact on their structure, with apoE4 having a more compact structure than apoE3 (5, 11, 36). Thus, based on our observation that HIV attachment and/or fusion occurred more readily in the presence of apoE4 than apoE3, we suggest that apoE4 may be a less efficient fusion inhibitor. This may be due to either inherent structural characteristics that make the fusion-blocking domains of apoE4 less accessible to the gp41–CD4 complex or to isoform-specific changes in inhibitory domain activity.
Alternatively, the observed differential effects of apoE isoforms may be exerted at the level of attachment to heparan sulfate proteoglycans, a rate-limiting step in HIV infection. It has been demonstrated that tandem repeat peptides synthesized based on the heparin-binding domain of apoE (residues 141 to 149) have notable antiviral activity when present in cell culture during HIV infection, and that this effect is likely due to blockade at the level of attachment (12, 37). Heparan sulfate proteoglycan-bound apoE is abundant on many cell types, including macrophages, and apoE is now known to be carried with HIV particles that bud from macrophages (38). ApoE may therefore block attachment of HIV to the cell-surface receptors or between cell-surface heparan sulfate proteoglycans and viral lipid, and there might be isoform-specific differences in these effects. Furthering the potential complexity of these interactions, it is known that, although both apoE3 and apoE4 have similar receptor-binding capacities (39), they have very different lipid-binding characteristics. It is possible that differing affinities for lipid may convey differential abilities to bind to the HIV envelop or to complex with cell-surface cholesterol-phospholipid rafts, through which HIV is believed to penetrate target cells (40, 41).
Additionally, it is also important to consider the possibility that the differential effects of apoE isoforms on lipid metabolism and cholesterol homeostasis may in turn affect both virus fusion/entry as well as assembly/release. Cholesterol is a crucial component of the HIV envelope, and it is now known to be essential for both viral entry and assembly/budding. Depletion of cholesterol from either the cell membrane or viral envelope results in loss of infectivity (42, 43). Furthermore, viral assembly and budding occur at cholesterol-rich lipid raft microdomains (44, 45). Compared with apoE3, apoE4 less efficiently promotes cholesterol efflux from cells, which correlates with its proatherogenic effects in vivo (46). This differential effect of the apoE isoforms on the mobilization of cellular cholesterol may allow accumulation of plasma membrane raft-associated cholesterol in ε4 homozygotes, resulting in enhancement of both the fusion/entry and assembly/release stages of the viral infection cycle. Further experimentation is clearly necessary to elucidate the mechanisms by which apoE isoforms convey contrasting effects on HIV attachment and fusion.
Although a reproducible and consistent effect on attachment/fusion was detected in this study, it is also important to consider other mechanisms by which apoE polymorphisms might result in the striking differences in HIV disease progression demonstrated in our study population. For example, a growing body of evidence suggests that apoE has multiple direct effects on the immune system, and that apoE isoforms have significant differences in their immunomodulatory behavior (5, 6, 47, 48).
Our findings have several broader implications. Foremost, they provide insights into host–virus interactions by demonstrating that apoE isoforms differentially modulate viral attachment and fusion, and that this might represent a mechanism by which APOE genotypes have contrasting effects on the steady-state viral load and HIV disease course. Second, although the prevalence of the ε4/ε4 genotype is small, the consistency of its association with disease acceleration in subjects of European and African descent suggests it might have value as a risk stratification tool, perhaps identifying a group of patients requiring more aggressive and earlier initiation of therapy. Finally, extensive efforts are being placed on development of novel pharmaceutical means to counteract the negative effects of apoE4 protein or mimic the beneficial effects of the apoE3 protein in cardiovascular disease and AD. Thus, when such apoE-targeted therapeutic agents become available, they might also have applicability to the treatment of HIV disease (49, 50). Among the most promising of these are small molecules that alter the intramolecular domain interaction of apoE4 to make its structural conformation and function more like that of apoE3 (11, 51).
Methods
Subjects Studied.
The characteristics of 1,267 HIV-seropositive adults and 1,132 ethnically similar HIV-seronegative controls from WHMC have been described extensively (21–24). For additional details, see SI Text.
Genotyping Assays.
Two SNPs at positions T2060C (rs429358) and C2198T (rs7412) in the APOE gene that result in the ε2, ε3, and ε4 alleles were genotyped. Methods for genotyping polymorphisms in CCR5 and the copy number of the gene encoding CCL3L1, its major agonist, and HIV-suppressive chemokine are as described, as are the methods for categorizing variations in CCL3L1 and CCR5 into low, moderate, and high CCL3L1-CCR5 genetic risk groups (22, 23). For additional details, see SI Text.
Cell Lines and Tissue Culture.
293T cells were used to produce HIV reporter virions expressing luciferase or containing BlaM-Vpr. For additional details, see SI Text.
HIV Virion-Based Fusion Assay.
Measurements of HIV fusion in SupT1-CCR5 cells were performed with the fluorescence resonance energy transfer-based fusion assay, as described (29). For additional details, see SI Text.
For details on virus production, infectivity assays in MAGI-R5 cells, and statistical analysis, see SI Text.
Supplementary Material
Acknowledgments.
We thank the people and funding agencies that made this work possible (see SI Text).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803526105/DCSupplemental.
References
- 1.Kaslow RA, Dorak T, Tang JJ. Influence of host genetic variation on susceptibility to HIV type 1 infection. J Infect Dis. 2005;191:S68–S77. doi: 10.1086/425269. [DOI] [PubMed] [Google Scholar]
- 2.Fatkenheuer G, et al. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat Med. 2005;11:1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- 3.Manes S, et al. CCR5 expression influences the progression of human breast cancer in a p53-dependent manner. J Exp Med. 2003;198:1381–1389. doi: 10.1084/jem.20030580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glass WG, et al. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J Exp Med. 2006;203:35–40. doi: 10.1084/jem.20051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahley RW, Rall SC., Jr Apolipoprotein E: Far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 6.Mahley RW. Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 7.Gerdes LU. The common polymorphism of apolipoprotein E: Geographical aspects and new pathophysiological relations. Clin Chem Lab Med. 2003;41:628–631. doi: 10.1515/CCLM.2003.094. [DOI] [PubMed] [Google Scholar]
- 8.Smith JD. Apolipoprotein E4: An allele associated with many diseases. Ann Med. 2000;32:118–127. doi: 10.3109/07853890009011761. [DOI] [PubMed] [Google Scholar]
- 9.Corder EH, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 10.Rocchi A, Pellegrini S, Siciliano G, Murri L. Causative and susceptibility genes for Alzheimer's disease: A review. Brain Res Bull. 2003;61:1–24. doi: 10.1016/s0361-9230(03)00067-4. [DOI] [PubMed] [Google Scholar]
- 11.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: A causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc Natl Acad Sci USA. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobson CB, et al. The receptor-binding region of human apolipoprotein E has direct anti-infective activity. J Infect Dis. 2006;193:442–450. doi: 10.1086/499280. [DOI] [PubMed] [Google Scholar]
- 13.Wozniak MA, et al. Does apolipoprotein E determine outcome of infection by varicella zoster virus and by Epstein Barr virus? Eur J Hum Genet. 2007;15:672–678. doi: 10.1038/sj.ejhg.5201812. [DOI] [PubMed] [Google Scholar]
- 14.Price DA, et al. Apolipoprotein ε3 allele is associated with persistent hepatitis C virus infection. Gut. 2006;55:715–718. doi: 10.1136/gut.2005.079905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wozniak MA, Riley EM, Itzhaki RF. Apolipoprotein E polymorphisms and risk of malaria. J Med Genet. 2004;41:145–146. doi: 10.1136/jmg.2003.014613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aucan C, Walley AJ, Hill AV. Common apolipoprotein E polymorphisms and risk of clinical malaria in the Gambia. J Med Genet. 2004;41:21–24. doi: 10.1136/jmg.2003.011981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itzhaki RF, et al. Herpes simplex virus type 1 in brain and risk of Alzheimer's disease. Lancet. 1997;349:241–244. doi: 10.1016/S0140-6736(96)10149-5. [DOI] [PubMed] [Google Scholar]
- 18.Corder E, Lannfelt L, Mulder M. Apolipoprotein E and herpes simplex virus 1 in Alzheimer's disease. Lancet. 1998;352:1312–1313. doi: 10.1016/S0140-6736(05)70525-0. [DOI] [PubMed] [Google Scholar]
- 19.Wozniak MA, et al. Apolipoprotein E-ε4 protects against severe liver disease caused by hepatitis C virus. Hepatology. 2002;36:456–463. doi: 10.1053/jhep.2002.34745. [DOI] [PubMed] [Google Scholar]
- 20.Corder EH, et al. HIV-infected subjects with the E4 allele for APOE have excess dementia and peripheral neuropathy. Nat Med. 1998;4:1182–1184. doi: 10.1038/2677. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez E, et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci USA. 1999;96:12004–12009. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 23.Dolan MJ, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat Immunol. 2007;8:1324–1336. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 24.Ahuja S, et al. CCL3L1-CCR5 genotype influences durability of immune recovery during antiretroviral therapy of HIV-1-infected individuals. Nat Med. 2008;14:413–420. doi: 10.1038/nm1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rausch DM, Davis MR. HIV in the CNS: Pathogenic relationships to systemic HIV disease and other CNS diseases. J Neurovirol. 2001;7:85–96. doi: 10.1080/13550280152058744. [DOI] [PubMed] [Google Scholar]
- 26.Minagar A, et al. The role of macrophage/microglia and astrocytes in the pathogenesis of three neurologic disorders: HIV-associated dementia, Alzheimer disease, and multiple sclerosis. J Neurol Sci. 2002;202:13–23. doi: 10.1016/s0022-510x(02)00207-1. [DOI] [PubMed] [Google Scholar]
- 27.Corder EH, et al. The role of APOE polymorphisms in late-onset dementias. Cell Mol Life Sci. 1998;54:928–934. doi: 10.1007/s000180050223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buttini M, et al. Dominant negative effects of apolipoprotein E4 revealed in transgenic models of neurodegenerative disease. Neuroscience. 2000;97:207–210. doi: 10.1016/s0306-4522(00)00069-5. [DOI] [PubMed] [Google Scholar]
- 29.Cavrois M, De Noronha C, Greene WC. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol. 2002;20:1151–1154. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- 30.Moretti EW, et al. APOE polymorphism is associated with risk of severe sepsis in surgical patients. Crit Care Med. 2005;33:2521–2526. doi: 10.1097/01.ccm.0000186368.96146.fb. [DOI] [PubMed] [Google Scholar]
- 31.Singh IP, et al. Lipoproteins account for part of the broad non-specific antiviral activity of human serum. Antiviral Res. 1999;42:211–218. doi: 10.1016/s0166-3542(99)00032-7. [DOI] [PubMed] [Google Scholar]
- 32.Owens BJ, et al. Apolipoprotein A-I and its amphipathic helix peptide analogues inhibit human immunodeficiency virus-induced syncytium formation. J Clin Invest. 1990;86:1142–1150. doi: 10.1172/JCI114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin I, Dubois MC, Saermark T, Ruysschaert JM. Apolipoprotein A-1 interacts with the N-terminal fusogenic domains of SIV (simian immunodeficiency virus) GP32 and HIV (human immunodeficiency virus) GP41: Implications in viral entry. Biochem Biophys Res Commun. 1992;186:95–101. doi: 10.1016/s0006-291x(05)80780-6. [DOI] [PubMed] [Google Scholar]
- 34.Briz V, Poveda E, Soriano V. HIV entry inhibitors: Mechanisms of action and resistance pathways. J Antimicrob Chemother. 2006;57:619–627. doi: 10.1093/jac/dkl027. [DOI] [PubMed] [Google Scholar]
- 35.McCune JM, et al. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53:55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- 36.Weisgraber KH. Apolipoprotein E: Structure-function relationships. Adv Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 37.Kelly BA, et al. Apolipoprotein E-derived antimicrobial peptide analogues with altered membrane affinity and increased potency and breadth of activity. FEBS J. 2007;274:4511–4525. doi: 10.1111/j.1742-4658.2007.05981.x. [DOI] [PubMed] [Google Scholar]
- 38.Chertova E, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahley RW, Ji ZS. Remnant lipoprotein metabolism: Key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J Lipid Res. 1999;40:1–16. [PubMed] [Google Scholar]
- 40.Nguyen DH, Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol. 2000;74:3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci USA. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pietiainen VM, Marjomaki V, Heino J, Hyypia T. Viral entry, lipid rafts and caveosomes. Ann Med. 2005;37:394–403. doi: 10.1080/07853890510011976. [DOI] [PubMed] [Google Scholar]
- 43.Manes S, et al. Membrane raft microdomains mediate lateral assemblies required for HIV-1 infection. EMBO Rep. 2000;1:190–196. doi: 10.1093/embo-reports/kvd025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell SM, Crowe SM, Mak J. Lipid rafts and HIV-1: From viral entry to assembly of progeny virions. J Clin Virol. 2001;22:217–227. doi: 10.1016/s1386-6532(01)00193-7. [DOI] [PubMed] [Google Scholar]
- 45.Maziere JC, et al. Lovastatin inhibits HIV-1 expression in H9 human T lymphocytes cultured in cholesterol-poor medium. Biomed Pharmacother. 1994;48:63–67. doi: 10.1016/0753-3322(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 46.Michikawa M, Fan QW, Isobe I, Yanagisawa K. Apolipoprotein E exhibits isoform-specific promotion of lipid efflux from astrocytes and neurons in culture. J Neurochem. 2000;74:1008–1016. doi: 10.1046/j.1471-4159.2000.0741008.x. [DOI] [PubMed] [Google Scholar]
- 47.Jofre-Monseny L, et al. Effects of apoE genotype on macrophage inflammation and heme oxygenase-1 expression. Biochem Biophys Res Commun. 2007;357:319–324. doi: 10.1016/j.bbrc.2007.03.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colton CA, Brown CM, Vitek MP. Sex steroids, APOE genotype and the innate immune system. Neurobiol Aging. 2005;26:363–372. doi: 10.1016/j.neurobiolaging.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 49.Huang Y. Molecular and cellular mechanisms of apolipoprotein E4 neurotoxicity and potential therapeutic strategies. Curr Opin Drug Discov Dev. 2006;9:627–641. [PubMed] [Google Scholar]
- 50.Poirier J. Apolipoprotein E: A pharmacogenetic target for the treatment of Alzheimer's disease. Mol Diagn. 1999;4:335–341. doi: 10.1016/s1084-8592(99)80010-1. [DOI] [PubMed] [Google Scholar]
- 51.Ye S, et al. Apolipoprotein (apo) E4 enhances amyloid β peptide production in cultured neuronal cells: ApoE structure as a potential therapeutic target. Proc Natl Acad Sci USA. 2005;102:18700–18705. doi: 10.1073/pnas.0508693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.