Abstract
Myosin is an actin-based motor protein that generates force by cycling between actin-attached (strong binding: ADP or rigor) and actin-detached (weak binding: ATP or ADP·Pi) states during its ATPase cycle. However, it remains unclear what specific conformational changes in the actin binding site take place on binding to actin, and how these structural changes lead to product release and the production of force and motion. We studied the dynamics of the actin binding region of myosin V by using fluorescence resonance energy transfer (FRET) to monitor conformational changes in the upper-50-kDa domain of the actin binding cleft in the weak and strong actin binding states. Steady-state and lifetime data monitoring the FRET signal suggest that the cleft is in a more open conformation in the weak actin binding states. Transient kinetic experiments suggest that a rapid conformational change occurs, which is consistent with cleft closure before actin-activated phosphate release. Our results have identified a pre-force-generation actomyosin ADP·Pi state, and suggest force generation may occur from a state not yet seen by crystallography in which the actin binding cleft and the nucleotide binding pocket are closed. Computational modeling uncovers dramatic changes in the rigidity of the upper-50-kDa domain in different nucleotide states, which suggests that the intrinsic flexibility of this domain allows myosin motors to accomplish simultaneous tight nucleotide binding (closed nucleotide binding pocket) and high-affinity actin binding (closed actin binding cleft).
Keywords: actin, energy transfer, motor proteins, myosin, structural dynamics
A current challenge in biophysics is to understand the mechanism of how motor proteins such as myosin convert chemical energy into mechanical work through a cyclic interaction with actin filaments. Numerous structure/function studies have converged on a structural model for force generation, where conformational changes in the active site are coupled to a large rotation of the light-chain binding region, also known as the lever arm hypothesis (1, 2). Myosin alters its affinity for actin in a nucleotide-dependent manner, from strong actin binding states (ADP or rigor state) to weak actin binding states (ATP or ADP·Pi state), and thus phosphate release is believed to be associated with a large increase in actin affinity. Based on the high-resolution x-ray structure of myosin II (3) it was predicted that the actin binding cleft may open during ATP-induced dissociation of actomyosin and close during the release of the hydrolysis products induced by actin binding. The crystal structure of myosin V in the absence of nucleotide (rigor) demonstrated a closed conformation of the actin binding cleft (4, 5), and this structure fit quite well into the electron microscopy image reconstructions of the actomyosin rigor complex (6). Further studies of myosin II (7) and myosin V (8) by electron microscopy image reconstruction have demonstrated conformational changes in the actin binding cleft in different nucleotide states. Recently, the closed-cleft conformation was also observed in crystallographic studies of molluscan myosin II, wherein a “counterclockwise” orientation of the cleft rather than the extent of its closure was proposed to be critical for forming the strong binding rigor conformation (9).
By placing fluorescent probes in the actin binding cleft it was directly demonstrated that the cleft opens during ATP-induced dissociation of actomyosin (10, 11). However, there is currently no direct evidence that describes the kinetics of actin binding cleft closure in relationship to actin-activated phosphate release. In addition, there is a lack of information about how conformational changes in the cleft are coupled to structural changes in the nucleotide binding region. Most models of the actomyosin cross-bridge cycle suggest that myosin binds to actin through both ionic and hydrophobic interactions that stabilize a structural change in the actin binding region, such as closure of the actin binding cleft (1, 2). This structural change activates phosphate release through a series of conformational changes involving one or more of the elements of the active site that coordinate nucleotide binding: switch I, switch II, and the P loop. Finally, the swing of the lever arm and force generation is thought to occur before phosphate release (12–14). In the current study we demonstrate our ability to measure conformational changes in the actin binding region of myosin V associated with the transition from a weak to a strong actin binding complex and to correlate the kinetics of these conformational changes with biochemical steps in the actomyosin ATPase cycle.
In previous work, we labeled the upper-50-kDa domain of myosin V at residues 292–297 with the Fluorescein biArscenical Hairpin-binding dye (MV FlAsH) (15). Fluorescence resonance energy transfer (FRET) between mant-labeled nucleotides and MV FlAsH was used to demonstrate that the nucleotide binding pocket is more closed in the presence of ATP than ADP (1- to 2-Å distance change) in the absence of actin (15), which was also observed in other spectroscopic studies (16, 17). In addition, we found there was no change during the actin-activated phosphate release step, suggesting that force generation occurs from a state in which the nucleotide binding pocket remains closed. A possible explanation for these results is the existence of a unique conformational change in the upper-50-kDa domain in which the nucleotide binding pocket remains closed while the actin binding cleft closes down or rotates to allow strong actin binding. To test this possibility, we labeled actin at Cys-374 with IAEDANS (5((((2-iodoacetyl)amino)ethyl)amino)-naphthalene-1-sulfonic acid) (I-actin), and used the FRET between I-actin and MV FlAsH to study the structural dynamics of the upper-50-kDa domain during the actomyosin ATPase cycle. The FRET between these two probes allowed us to demonstrate a rapid temperature-dependent conformational change in myosin that occurs on binding to actin in the ADP·Pi state. We conclude that a pre-force generation ADP·Pi state is populated in the actomyosin ATPase cycle, which may be critical for the transition from weak to strong actin binding.
Results
IAEDANS-FlAsH FRET in the Weak and Strong Actin Binding States.
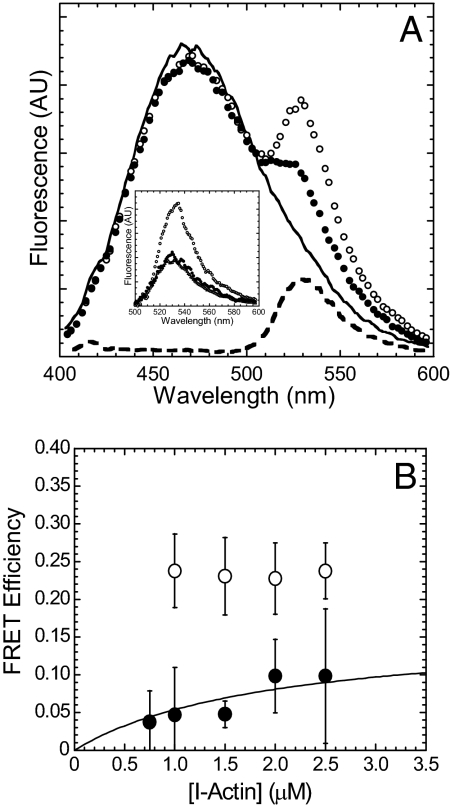
To investigate the weak binding states we generated myosin V with a point mutation in the switch II region (E442A MV) that essentially prevents ATP hydrolysis and therefore traps myosin V in the prehydrolysis M·ATP state (18, 19). Wild-type myosin V (WT MV) was also used to examine the strong binding states and examine the transition from weak to strong binding in the transient kinetic experiments described below. The degree of energy transfer from I-actin to E442A MV FlAsH in the strong binding (nucleotide-free or rigor) and weak binding (1 mM ATP) states was monitored by the enhancement of FlAsH fluorescence after subtraction of the contribution from I-actin fluorescence (Fig. 1). The efficiency of energy transfer (E) with the I-actin (1 μM) and MV FlAsH (0.5 μM) pair in the rigor state was similar for WT and E442A MV FlAsH (data not shown). In the presence of ATP, the FRET efficiency was determined from the titration curve with I-actin, which allowed us to extrapolate the efficiency at 100% bound E442A MV FlAsH:I-actin (Emax) (Fig. 1B) (15). The data were fit to a hyperbolic equation {E = Emax*[I-actin]/(Kd + [I-actin]), where Kd = 2 was determined from actin cosedimentation assays and a previous study (19)}. In the presence of ATP, the FRET efficiency was 7% lower for E442A MV FlAsH compared with rigor. The calculated distances (r) between the FlAsH site and I-actin in the absence and presence of ATP are shown in Table 1.
Fig. 1.
Steady-state energy transfer of E442A MV FlAsH bound to I-actin. (A) The fluorescence spectra [arbitrary units (AU)] of I-actin (1 μM) in the presence of unlabeled E442A MV (0.5 μM) (black line), and E442A MV FlAsH in the absence (open circles) or presence of 1 mM ATP (filled circles). (Inset) The FlAsH fluorescence component of the spectra, corrected by subtracting the I-actin fluorescence component. The fluorescence spectrum of MV FlAsH alone is also shown (open triangles). (B) The FRET efficiency of E442A MV FlAsH in the absence (open circles) and presence of ATP (filled circles) were plotted as a function of I-actin concentration. The data in the presence of ATP are fit to a hyperbola to determine the efficiency extrapolated to 100% bound MV FlAsH. The error bars represent the S.D. from at least three separate experiments.
Table 1.
Summary of I-actin: MV FlAsH energy transfer results
| Nucleotide states | R0, Å* | Steady state |
Lifetime |
||
|---|---|---|---|---|---|
| FRET efficiency† | r, Ň(rmin–rmax)§ | FRET efficiency¶ | r, Ň(rmin–rmax)§ | ||
| MV E442A FlAsH: I-actin (Rigor) | 51 | 0.23 ± 0.02 | 62.4 ± 1.2(43.4–81.9) | 0.36 ± 0.05 | 56.1 ± 2.2(40.0–73.7) |
| MV E442A FlAsH: I-actin (ATP) | 51 | 0.16 ± 0.01 | 67.2 ± 0.6(46.3–88.3) | 0.24 ± 0.05 | 61.8 ± 3.1(42.6–81.2) |
*Förster distance at which the efficiency of energy transfer is 50%.
†Steady-state FRET efficiency calculated from the titration curve with I-actin in the presence of ATP (errors represent standard error of the fit). In the rigor state errors represent standard deviation of the mean FRET efficiency.
‡Distance (r) between the donor and acceptor probes calculated by using a κ2 value of 2/3.
§The range of distances (rmin–rmax) possible based on the maximum and minimum value for κ2 determined from the steady-state and limiting anisotropy (20, 21).
¶FRET efficiency calculated from the decrease in donor lifetime (E = 1 − τ DA/τ D). The average lifetime 〈τ〉 of the donor was determined in the presence and absence of acceptor. The errors represent the standard deviation of the mean from 3 to 4 separate experiments that included different protein preparations.
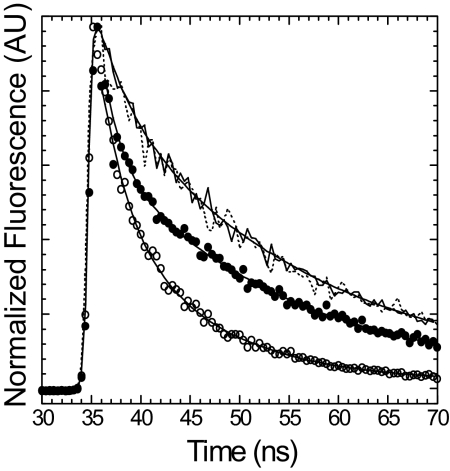
The efficiency of energy transfer was also measured by examining the fluorescence lifetime of I-actin in the presence of both unlabeled and FlAsH labeled E442A MV (Fig. 2 and Table 1). Compared with the steady-state fluorescence results the energy transfer efficiency determined by monitoring the lifetime of the donor was found to be slightly higher in both the rigor (0.5 μM I-actin/1.0 μM E442A MV FlAsH) and ATP states (0.5 μM I-actin/2.8 μM MV FlAsH, 1 mM ATP) (Fig. 2, Table 1). The lifetime decays were best fit by a two-exponential function for the rigor complex and a three-exponential function in the presence of ATP (two bound components and one unbound component) (Fig. 2, supporting information (SI) Table S1). In the presence of ATP, the lifetime of the unbound component was similar to that of I-actin alone and the fractional amplitude of this component was similar that expected based on the affinity of E442A MV·ATP for I-actin.
Fig. 2.
Energy transfer between E442A MV FlAsH and I-actin examined by fluorescence lifetime measurements. Time-resolved fluorescence decays of 0.5 μM I-actin were examined in the presence of 1.0 μM unlabeled (dashed line) or FlAsH labeled (open circles) E442A MV in the rigor state and 2.8 μM unlabeled (solid line) or FlAsH labeled (closed circles) E442A MV in the presence of 1 mM ATP. The data were fit to a two-exponential function in the rigor state and a three-exponential function in the presence of ATP. The average lifetime was calculated and used for the FRET efficiency measurements (see Table S1).
Transient Kinetic Measurements.
We examined the kinetics of the FRET signal with WT MV FlAsH binding to I-actin, or the pyrene signal with binding to pyrene actin in the strong binding rigor state (Fig. S1). The association rates of MV FlAsH binding to I-actin monitored by FRET were determined by examining the fluorescence increase in the acceptor (FlAsH) after mixing MV FlAsH (0.4 μM or 0.8 μM) with increasing concentrations of I-actin (5- to 10-fold excess I-actin) in the stopped flow. In the rigor state the fluorescence transients were fit to a single-exponential function at each actin concentration measured. The donor-only control demonstrates that the I-actin fluorescence is minimal and does not change with unlabeled MV binding at the emission wavelengths examined (Fig. S1B). There was also no change in acceptor fluorescence associated with binding to unlabeled actin (Fig. S1B). The rates of binding to actin monitored by FRET were linearly dependent on actin concentration, which allowed us to determine the second-order rate constant associated with actin binding (39.1 ± 2.1 μM−1 s−1) (Fig. S1A). The binding of MV FlAsH to pyrene actin was monitored by pyrene fluorescence quenching (Fig. S1A). In the absence of nucleotide, the fluorescence transients followed a single-exponential function, and the second-order binding constant determined from the linear fit of the binding rates was similar to the FRET signal (48.8 ± 0.8 μM−1 s−1).
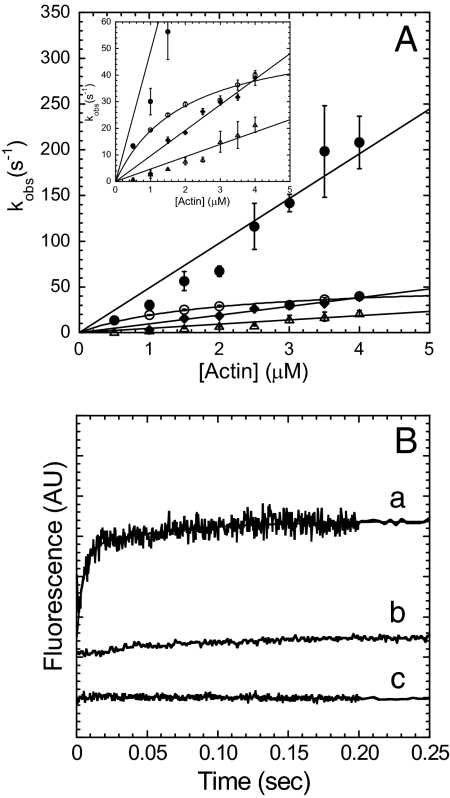
We examined the FRET signal during the weak-to-strong transition that occurs with MV FlAsH ADP·Pi binding to I-actin. MV FlAsH (0.4–0.8 μM) was mixed with 20 μM ATP, aged for 1 s, and then mixed with varying concentrations of I-actin (5- to 10-fold excess). The FlAsH fluorescence increase followed a biexponential function (Fig. 3). Both rates were linearly dependent on actin concentration and the amplitude was dominated by the fast phase, which had an actin dependence of 49.0 ± 3.0 μM−1 s−1. The slow phase (4.7 ± 0.3 μM·1 s−1) was <20% of the total amplitude and the relative amplitudes were similar at each actin concentration measured. The donor-only control demonstrates no change with unlabeled MV binding to I-actin in ADP·Pi state. However, the acceptor-alone control shows a small rise (actin dependence ≈5 μM−1 s−1) on mixing of MV FlAsH with unlabeled actin in the ADP·Pi state, which is similar to the slow phase in the FRET signal.
Fig. 3.
MV FlAsH binding to actin in the ADP·Pi state. The kinetics of the FRET signal was compared with the pyrene actin and phosphate release signals. (A) The FRET signal measured on MV FlAsH ADP·Pi binding to I-actin monitored by the increase in FlAsH fluorescence was fit to a biexponential function at each I-actin concentration. The fast rate (filled circles) and the slow rate (open triangles) were linearly dependent on I-actin concentration. MV FlAsH ADP·Pi binding to pyrene actin was hyperbolically dependent on actin concentration (open circles). The phosphate release rate was linearly dependent on I-actin concentration (filled diamonds). (Inset) Close-up of the phosphate release and pyrene actin plots. (B) Fluorescence traces of the FRET signal (a) monitored by the enhancement of FlAsH fluorescence and fit to a two-exponential function (final conditions: 0.2 μM MV FlAsH, 10 μM ATP, 2 μM I-actin; kobs = 143 ± 17 and 12 ± 2 s−1), compared with the donor-alone (b) (final conditions: 0.2 μM unlabeled MV, 10 μM ATP, and 2 μM I-actin) and acceptor-alone (c) (final conditions: 0.2 μM MV FlAsH,10 μM ATP, 2 μM unlabeled actin).
For comparison to our FRET signal, actin-activated phosphate release was measured. We examined the rate of actin-activated phosphate release by using a sequential-mix experiment identical to that described above, but with the phosphate-binding protein (PBP) in the final mix (Fig. 3A). The PBP fluorescence increase was fit to a burst followed by a linear phase. The burst rate was plotted as a function of I-actin concentration, and a linear fit was used to determine the second-order rate constant for actin-activated phosphate release (9.6 ± 0.4 μM−1 s−1).
To compare our FRET signal with the pyrene actin signal, a sequential mix experiment was also performed to monitor MV ADP·Pi binding to pyrene actin (Fig. 3A). Using the experimental design described above we found the rates of pyrene fluorescence quenching followed a single exponential function. The rate of the pyrene fluorescence decrease was plotted as a function of pyrene actin concentration and fit to a hyperbola with a maximum binding rate of 56.4 ± 6.9 s−1. The linear phase of the curve demonstrated a second-order binding constant of 28.6 ± 4.8 μM−1 s−1.
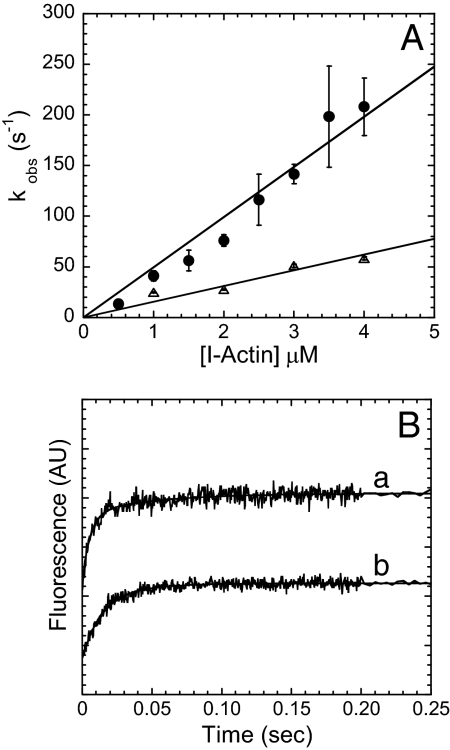
The temperature dependence of the FRET signal observed with MV FlAsH ADP·Pi binding to I-actin was determined by performing the sequential-mix experiments described above at 15°C (Fig. 4). At 15°C, the rate of the MV FlAsH fluorescence enhancement was fit to a single-exponential function at each I-actin concentration. The second-order rate constant calculated from the linear fit of the FRET transients was 15.5 ± 1.3 μM−1 s−1 at 15°C, which is ≈3-fold slower than that measured at 25°C.
Fig. 4.
The temperature dependence of the FRET signal. (A) The rate of the FlAsH signal increase (kobs) on MV FlAsH ADP·Pi binding (0.4–0.8 μM) to increasing concentrations of I-actin was plotted as a function of I-actin concentration at 25°C (filled circles) or 15°C (open triangles). The second-order binding constants were determined from the linear dependence on actin concentration. (B) The rates of the FlAsH fluorescence transients on 0.8 μM MV FlAsH ADP·Pi binding to 4 μM I-actin at 25 and 15°C (a, kobs = 198 ± 15 and 20 ± 2 s−1 at 25°C and b, kobs = 57 ± 3 s−1for 15°C).
Discussion
Our data provide critical information about the conformational changes in myosin that are associated with the transition from weak to strong actin binding. The FRET efficiency with the I-actin:MV FlAsH pair is ≈7% reduced in the weak binding states compared with the strong binding rigor state in the steady-state experiments, and ≈12% reduced in the lifetime measurements. These results suggest that there is a ≈5- to 6-Å distance change between the weak actin binding and strong actin binding states. Two possible conformational changes that could explain the distance between these two complexes are the opening/closing of the actin binding cleft, or some other conformational change in the actin binding region that prevents a tight stereospecific interaction between actin and myosin. Examination of the limiting and steady-state anisotropies of the donor and acceptor fluorophores (Table S2) in each nucleotide state allowed us to place limits on the orientation factor used to calculate the FRET distances (20, 21). The corresponding range of possible distances is quite large with this method, which is an upper limit because it does not take into account the further depolarization caused by the energy transfer process itself (Table 1). The limiting and steady-state anisotropies of the donor and acceptor probes are quite similar in each nucleotide state, suggesting that our probes detect a true change in distance associated with the weak and strong actin binding states. However, because the FlAsH anisotropies are quite high, change in orientation may not change the overall anisotropy values. Nevertheless, our results provide evidence for a conformational change between these states. Interestingly, our results are in reasonable agreement with the distance changes (Cys-374 of actin to Pro-294 of MV) observed in the cryo-EM structures of myosin V. The IAEDANS probe on two adjacent actin monomers can participate in FRET with MV FlAsH. Proceeding from the rigor to the ATP state, there is a 6-Å increase (73 → 79 Å) in the distance between Pro-294 of MV and Cys-374 of one actin monomer and a 2-Å increase (63 → 65 Å) with the adjacent monomer (8).
Rapid Attachment to Actin in the ADP·Pi State.
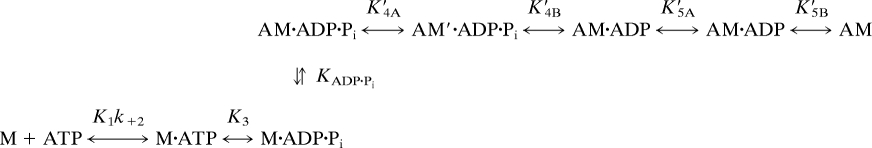
We discuss our transient kinetic results in the context of Scheme 1, which adds to the well established models (22–25) by including both a weak and a strong binding actomyosin ADP·Pi state (AM·ADP·Pi and AM′·ADP·Pi, respectively). Our previous study with the mant nucleotide/MV FlAsH donor/acceptor pair demonstrated that the nucleotide binding pocket remains closed during the actin-activated phosphate release step (K′4A and K′4B in Scheme 1) (15). Our results from mixing MV FlAsH with I-actin in the ADP·Pi state demonstrate that the fluorescent transient from the FRET signal contains two phases, and is dominated by the fast component that has an actin dependence of 49 μM−1 s−1 [1/KADP·Pi*(k′+4A + k′−4A)]. No maximum rate was obtained, but because the actin affinity is known in the M·ADP·Pi state (1/KADP·Pi = 10 μM) (19, 22), we can estimate that the maximum rate of the conformational change is quite rapid (k′+4A + k′−4A ≥ 500 s−1). The slow phase is ≈20% of the total amplitude, and we observed a similar fluorescence change with MV FlAsH binding to unlabeled actin in the ADP·Pi state. This result suggests that the slow rate in the FRET signal is a local conformational change in the FlAsH site, possibly associated with ADP release (K′5). Phosphate release from MV FlAsH measured with the PBP was found to have a second-order binding constant of 9.6 μM−1 s−1, which is similar to previous studies (22–25). These results demonstrate that our FRET signal is similar to the rate of MV binding to actin in the rigor state, which is 4- to 5-fold faster than the phosphate release rate. We examined the possibility that the FRET signal only monitors the initial binding of MV to actin, by determining the temperature dependence of the FRET signal. We observed a 3.2-fold reduction in the second-order binding constant when the temperature was reduced to 15°C, indicating that the FRET signal monitors a fast conformational change instead of a diffusion-limited binding process. This conformational change may be closure of the actin binding cleft, or another conformational change in some other element of the actin binding region, such as loop 2. Our observed results with pyrene actin are similar to a study by Rosenfeld and Sweeney (24), suggesting that the FlAsH probe does not significantly alter the actin binding process.
Scheme 1.
Conformational Change in the Actin Binding Cleft.
It has been proposed that the force generation step is closely associated with phosphate release in myosin (14). However, the specific conformational changes associated with phosphate release remain unclear. The relationship between force generation and phosphate release has been intensely examined with intact muscle fiber studies (12–14, 26, 27), solution biochemistry experiments with purified proteins (28–30), and single-molecule studies (31, 32). Currently the most widely accepted model suggests that myosin in the M·ADP·Pi state binds weakly to actin, undergoes a weak-to-strong transition forming a strongly bound actomyosin complex, which is followed by force generation and phosphate release (12–14). Key results from optical trap studies, and tension-recovery modeling on the timing of phosphate release in skeletal muscle myosin demonstrate that force generation occurs before Pi release (12–14). In this study, we observed a new AM·ADP·Pi intermediate during the actomyosin ATPase cycle. This intermediate is associated with a fast conformational change on MV·ADP·Pi binding to actin and may allow for rapid attachment to actin before force generation.
If the conformational change monitored by FRET is closure of the actin binding cleft, myosin must close down the cleft immediately on M·ADP·Pi binding to actin (k′+4A), and adopt a strongly bound conformation in the AM′·ADP·Pi state. Our results suggest that M·ADP·Pi binds to actin, followed by a fast rotation of the upper-50-kDa domain that results in closure of the actin binding cleft. This conformational change may cause distortion of the transducer region (β-sheet), generating strain-dependent tension that subsequently results in actin-activated phosphate release through the “back door” (33). A strongly bound AM·ADP·Pi state has been proposed based on muscle fiber studies (14), single-molecule laser trap studies (32), and rapid freezing EM studies (34). The large temperature dependence of the conformational change (Q10 = 3.2) is consistent with a significant domain motion such as cleft closure that proceeds through a large activation energy barrier (Ua ≈34 kBT). Structural and biochemical analysis of the nucleotide-free state in different myosin isoforms demonstrates that temperature-dependent rigor binding is an indication that cleft closure occurs during the binding process (i.e., a large temperature dependence for skeletal muscle myosin, Ua ≈28 kBT, and little or no temperature dependence for myosin V) (4, 9, 23). Thus, our temperature-dependent results are consistent with previous reports describing the energetics of cleft closure.
Structural Rearrangement of Loop 2.
Alternatively, the fast conformational change monitored by our FRET signal could be a conformational change in some other element of the actin binding region, such as within loop 2. A cryo-EM study suggests that loop 2 may adopt different conformations in different nucleotide states (AM·ADP/rigor, AM·ADP·Pi, and AM·ATP), and might be important for allowing myosin to stay loosely attached to actin in the weak binding states (8). If our data represent a conformational change in loop 2, this would suggest that there is a fast structural rearrangement in loop 2 during the weak-to-strong transition. Our results may fit in with the cryo-EM studies, which suggest that loop 2 adopts a more ordered conformation in the strongly bound state (rigor, ADP). These studies suggest that loop 2 rearranges in the ATP state, and rearranges again in the transition state (M·ADP·Pi). If the structural rearrangement of loop 2 is the fast conformational change we observed by our FRET signal, myosin must undergo another conformational change to close down the actin binding cleft. Because we did not observe a second slower transition in our FRET signal we feel that this interpretation is not likely. However, it is possible that the cleft closes by a unique rotation of the upper-50-kDa domain that does not change the distance between our MV FlAsH/I-actin donor/acceptor pair.
Dynamics of the Upper-50-kDa Domain in Myosin.
The recent rigor structures of molluscan myosins demonstrated that different conformations of the “inner” and “outer” actin binding cleft are possible (9), which implies that the cleft has some intrinsic flexibility. We investigated the possibility that a change in the flexibility of the upper-50-kDa domain allows myosin to adopt a closed nucleotide binding pocket and closed actin binding cleft conformation. We examined the RMSD of alpha carbon atoms in the myosin V rigor, ADP, and ADP-BeFX crystal structures (4, 5) by performing geometric simulations (35, 36) (see supplemental data in Figs. S2 and S3). We find that the upper-50-kDa domain is more flexible in the ADP state than in the ADP-BeFX and rigor states. These results show that the intrinsic flexibility of the upper-50-kDa domain may play a role in allowing myosin to rapidly adopt the strongly bound pre-force conformation. Interestingly, the increased flexibility of the upper-50-kDa domain in the ADP state may allow myosin motors to simultaneously bind tightly to actin and ADP. Because there is a large amount of variability within the myosin superfamily in the degree of coupling between the nucleotide and actin binding regions, the flexibility of the upper-50-kDa domain may be an important determinant of degree of nucleotide/actin coupling in myosin motors.
In conclusion, we observed a pre-force generation actomyosin ADP·Pi intermediate during the myosin cross-bridge cycle by monitoring the FRET signal between I-actin and MV FlAsH. We demonstrate a temperature-dependent conformational change before phosphate release associated with M·ADP·Pi binding to actin, which may be important for rapid attachment to actin before phosphate release and force generation. Our results suggest that force generation occurs in a structural state of myosin not yet seen by crystallographic studies, which has both a closed actin binding cleft and closed nucleotide binding pocket.
Materials and Methods
Myosin V cDNA Construction, Expression, and Purification.
Myosin V 1IQ constructs (wild-type and E442A MV) containing the upper-50-kDa tetracysteine motif, were coexpressed with calmodulin by using the baculovirus system (15). FlAsH labeling was very efficient (≥95%) as determined by absorbance measurements (15). Actin was purified from rabbit skeletal muscle by using an acetone powder method (37) and labeled with pyrene iodoacetamide (38) or (5((((2-iodoacetyl) amino) ethyl) amino)-naphthalene-1-sulfonic acid) (IAEDANS) (39). All experiments were performed in KMg50 TCEP buffer (50 mM KCl, 1 mM EGTA, 1 mM MgCl2, 1 mM TCEP, and 10 mM imidazole-HCl, pH 7.0).
FRET Measurements.
A Quantamaster fluorometer (Photon Technology International), equipped with a 75-watt xenon arc lamp as an excitation source and excitation/emission monochromaters, was used to measure steady-state fluorescence. Steady-state energy transfer measurements were performed by exciting I-actin at 365 nm, and the emission was measured from 400 to 600 nm with a 0.5-nm band pass. The enhancement in acceptor fluorescence was used to calculate the energy transfer efficiency after subtraction of the donor fluorescence (15, 21, 40). Fluorescence lifetime experiments were performed with a Timemaster Fluorescence Spectrometer (PTI), equipped with a picosecond pulse N2 dye laser. I-actin fluorescence was excited at 366 nm with a N2 dye laser and the emission measured through a single-grating monochromator at 471 nm. Fluorescence decays were analyzed with Global analysis software provided with the instrument. The quantum yield (QD) of the donor, extinction coefficients of the donor (εD) and acceptor (εA) at the donor excitation wavelength (365 nm), and overlap integral for the I-actin:MV FlAsH pair [J(λ)] were, QD = 0.38 (<5% change was observed in the ATP and rigor states), εA = 1,259 M−1 cm−1 (15), εD = 5,700 M−1 cm−1 (39), J(λ) = 3.2 × 10−13 cm3 M−1 and used in our FRET calculations. The steady-state (r) (measured in KMg50 at 25°C) and limiting (r0) anisotropy (measured in KMg50 in a glycerol solution at −14°C) was measured for the donor and acceptor fluorophore in the rigor and ATP states (see Table S2).
Kinetic Measurements.
Transient kinetic experiments were performed in an Applied Photophysics stopped-flow apparatus with a dead time of 1.2 ms. Pyrene actin was excited at 365 nm, and the fluorescence emission was measured by using a 400-nm long pass filter. FlAsH fluorescence was measured by energy transfer from I-actin by exciting at 365 nm and the emission was measured with a 515-nm long pass filter. Nonlinear least-squares fitting of the data was done with software provided with the instrument or Kaleidagraph (Synergy Software). Phosphate release experiments were performed by exciting the PBP at 400 nm and measuring the emission with a 420-nm long pass filter (22). Uncertainties reported are standard error of the fits unless stated otherwise. Kinetic modeling and simulations were performed with Pro-K software (Applied Photophysics) using a standard myosin V reaction scheme (15, 22–25) or Scheme 1. All concentrations mentioned in the stopped-flow experiments are final concentrations unless stated otherwise.
Computational Modeling.
Three x-ray crystal structures from the Protein Data Bank (PDB ID codes 1OE9, 1W7J, and 1W7I) were used respectively to represent the Rigor, ADP.BeFx, and ADP conformational states of myosin V. Floppy Inclusions and Rigid Substructure Topography [FIRST (35)] was used to determine the number of independent degrees of freedom and identify flexible and rigid regions within each structure. We used the Framework Rigidity Optimized Dynamic Algorithm (FRODA) to perform a geometric simulation (36). The root-mean-square displacement (rmsd) for carbon-alpha atoms was calculated for each structure (see Fig. S2 for detailed methods).
Supplementary Material
Acknowledgments.
We thank Dr. Howard White for the phosphate-binding protein, Andrei Istomin for the homology modeling on myosin V to incorporate loop 2 into the structure, Dorit Hanein and Niels Volkman for sharing the coordinates for the myosin V cryo-EM structures and Stephen Adams and Roger Tsieu for providing FlAsH. This work was supported in part by an American Heart Association Scientist Development Grant (to C.M.Y.), National Institutes of Health R01 GM073082-01A1 (to D.J.J.), the Center for Biomedical Engineering Systems, and the UNC Charlotte Graduate School.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710793105/DCSupplemental.
References
- 1.Holmes KC, Geeves MA. Structural mechanism of muscle contraction. Annu Rev Biochem. 1999;68:687–728. doi: 10.1146/annurev.biochem.68.1.687. [DOI] [PubMed] [Google Scholar]
- 2.Sweeney HL, Houdusse A. The motor mechanism of myosin V: Insights for muscle contraction. Philos Trans R Soc B. 2004;359:1829–1841. doi: 10.1098/rstb.2004.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 4.Coureux P, et al. A structural state of the myosin V motor without bound nucleotide. Nature. 2003;425:419–423. doi: 10.1038/nature01927. [DOI] [PubMed] [Google Scholar]
- 5.Coureux P, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes KC, Schroder RR, Sweeney HL, Houdusse A. The structure of the rigor complex and its implications for the power stroke. Philos Trans R Soc Lond B Biol Sci. 2004;359:1819–1828. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volkmann N, et al. Evidence for cleft closure in actomyosin upon ADP release. Nat Struct Biol. 2000;7:1147–1155. doi: 10.1038/82008. [DOI] [PubMed] [Google Scholar]
- 8.Volkmann N, et al. The structural basis of myosin V processive movement as revealed by electron cryomicroscopy. Mol Cell. 2005;19:595–605. doi: 10.1016/j.molcel.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y, et al. Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor. Structure. 2007;15:553–564. doi: 10.1016/j.str.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Yengo CM, De La Cruz EM, Chrin LR, Gaffney DP, II, Berger CL. Actin-induced closure of the actin-binding cleft of smooth muscle myosin. J Biol Chem. 2002;277:24114–24119. doi: 10.1074/jbc.M111253200. [DOI] [PubMed] [Google Scholar]
- 11.Conibear PB, Bagshaw CR, Fajer PG, Kovacs M, Malnasi-Csizmadia A. Myosin cleft movement and its coupling to actomyosin dissociation. Nat Struct Biol. 2003;10:831–835. doi: 10.1038/nsb986. [DOI] [PubMed] [Google Scholar]
- 12.Dantzig JA, Goldman YE, Millar NC, Lacktis J, Homsher E. Reversal of the cross-bridge force-generating transition by photogeneration of phosphate in rabbit psoas muscle fibers. J Physiol. 1992;451:247–278. doi: 10.1113/jphysiol.1992.sp019163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith DA, Sleep J. Mechanokinetics of rapid tension recovery in muscle: The myosin working stroke is followed by a slower release of phosphate. Biophys J. 2004;87:442–456. doi: 10.1529/biophysj.103.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takagi Y, Shuman H, Goldman YE. Coupling between phosphate release and force generation in muscle actomyosin. Philos Trans R Soc Lond B Biol Sci. 2004;359:1913–1920. doi: 10.1098/rstb.2004.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun M, et al. Dynamics of the upper 50 kDa domain of myosin V examined with fluorescence resonance energy transfer. J Biol Chem. 2006;281:5711–5717. doi: 10.1074/jbc.M508103200. [DOI] [PubMed] [Google Scholar]
- 16.Robertson I, Gaffney DP, Chrin LR, Berger CL. Structural rearrangements in the active site of smooth-muscle myosin. Biophys J. 2005;89:1882–1892. doi: 10.1529/biophysj.105.059840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naber N, Purcell TJ, Pate E, Cooke R. Dynamics of the nucleotide pocket of myosin measured with spin-labeled nucleotides. Biophys J. 2007;92:172–184. doi: 10.1529/biophysj.106.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki N, Shimada T, Sutoh K. Mutational analysis of the switch II loop of Dictyostelium myosin II. J Biol Chem. 1998;273:20334–20340. doi: 10.1074/jbc.273.32.20334. [DOI] [PubMed] [Google Scholar]
- 19.Yengo CM, De La Cruz EM, Safer D, Ostap EM, Sweeney HL. Kinetic characterization of the weak binding states of myosin V. Biochemistry. 2002;41:8508–8517. doi: 10.1021/bi015969u. [DOI] [PubMed] [Google Scholar]
- 20.Dale RE, Eisinger J. Intramolecular distances determined by energy transfer: Dependence on orientational freedom of donor and acceptor. Biopolymers. 1974;13:1573–1605. [Google Scholar]
- 21.Lakowicz JR. 3rd Ed. New York, NY: Springer; 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 22.Yengo CM, Sweeney HL. Functional role of loop 2 in myosin V. Biochemistry. 2004;43:2605–2612. doi: 10.1021/bi035510v. [DOI] [PubMed] [Google Scholar]
- 23.De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc Natl Acad Sci USA. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenfeld SS, Sweeney HL. A model of myosin V processivity. J Biol Chem. 2004;279:40100–40111. doi: 10.1074/jbc.M402583200. [DOI] [PubMed] [Google Scholar]
- 25.Hannemann DE, Cao W, Olivares AO, Robblee JP, De La Cruz EM. Magnesium, ADP, and actin binding linkage of myosin V: Evidence for multiple myosin V-ADP and actomyosin V-ADP states. Biochemistry. 2005;44:8826–8840. doi: 10.1021/bi0473509. [DOI] [PubMed] [Google Scholar]
- 26.Hibberd MG, Dantzig JA, Trenthan DR, Goldman YE. Phosphate release and force generation in skeletal muscle fibers. Science. 1985;228:1317–1319. doi: 10.1126/science.3159090. [DOI] [PubMed] [Google Scholar]
- 27.Hibberd MG, Webb MR, Goldman YE, Trentham DR. Oxygen exchange between phosphate and water accompanies calcium-regulated ATPase activity of skinned fibers from rabbit skeletal muscle. J Biol Chem. 1985;260:3496–3500. [PubMed] [Google Scholar]
- 28.Lymn RW, Taylor EW. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- 29.White HD, Taylor EW. Energetics and mechanism of actomyosin adenosine triphosphatase. Biochemistry. 1976;15:5818–5826. doi: 10.1021/bi00671a020. [DOI] [PubMed] [Google Scholar]
- 30.White HD, Belknap B, Webb MR. Kinetics of nucleoside triphosphate cleavage and phosphate release steps by associated rabbit skeletal actomyosin, measured using a novel fluorescent probe for phosphate. Biochemistry. 1997;36:11828–11836. doi: 10.1021/bi970540h. [DOI] [PubMed] [Google Scholar]
- 31.Takagi Y, Homsher EE, Goldman YE, Shuman H. Force generation in single conventional actomyosin complexes under high dynamic load. Biophys J. 2006;90:1295–1307. doi: 10.1529/biophysj.105.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker JE, Brossea C, Joel PB, Warshaw DM. The biochemical kinetics underlying actin movement generated by one and many skeletal muscle myosin molecules. Biophys J. 2002;82:2134–2147. doi: 10.1016/S0006-3495(02)75560-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawson JD, Pate E, Rayment I, Yount RG. Molecular dynamics analysis of structural factors influencing back door pi release in myosin. Biophys J. 2004;86:3794–3803. doi: 10.1529/biophysj.103.037390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker M, Zhang XZ, Jiang W, Trinick J, White HD. Observation of transient disorder during myosin subfragment-1 binding to actin by stopped-flow fluorescence and millisecond time resolution electron croyomicroscopy: Evidence that the start of the crossbridge power stroke in muscle has variable geometry. Proc Natl Acad Sci USA. 1999;95:8034–8039. doi: 10.1073/pnas.96.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacobs DJ, Rader A, Kuhn LA, Thorpe MF. Protein flexibility predictions using graph theory. Proteins. 2001;44:150–165. doi: 10.1002/prot.1081. [DOI] [PubMed] [Google Scholar]
- 36.Wells S, Menor S, Hespenheide B, Thorpe MF. Constrained geometric simulation of diffusive motion in proteins. Phys Biol. 2005;2:S127–S136. doi: 10.1088/1478-3975/2/4/S07. [DOI] [PubMed] [Google Scholar]
- 37.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 38.Pollard TD. Polymerization of ADP-actin. J Cell Biol. 1984;99:769–777. doi: 10.1083/jcb.99.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yengo CM, Chrin LR, Berger CL. Interaction of myosin LYS-553 with the C-terminus and DNase I-binding loop of actin examined by fluorescence resonance energy transfer. J Struct Biol. 2000;131:187–196. doi: 10.1006/jsbi.2000.4296. [DOI] [PubMed] [Google Scholar]
- 40.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.