Abstract
Defined mutant libraries allow for efficient genome-scale screening and provide a convenient collection of mutations in almost any nonessential gene of interest. Here, we present a near-saturating transposon insertion library in Vibrio cholerae strain C6706, a clinical isolate belonging to the O1 El Tor biotype responsible for the current cholera pandemic. Automated sequencing analysis of 23,312 mutants allowed us to build a 3,156-member subset library containing a representative insertion in every disrupted ORF. Because uncharacterized mutations that affect motility have shown utility in attenuating V. cholerae live vaccines, we used this genome-wide subset library to define all genes required for motility and to further assess the accuracy and purity of the library. In this screen, we identified the hypothetical gene VC2208 (flgT) as essential for motility. Flagellated cells were very rare in a flgT mutant, and transcriptional analysis showed it was specifically stalled at the class III/IV assembly checkpoint of the V. cholerae flagellar regulatory system. Because FlgT is predicted to have structural homology to TolB, a protein involved in determining outer membrane architecture, and the sheath of the V. cholerae flagellum appears to be derived from the cell's outer membrane, FlgT may play a direct role in flagellar sheath formation.
Keywords: cholera, flagella, flgT, mariner
Forward genetic screens have served as powerful tools for studying gene function in Vibrio cholerae, a water-borne human pathogen responsible for an estimated 120,000 deaths annually (1). Many of the virulence factors that allow the bacterium to infect the small intestine and induce massive diarrhea were discovered by using classical in vitro screening methods (2–4), and several new screening methods including signature tagged mutagenesis and in vivo expression technology have been developed to study gene function during infection (5, 6). A central characteristic of all of these forward genetic screens is that the genotype of the assayed mutants is unknown and is determined only in mutants where the desired phenotype is observed. As a result, large numbers of mutants need to be screened to achieve high genomic saturation, and it is impossible to be sure that mutations in all genes of interest were tested.
In contrast, large-scale sequence-defined mutant libraries contain mutants with identified disruptions throughout the genome (7). Because the genotype of all of these mutants is known, screens can be conducted on a small subset of the library containing only representative mutants for every gene or region of interest. This not only greatly reduces the time and cost of genome-wide saturating screens but also allows these screens to target specific subsets of genes (e.g., putative DNA-binding domains or ORFs in known pathogenicity islands). When transposons are used to generate these sequence-defined libraries, they can contain additional screening tools like transcriptional reporters or protein epitope tags and can be designed to support site-specific genomic targeting, allowing each insertion site to serve as a platform for any future genetic construct (7). Finally, defined insertion libraries serve as a convenient source of individual mutants and help identify genes essential for survival by defining ORFs that cannot tolerate transposon insertions.
Here, we describe the creation of a large sequence-defined transposon insertion library in V. cholerae strain C6706, a seventh pandemic O1 El Tor isolate (8) that is closely related to the sequenced strain N16961 (9, 10), and that contains a functional quorum-sensing system (11). Strains from the clonal outbreak where C6706 was isolated (12) are missing only a single gene from N16961 by genomic microarray (13), allowing us to use the annotated N16961 genome as a template to determine transposon insertion locations in C6706. In this report, we use a mariner transposon to create a 23,312-member insertion library and then demonstrate its utility in a genome-wide subset screen for chemotactic motility that identified VC2208 (flgT) as an essential factor in V. cholerae flagellar assembly.
Results and Discussion
Transposon Construction.
We used TnFGL3, a Himar1 mariner transposon based on TnSC189 (14), to generate a large library of insertional mutants in V. cholerae strain C6706lacZ. The transposon was constructed to contain promoterless lacZ and gfp transcriptional reporters and a constitutively expressed kanamycin resistance cassette (Fig. 1). FLP-mediated recombination at the flanking FLP recombinase target (FRT) sites can excise these genes, leaving behind a 192-bp scar whose remaining FRT site enables subsequent genomic targeting. The scar also contains a triple-FLAG epitope that can serve as a tag for protein identification and purification if it is in the proper frame and orientation (14). Finally, placement of the C9 Himar1 transposase (15) outside of the TnFGL3 coding region ensured stability of the transposon after genomic insertion and loss of the suicide delivery vector.
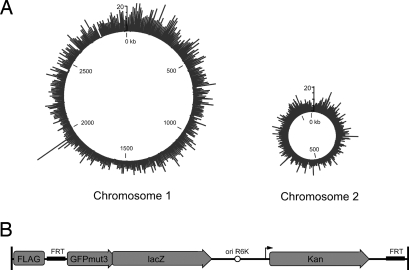
Fig. 1.
Transposon TnFGL3 and its insertion-site distribution in V. cholerae. (A) Number of insertions per kb in chromosome 1 (2.96 Mb) and chromosome 2 (1.07 Mb). The circumference of each depicted chromosome is proportional to its size and the origin of replication is at coordinate 0 (kb). (B) Schematic of TnKGL3 (6.1 kb). The transposon contains a triple FLAG epitope, promoterless gfp and lacZ transcriptional reporters, and a constitutively active kanamycin resistance cassette (kan). Two FRT sites enable FLP-mediated excision and subsequent genomic targeting. The functionality of the FLAG epitope and FRT sites was confirmed in several mutants by Western blot and sequencing after FlpE expression from a multicopy plasmid (data not shown).
Library Production.
TnFGL3 was introduced into C6706lacZ by conjugation, and 23,312 insertion mutants were isolated. After PCR amplification and direct sequencing of the transposon–genome junction, a Java application using BLAST was able to assign definitive insertion locations for 19,526 of these mutants using strain N16961 as the template genome (see Materials and Methods). Of these defined insertion mutants, 18,126 (92.8%) were in unique locations (Table 1). The transposon insertions were well distributed across both chromosomes (Fig. 1) but showed a small insertional bias toward chromosome 1 (Table 1). Insertions were also slightly biased against predicted ORFs, which represent 88.3% of the genome but contain only 86.1% of the transposon insertions, perhaps reflecting the essential nature of some coding regions (see below). Intragenic insertions did not demonstrate any directionality, as 50.2% of insertions were in the “+” direction in which gfp and lacZ are in the same orientation as the disrupted ORF. This suggests that selection for transposon insertions was independent of genomic promoter activity, and that expression of the lacZ and gfp reporters did not inhibit growth in a significant number of mutants.
Table 1.
TnFGL3 transposon library summary
| Transposon mutants isolated | 23,312 |
| Mutants with well defined insertion locations | 19,526 |
| Mutants in unique genomic locations | 18,126 |
| Chromosome 1 insertions (73.4% of genome) | 14,875 (76.2%) |
| Chromosome 2 insertions (26.6% of genome) | 4,654 (23.8%) |
| Insertions per kilobase in genome | 4.84 |
| Intragenic insertions | 16,810 |
| Insertions in + direction | 8,436 (50.2%) |
| Insertions in − direction | 8,374 (49.8%) |
| Intergenic insertions | 2,716 |
| Annotated ORFs in V. cholerae | 3,885 |
| ORFs disrupted in the defined library | 3,096 |
| ORFs never disrupted | 789 |
| Intragenic insertions per annotated ORF | 4.33 |
| Intragenic insertions per disrupted ORF | 5.43 |
Quality Control.
To confirm the accuracy of the assigned transposon locations, the insertion sites of 167 mutants were reamplified and sequenced. Manual inspection of the 163 readable sequencing results determined that 158 (96.9%) contained insertion locations at the exact base pair assigned in the library database. Of the remaining five mutants that gave alternative insertion locations, one was very likely the result of bacterial contamination from a neighboring well, and the others likely originated from “double picks” during robotic colony picking. In each case, we were able to isolate the originally annotated mutant from the contaminated well. Phenotypic screening of a subset library displayed a similar level of contamination (see Motility Screen below).
Essential Gene Analysis.
Of the 3,885 ORFs annotated for V. cholerae N16961 (10), 3,096 were interrupted at least once in the library (Fig. 2). The remaining 789 ORFs may have eluded disruption because they are required for growth in the nutrient-rich conditions used to generate the library [supporting information (SI) Dataset S1]. To estimate how many ORFs in this set are truly essential and how many were not hit because of chance alone, we used a neutral base-pair model, which assumes that every base pair in the genome has the same probability of containing a TnFGL3 insertion (16). The relatively even distribution of insertions across both chromosomes (Fig. 1, Table 1) and the nonspecific “TA” insertion site requirement of the Himar1 transposase (17) suggest that this assumption is largely valid. Given the current saturation level of the library, the neutral base-pair model estimates that at least 417 ORFs were not disrupted because of chance and are therefore unlikely to be essential (see Materials and Methods). When this model is further applied to ORFs that are grouped by size, it is clear that a large percentage of the ORFs that were likely missed by chance are <500 bp (Fig. 3). This is due to both the large number of small ORFs annotated in the V. cholerae genome and the relatively low probability of hitting ORFs of this size. In all, 349 ORFs shorter than 500 bp are predicted to be missed by chance, representing 74% of the 472 ORFs of this size that were not hit in the library. On the other hand, ORFs >1 kbp that remain uninterrupted in the library are much more likely to contain essential functions, as the neutral base-pair model predicts that nonessential ORFs of this size have at most a 0.8% chance of not being hit in the library. This is further reflected in the fact that 110 of the 143 V. cholerae ORFs in this group have orthologs in Escherichia coli K12, and 78 (71%) of these are essential under similar growth conditions (18) (Dataset S1).
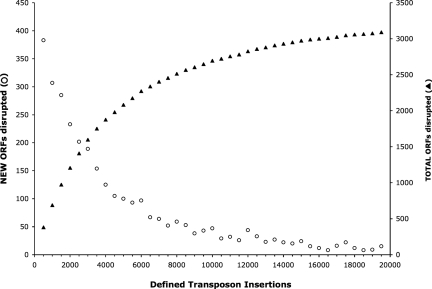
Fig. 2.
Saturation level of the transposon insertion library. The number of new ORFs disrupted in each group (open circles) and the total number of ORFs disrupted in the library (closed triangles) are shown. Mutants with defined insertion locations were analyzed in groups of 500 in the order they were sequenced.
Fig. 3.
Essential gene analysis. The neutral base pair model was used to estimate the number of ORFs missed in the library due to chance alone. The difference between the number of ORFs that were not hit in the library (gray bars) and the number of ORFs likely not hit by chance (striped bars) represent ORFs that are likely essential in V. cholerae (white bars). Of the 48 ORFs larger than 3 kb that are not shown on the graph, 3 were not hit in the library (the RNA polymerase subunits rpoB and rpoC and the DNA polymerase dnaE). These ORFs are very likely to be essential. ORFs are grouped by size in 250-bp increments.
It is important to consider whether any ORFs are protected from disruption due to polar effects on downstream essential genes. To address this issue, we identified all putatively operonic ORFs that were not disrupted in the library and then determined the frequency and orientation of transposon insertions in the other ORFs in these operons (see Materials and Methods). If polarity played a significant factor in determining transposon insertion locations, we would expect to observe a clear bias against insertions in genes upstream of putatively essential ORFs. Surprisingly, this was not the case; 44.0% of co-operonic genes upstream of unhit ORFs contained insertions, whereas 43.6% of downstream genes were hit. Furthermore, there was very little if any orientation bias for insertions in ORFs directly upstream of unhit ORFs (53.3% in the “+” direction). These data indicate that TnFGL3 allows sufficient transcriptional readthrough in both directions to avoid significant polar interference.
A confounding factor that could lead us to underestimate the number of essential genes in V. cholerae is that transposon insertions in the extreme ends of an ORF may not fully disrupt gene function. As observed in other sequence-defined libraries (16, 19), insertions in ORFs that have been disrupted only once in the library are disproportionately skewed to both the 5′ and 3′ ends of the coding sequence (Fig. S1). For example, 11.6% of all intragenic insertions in the library are found in the 3′ 10% of coding sequence, but this increases to 20.9% for ORFs that were hit only once in the library. This observation suggests that some of these insertions are not functional knockouts and that a subset of these ORFs may be essential for growth. If insertions in the first 5% and last 10% of each ORF are not considered to be functional knockouts, 984 ORFs were not fully disrupted in the library and are thus putatively essential for growth (Dataset S2). Experiments are currently underway to conditionally disrupt each ORF in this list to determine which are truly essential in V. cholerae.
This expanded list includes 228 of the 268 essential genes in E. coli that have orthologs in V. cholerae (85.1%), suggesting that a high percentage of essential E. coli genes have orthologs in V. cholerae that are also essential. Interestingly, only 5 of these 228 shared putatively essential genes are located on chromosome 2 (Dataset S2): the GTP cyclohydrolase folE; the NAD+ synthase nadE; and a gene cluster containing threonyl-tRNA synthetase (thrS), initiation factor IF3 (infC), and ribosomal protein L20 (rplT). Of these, only nadE (VCA0207) is found on chromosome 2 of all four Vibrio species whose genomes have been fully sequenced and annotated (www.jcvi.org). Because chromosome 2 is thought to have originated as a megaplasmid that became a permanent component of the ancestoral Vibrio genome after gaining essential genes from chromosome 1 (10, 20), it is tempting to speculate that nadE was the essential gene responsible for this megaplasmid capture event. Alternatively, it is possible that the responsible gene(s) have subsequently relocated to chromosome 1 in some Vibrio species or are essential in Vibrio species but not in E. coli.
Nonredundant Library.
To expedite genome-wide screening, we created a subset library containing a single mutant for each ORF disrupted in the library (Dataset S3). Priority was given to mutants with well defined insertions in the most 5′ location in each ORF while excluding transposon insertions in the first 5% of coding sequence. These extreme 5′ mutants and those containing ambiguous insertion locations were included when necessary to achieve maximum coverage of the genome (see Materials and Methods for details). The resulting nonredundant library contained 3,156 mutants.
Motility Screen.
To validate the nonredundant library and demonstrate its utility, the library was screened for chemotactic motility in soft agar under nutrient-rich conditions. This screen was chosen for several reasons. First, the large number of known flagellar and chemotaxis genes in V. cholerae served as convenient controls to assess the annotation accuracy of the library. Second, the ability of any contaminating bacteria to produce easily visible motility zones in these nonmotile mutants served as a powerful test for purity of the library. Third, there are several unique features of the V. cholerae flagellar apparatus (21), including a membranous sheath surrounding the flagella and a large array of auxiliary chemotaxis genes (22, 23) that remain largely uncharacterized. Finally, motility mutants are of special interest, because they have been useful in the development of oral cholera vaccines (24–26).
Of the 40 known flagellar biosynthesis genes disrupted in this library, 36 showed the predicted nonchemotactic phenotype (Dataset S4), including insertions in flhF and flhG, which demonstrated moderate motility defects as seen in previous studies (27). Interestingly, insertions in flgA and flgJ were fully motile (Fig. 4), despite their essential role in flagellar assembly in other organisms (28–30). Independent insertions throughout the flgA coding region show that its proposed function as a periplasmic chaperone for the P-ring protein FlgI is not essential for flagellar function in V. cholerae. In flgJ, the C-terminal location of the transposon in the muramidase domain suggests that FlgJ's peptidoglycan-degrading activity is not necessary for flagellar assembly, but it is not known whether the N-terminal rod-binding domain of FlgJ remains functional in this mutant. Finally, the motility observed in the flgG and fliG mutants resulted from contamination, because colony-purified strains of each were nonmotile, as were independent transposon insertions in both genes. From this observation and the resequencing analysis described above, we estimate that 3–5% of the library contains contaminating bacteria.
Fig. 4.
Chemotactic motility of library mutants. (A) Transposon mutants from the nonredundant library were tested for motility in nutrient rich plates containing 0.3% agar. C6706 and flgK serve as the wild type and nonmotile controls, respectively. (B) The motility defect in a flgT mutant can be complemented in trans by expression of full length flgT from the inducible PBAD promoter (0.01% arabinose). Strains were stabbed into motility plates and left at 30°C for 12 h.
In addition, V. cholerae contains three chemotaxis-like signal transduction clusters, but only cluster II appears to be important in regulating flagellar motility (31–33). In agreement, insertions in cheW-1, cheA-2, cheB-2, and cheY-3 in cluster II did not form motility zones, whereas insertions in all disrupted ORFs in clusters I and III were fully chemotactic in the nutrient-rich motility agar. It is interesting to note that an insertion in cheZ, which is not essential for chemotaxis in V. cholerae (31), was fully chemotactic in our assay and therefore did not block expression of the chemotaxis genes directly downstream in the cluster II operon (34). This was also true for insertions in VC2060 and VC2061 in this operon (data not shown), underscoring the nonpolar nature of TnFGL3 insertions at this locus. Interestingly, none of the 41 methyl-accepting chemotaxis genes (MCPs) disrupted in the V. cholerae genome exhibited a strong defect in motility/chemotaxis. This may be the result of redundancy among MCPs for specific chemoattractants, or it may reflect the large number of chemoeffectors that are likely present in LB motility agar. Alternatively, many of these MCPs may signal through clusters I and III, whose functions are unknown.
Several hypothetical ORFs exhibited motility defects. An insertion in VC2058, which lies directly downstream of chemotaxis cluster II, had a moderate motility defect (data not shown), whereas disruption of VC2206 (flgP), which is also adjacent to a large flagellar gene cluster, had a strong effect on flagellar motility as recently described (35), although in C6706 the flagella produced in this mutant appear normal by electron microscopy (data not shown). Several independent insertions in VC2207 [flgO (35)] directly upstream displayed only a small motility defect (Fig. 4 and data not shown).
FlgT Characterization.
A transposon insertion in VC2208 (here named flgT) displayed the strongest motility defect among uncharacterized ORFs in the nonredundant library (Fig. 4). An unmarked deletion of its predicted coding region (ΔflgT) resulted in the same strong motility defect, and the phenotype could be complemented in trans. When grown in liquid culture, very few bacteria were flagellated by electron microscopy (Fig. 5) and motile bacteria were rare under light microscopy (data not shown). Orthologs of flgT are found throughout the Vibrionaceae family of Gamma-Proteobacteria.
Fig. 5.
The flgT mutant is defective in flagellar assembly. Full length flagella are readily visible in representative electron micrographs of C6706 (wild type, Left) as are truncated flagella in the fliA insertional mutant (Right). The flgT mutant does not form visible flagella in a vast majority of cells (Middle). Bar represents 1 μm.
To further examine flgT's role in flagellar biosynthesis, we constructed lacZ transcriptional fusions to several flagellar promoters that are known to be sequentially expressed in V. cholerae's four-tiered transcriptional hierarchy (34). β-Galactosidase assays in wild-type and ΔflgT strains demonstrated that the ΔflgT mutant is specifically stalled at the class III/IV assembly checkpoint in V. cholerae's flagellar regulatory system (Fig. 6). Transcription of class II and III genes, which encode the hook and basal body (HBB) complex and the core flagellin FlaA (27, 34), was normal or elevated in the ΔflgT mutant, whereas expression from FliA (σ28)-dependent class IV promoters was strongly repressed compared with wild type. The class IV genes down-regulated in this mutant include the alternative flagellins flaB-D, which are dispensible for motility [Dataset S4 (36)] and the motor genes pomAB that are essential for flagellar rotation. The class III/IV checkpoint is controlled by FlgM, an anti-σ factor known to repress FliA activity until its secretion after HBB formation (37). However, FlgM may play a more complex role in V. cholerae given its inhibitory and apparent excitatory effect at different class IV promoters (38).
Fig. 6.
Flagellar transcription profile of the ΔflgT mutant. C6706lacZ (wild type) and the ΔflgT mutant (flgT) carry multicopy plasmids containing the indicated promoter in front of lacZ. Beta-galactosidase activity was measured from bacteria in midlog growth phase. Expression from class I and class II promoters (flrAp, flrBp, fliEp) remain at wild type levels in the ΔflgT mutant and expression from class III promoters is normal (flhAp) or elevated (flgKp, flaAp). Transcription from class IV promoters (flaBp, flaCp, flaDp) is strongly repressed in the flgT mutant.
The observation that very few flagella are formed in the ΔflgT mutant suggests that flgT plays a role in flagellar assembly upstream of this regulatory checkpoint. It is clear from electron microscopy images of a fliA mutant that expression of class II and III flagellar genes alone is sufficient to form the HBB complex and assemble a short-sheathed filament that is readily visible on a large percentage of cells (Fig. 5). However, flagella of any size are rarely formed in a ΔflgT mutant despite high transcription levels of these genes. This suggests that either HBB formation or sheathed filament assembly is impaired, but FlgT's precise role in this process remains undefined. FlgT contains an N-terminal signal sequence (39), and the protein structure metaserver Phyre (40) predicts that FlgT has strong structural homology to the N-terminal domain of TolB, a periplasmic member of the Tol system involved in maintaining outer-membrane integrity (41). Because this TolB domain is necessary and sufficient for binding to the inner membrane anchor TolA (42), FlgT may serve to disrupt this interaction at the flagellar pole to locally release the outer membrane and allow flagellar sheath formation. Future experiments may explore this possibility.
Conclusion
In summary, we have created a near-saturating transposon insertion library in V. cholerae strain C6706, a quorum-sensing competent clinical isolate belonging to the O1 El Tor biotype that is responsible for the ongoing seventh pandemic. Automated sequencing analysis of 23,312 mutants allowed us to build a 3,156-member subset library containing a single representative insertion for every disrupted ORF in the genome.
For several decades, live attenuated V. cholerae strains have been explored as oral vaccines against cholera but have suffered setbacks associated with high levels of unwanted side effects in human subjects (43, 44). However, uncharacterized spontaneous mutations that affect motility of V. cholerae have been correlated with an improved safety profile (24–26), and this has been attributed to decreased shedding of flagellin, a potent proinflammatory Toll-like receptor agonist (45). V. cholerae can make five different flagellins (36), and thus defining mutations that block flagellar assembly and flagellin secretion has utility in constructing live attenuated cholera vaccines. We applied our nonredundant transposon insertion library to this problem and defined almost all genes required for chemotactic motility in soft agar. This genome-wide screen identified VC2208 (flgT) as an essential factor in flagellar assembly and class IV gene expression during V. cholerae flagellar biosynthesis. A full analysis of the ability of these mutations to block the production of proinflammatory signaling molecules such as flagellins is currently in progress. Thus, the transposon insertion library described here represents a tool in cholera vaccinology and phenomics.
Materials and Methods
Bacterial Strains.
V. cholerae El Tor biotype strain C6706lacZ, a streptomycin-resistant spontaneous lacZ− derivative of C6706, was used as the parental strain for the library. E. coli DH5α λpir and Sm10 λpir were used for cloning and conjugation, respectively. Antibiotic concentrations used were streptomycin (800 μg/ml), kanamycin (75 μg/ml), and carbenicillin (75 μg/ml). LB was used for all growth conditions [10 g/liter of tryptone (Bacto), 5 g/liter of yeast extract (Bacto), and 5 g/liter of NaCl] and was supplemented with 16 g/liter of agar (Bacto) for growth on plates. All insertional mutants described in text and figures are the representative strains of each ORF in the nonredundant subset library (Dataset S3), unless otherwise noted.
DNA Manipulations.
pTnFGL3 was constructed by amplifying GFPmut3 and the translationally coupled lacZ gene from TnTGL3 (46) and cloning the PCR product into the AscI site in pSC189 (14). To generate an in-frame deletion in flgT, genomic DNA surrounding flgT was amplified by crossover PCR and cloned into pWM91 (47) for subsequent sacB-mediated allelic exchange in C6706lacZ, as described (48). For construction of pBAD24-flgT, full-length flgT was amplified from chromosomal DNA and cloned into plasmid pBAD24 (49) after digestion with KpnI and PstI. Transcriptional lacZ fusions of all flagellar promoters were constructed by cloning an ≈400-bp region upstream of each gene into pTL61T (50) between the XmaI and XbaI sites. All cloning products were sequence-verified, and the nucleotide sequence of all primers used is listed in Table S1.
Transposon Mutagenesis Protocol.
TnFGL3 insertions were generated by introducing the suicide plasmid pTnFGL3 into C6706lacZ by conjugation from Sm10 λpir. The donor and recipient strains were coincubated on LB agar for 45 min at 37°C. V. cholerae containing transposon insertions were selected on LB agar plates containing streptomycin and kanamycin. Colonies were robotically picked into 96-well microtiter plates and grown statically overnight at 37°C under streptomycin and kanamycin selection. Cultures were transferred by robot to duplicate 384-well microtiter plates for storage in 15% glycerol (vol/vol) and to 384-well PCR plates for insertion site identification.
Insertion Site Identification.
The location of each transposon insertion was determined by amplifying the insertion junctions using two-round semiarbitrary PCR (see SI Methods for a detailed protocol). Second-round PCR products were directly sequenced using a transposon-specific primer 83 bp from the insertion junction. A Java application adapted from published methods (19) was used to automate sequence base calling and quality assessment with Phred (www.phrap.org) and initial insertion location identification with BLAST. To further refine the insertion location, a Perl script used a variant of the Smith–Waterman algorithm (51) to compare the 20 bases that follow the transposon junction in the sequencing read to the corresponding genomic bases at the identified insertion site. The optimal local alignment, defined as the lowest Levenshtein distance (52) closest to the initially identified insertion site, was used to determine the transposon insertion site and the accompanying Levenshtein distance was used as a measure of insertion site accuracy. The script considered only genomic sites within a distance defined by the Levinshtein distance of the initial insertion site. Transposon mutants were included in the defined insertion library if their best BLAST score >200 or if they had a BLAST score >50 and a Levenshtein distance <4. Mutants with ambiguous insertion locations, i.e., those with two or more best BLAST scores of equal value from different areas of the genome, were excluded from the defined library. V. cholerae strain N16961 served as the template genome (10), and all data were stored in a MySQL relational database for subsequent analysis. For intragenic insertions, the direction of the transposon was defined as + if the transposon's gfp and lacZ transcriptional reporters were in the same orientation as the disrupted ORF.
Library Analysis.
Orthology between V. cholerae and E. coli K12 genes was defined as gene pairs that were reciprocal best BLASTP hits and contained >50% alignment of the gene and >15% identity within the alignment. Operonic ORFs were defined as those that are transcribed in the same direction as surrounding ORFs and are separated by <100 bp. Putatively essential ORFs that were predicted to be the first or last in the operon were excluded from consideration in this analysis. In the neutral base-pair model, each base is considered equally likely to contain a transposon insertion (16). The number of ORFs predicted to contain insertions was calculated as:
 |
where k is the number of ORFs in the genome, n is the number of defined transposon insertions, lengthj is the size of the jth ORF (in base pairs), and genome is the total genome length (in base pairs).
Nonredundant Library.
Strains from the defined insertion library were chosen for inclusion in the nonredundant set according to several criteria. For each annotated ORF (10), the strain with the most 5′ insertion location not within the first 5% of the ORF was chosen for inclusion in the nonredundant set. Only insertions with a BLAST score >60 were considered at this step. Preference was then given to strains with insertions oriented such that the GFP-lacZ cassette serves as a transcriptional reporter for the interrupted ORF. Subsequently, strains that contained insertions in the first 5% of the ORF were considered, and those with ambiguously defined insertion locations were then used to complete the nonredundant set. Strains were robotically cherry-picked into 96-well microtiter plates containing 200 μl of LB per well, grown statically overnight, and frozen at −80°C in 15% glycerol.
Motility Assays.
For the nonredundant library screen, strains were grown statically overnight at 37°C in 96-well microtiter plates and were robotically stabbed into motility plates containing LB and 0.3% agar. After incubation at 30°C for 12 h, motility zones were recorded with a digital camera, and motility was scored as a percentage area of wild type. When individual strains were assayed, they were grown to exponential phase before being manually stabbed into motility agar. Carbenicillin and 0.01% arabinose were included when indicated.
Electron Microscopy.
Strains were grown to exponential phase in LB, pelleted at 2,500 × g, and gently resuspended in 20 mM NaCl. Bacteria were allowed to adhere to carbon-coated grids, stained with 0.25% uranyl formate, and visualized with a Tecnai G2 Spirit BioTWIN microscope.
β-Galactosidase Assays.
Strains containing lacZ-promoter fusions in pTL61T were diluted 1:1,000 from overnight cultures and grown at 37°C shaking in LB plus carbenicillin to exponential phase (OD600 ≈0.4). The cells were permeabilized with chloroform and assayed in triplicate for β-galactosidase activity using the method described by Miller (53).
Supplementary Material
Acknowledgments.
We thank Frederick Ausubel and Nicole Liberati for helpful discussions and for sharing protocols, Maria Ericsson for assistance with electron microscopy, and Eric Rubin for use of his laboratory's liquid-handling robot. This work was supported by National Institutes of Health Grants AI-18045 and AI-26289 (to J.J.M.) and AI-064332 and DK-040561 (to F. Ausubel).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803281105/DCSupplemental.
References
- 1.Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiRita VJ, Parsot C, Jander G, Mekalanos JJ. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller VL, Mekalanos JJ. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang SL, Mekalanos JJ, Holden DW. In vivo genetic analysis of bacterial virulence. Annu Rev Microbiol. 1999;53:129–154. doi: 10.1146/annurev.micro.53.1.129. [DOI] [PubMed] [Google Scholar]
- 6.Slauch JM, Camilli A. IVET and RIVET: Use of gene fusions to identify bacterial virulence factors specifically induced in host tissues. Methods Enzymol. 2000;326:73–96. doi: 10.1016/s0076-6879(00)26047-3. [DOI] [PubMed] [Google Scholar]
- 7.Salama NR, Manoil C. Seeking completeness in bacterial mutant hunts. Curr Opin Microbiol. 2006;9:307–311. doi: 10.1016/j.mib.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wachsmuth IK, Olsvik Ø, Evins GM, Popovic T. In: Vibrio cholerae and Cholera: Molecular to Global Perspectives. Wachsmuth IK, Blake PA, Olsvik Ø, editors. Washington, DC: Am Soc Microbiol; 1994. pp. 357–370. [Google Scholar]
- 10.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J, et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wachsmuth IK, et al. The molecular epidemiology of cholera in Latin America. J Infect Dis. 1993;167:621–626. doi: 10.1093/infdis/167.3.621. [DOI] [PubMed] [Google Scholar]
- 13.Dziejman M, et al. Comparative genomic analysis of Vibrio cholerae: Genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci USA. 2002;99:1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang SL, Rubin EJ. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene. 2002;296:179–185. doi: 10.1016/s0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]
- 15.Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci USA. 1999;96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs MA, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lampe DJ, Grant TE, Robertson HM. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberati NT, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egan ES, Fogel MA, Waldor MK. Divided genomes: Negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol Microbiol. 2005;56:1129–1138. doi: 10.1111/j.1365-2958.2005.04622.x. [DOI] [PubMed] [Google Scholar]
- 21.McCarter LL. Polar flagellar motility of the Vibrionaceae. Microbiol Mol Biol Rev. 2001;65:445–462. doi: 10.1128/MMBR.65.3.445-462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boin MA, Austin MJ, Hase CC. Chemotaxis in Vibrio cholerae. FEMS Microbiol Lett. 2004;239:1–8. doi: 10.1016/j.femsle.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 23.Butler SM, Camilli A. Going against the grain: Chemotaxis and infection in Vibrio cholerae. Nat Rev Microbiol. 2005;3:611–620. doi: 10.1038/nrmicro1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coster TS, et al. Safety, immunogenicity, and efficacy of live attenuated Vibrio cholerae O139 vaccine prototype. Lancet. 1995;345:949–952. doi: 10.1016/s0140-6736(95)90698-3. [DOI] [PubMed] [Google Scholar]
- 25.Kenner JR, et al. Peru-15, an improved live attenuated oral vaccine candidate for Vibrio cholerae O1. J Infect Dis. 1995;172:1126–1129. doi: 10.1093/infdis/172.4.1126. [DOI] [PubMed] [Google Scholar]
- 26.Qadri F, et al. Peru-15, a live attenuated oral cholera vaccine, is safe and immunogenic in Bangladeshi toddlers and infants. Vaccine. 2007;25:231–238. doi: 10.1016/j.vaccine.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 27.Correa NE, Peng F, Klose KE. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J Bacteriol. 2005;187:6324–6332. doi: 10.1128/JB.187.18.6324-6332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirano T, Minamino T, Macnab RM. The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J Mol Biol. 2001;312:359–369. doi: 10.1006/jmbi.2001.4963. [DOI] [PubMed] [Google Scholar]
- 29.Nambu T, Minamino T, Macnab RM, Kutsukake K. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J Bacteriol. 1999;181:1555–1561. doi: 10.1128/jb.181.5.1555-1561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nambu T, Kutsukake K. The Salmonella FlgA protein, a putative periplasmic chaperone essential for flagellar P ring formation. Microbiology. 2000;146:1171–1178. doi: 10.1099/00221287-146-5-1171. [DOI] [PubMed] [Google Scholar]
- 31.Butler SM, Camilli A. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc Natl Acad Sci USA. 2004;101:5018–5023. doi: 10.1073/pnas.0308052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gosink KK, Kobayashi R, Kawagishi I, Hase CC. Analyses of the roles of the three cheA homologs in chemotaxis of Vibrio cholerae. J Bacteriol. 2002;184:1767–1771. doi: 10.1128/JB.184.6.1767-1771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyakutake A, et al. Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation. J Bacteriol. 2005;187:8403–8410. doi: 10.1128/JB.187.24.8403-8410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prouty MG, Correa NE, Klose KE. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol Microbiol. 2001;39:1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x. [DOI] [PubMed] [Google Scholar]
- 35.Morris DC, Peng F, Barker JR, Klose KE. Lipidation of an FlrC-dependent protein is required for enhanced intestinal colonization by Vibrio cholerae. J Bacteriol. 2008;190:231–239. doi: 10.1128/JB.00924-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klose KE, Mekalanos JJ. Differential regulation of multiple flagellins in Vibrio cholerae. J Bacteriol. 1998;180:303–316. doi: 10.1128/jb.180.2.303-316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aldridge P, Hughes KT. Regulation of flagellar assembly. Curr Opin Microbiol. 2002;5:160–165. doi: 10.1016/s1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- 38.Correa NE, Barker JR, Klose KE. The Vibrio cholerae FlgM homologue is an anti-sigma28 factor that is secreted through the sheathed polar flagellum. J Bacteriol. 2004;186:4613–4619. doi: 10.1128/JB.186.14.4613-4619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 40.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 41.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walburger A, Lazdunski C, Corda Y. The Tol/Pal system function requires an interaction between the C-terminal domain of TolA and the N-terminal domain of TolB. Mol Microbiol. 2002;44:695–708. doi: 10.1046/j.1365-2958.2002.02895.x. [DOI] [PubMed] [Google Scholar]
- 43.Tacket CO, et al. Volunteer studies investigating the safety and efficacy of live oral El Tor Vibrio cholerae O1 vaccine strain CVD 111. Am J Trop Med Hyg. 1997;56:533–537. doi: 10.4269/ajtmh.1997.56.533. [DOI] [PubMed] [Google Scholar]
- 44.Woodward WE, Gilman RH, Hornick RB, Libonati JP, Cash RA. Efficacy of a live oral cholera vaccine in human volunteers. Dev Biol Stand. 1976;33:108–112. [PubMed] [Google Scholar]
- 45.Yoon SS, Mekalanos JJ. Decreased potency of the Vibrio cholerae sheathed flagellum to trigger host innate immunity. Infect Immun. 2008;76:1282–1288. doi: 10.1128/IAI.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rietsch A, Wolfgang MC, Mekalanos JJ. Effect of metabolic imbalance on expression of type III secretion genes in Pseudomonas aeruginosa. Infect Immun. 2004;72:1383–1390. doi: 10.1128/IAI.72.3.1383-1390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metcalf WW, et al. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 48.Bina JE, Mekalanos JJ. Vibrio cholerae tolC is required for bile resistance and colonization. Infect Immun. 2001;69:4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Linn T, St Pierre R. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol. 1990;172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 52.Levenshtein VI. Binary codes capable of correcting deletions, insertions, and reversals. Soviet Physics Doklady. 1966;10:707–710. [Google Scholar]
- 53.Miller JH. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Lab Press; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.