Abstract
The plastid-localized methylerythritol phosphate (MEP) pathway synthesizes the isoprenoid precursors for the production of essential photosynthesis-related compounds and hormones. We have identified an Arabidopsis thaliana mutant, rif1, in which posttranscriptional upregulation of MEP pathway enzyme levels is caused by the loss of function of At3g47450, a gene originally reported to encode a mitochondrial protein related to nitric oxide synthesis. However, we show that nitric oxide is not involved in the regulation of the MEP pathway and that the encoded protein is a plastid-targeted homolog of the Bacillus subtilis YqeH protein, a GTPase required for proper ribosome assembly. Consistently, in rif1 seedlings, decreased levels of plastome-encoded proteins were observed, with the exception of ClpP1, a catalytic subunit of the plastidial Clp protease complex. The unexpected accumulation of ClpP1 in plastids with reduced protein synthesis suggested a compensatory mechanism in response to decreased Clp activity levels. In agreement, a negative correlation was found between Clp protease activity and MEP pathway enzyme levels in different experiments, suggesting that Clp-mediated degradation of MEP pathway enzymes might be a mechanism used by individual plastids to finely adjust plastidial isoprenoid biosynthesis to their functional and physiological states.
INTRODUCTION
Plastids are arguably the most distinctive organelles of plant cells. Besides the central importance of chloroplasts for photosynthesis, and therefore for life on earth as we know it, plastids harbor essential metabolic pathways that occur uniquely in plants among eukaryotes. A good example is the 2-methylerythritol 4-phosphate (MEP) pathway for the biosynthesis of isoprenoid precursors (Rodríguez-Concepción and Boronat, 2002; Eisenreich et al., 2004). Isoprenoids with primary (essential) roles in cell architecture, respiration, and regulation of growth and development are synthesized in all living organisms, but plant cells also produce an astonishing diversity of isoprenoid compounds for photosynthesis-related processes and as secondary metabolites that influence interactions with their environment. Most organisms have only one of the two currently known pathways for the biosynthesis of the prenyl diphosphate precursors of all isoprenoids, isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP). Thus, IPP and DMAPP are made exclusively from mevalonic acid (MVA) in archaebacteria, fungi, and animals, whereas most eubacteria (including cyanobacteria, the ancestors of plant plastids) only use the MEP pathway. By contrast, plants use both the MVA and MEP pathways, although in different cellular compartments (Lichtenthaler, 1999). Sterols, brassinosteroids, triterpenes, some sesquiterpenes, polyterpenes, and dolichol are formed from cytosolic prenyl diphosphates derived from the MVA pathway. On the other hand, the plastid-localized MEP pathway synthesizes the precursors for photosynthesis-related compounds (carotenoids and the side chain of chlorophylls, tocopherols, phylloquinones, and plastoquinones), hormones (gibberellins and abscisic acid), isoprene, monoterpenes, and some sesquiterpenes (Figure 1). Although some exchange of prenyl diphosphates can take place between the cytosol and the plastid in at least some plants, including Arabidopsis thaliana (Kasahara et al., 2002; Nagata et al., 2002; Laule et al., 2003), the block of one of the two pathways in seedlings cannot be rescued with common isoprenoid precursors synthesized by the other pathway under normal growth conditions (Gutierrez-Nava et al., 2004; Rodríguez-Concepción et al., 2004). Pathway- and compartment-specific regulatory mechanisms, therefore, must be present in plant cells to ensure that isoprenoid precursors are supplied when needed in each subcellular location.
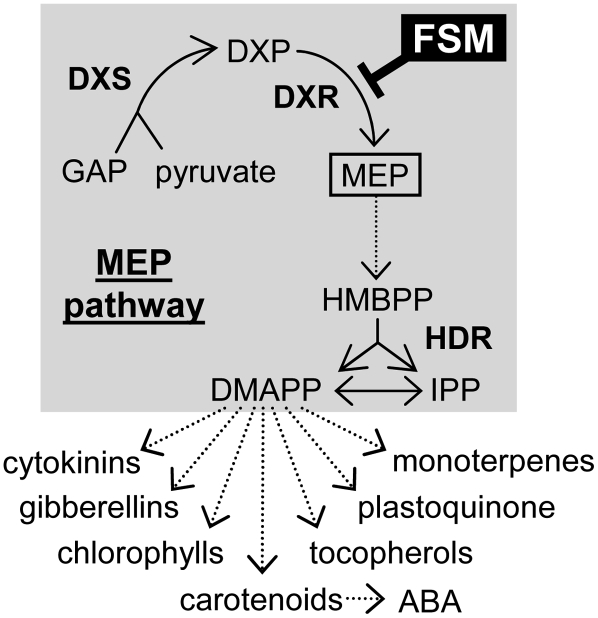
Figure 1.
Isoprenoid Biosynthesis in Plastids.
Dashed arrows represent multiple enzymatic steps. ABA, abscisic acid; GAP, glyceraldehyde 3-phosphate. MEP pathway enzymes (in boldface) are DXS, DXR, and HDR. The step inhibited by FSM is indicated.
Plant MEP pathway enzymes are encoded by nuclear genes and imported into plastids (Rodríguez-Concepción and Boronat, 2002; Eisenreich et al., 2004). 1-Deoxyxylulose 5-phosphate (DXP) synthase (DXS) catalyzes the first reaction of the pathway, the production of DXP from the central metabolic intermediates glyceraldehyde 3-phosphate and pyruvate. MEP is synthesized from DXP in the next step of the pathway, catalyzed by DXP reductoisomerase (DXR). Several enzymatic steps transform MEP into 1-hydroxy 2-methylbutenyl 4-diphosphate (HMBPP), which is finally converted by the enzyme HMBPP reductase (HDR) into a mixture of IPP and DMAPP (Figure 1). DXS, DXR, and HDR activities have been shown to share control over the metabolic flux of the MEP pathway (Estévez et al., 2001; Mahmoud and Croteau, 2001; Botella-Pavía et al., 2004; Enfissi et al., 2005; Carretero-Paulet et al., 2006). Besides the coarse control exerted by changes in the expression of genes encoding the biosynthetic enzymes in response to developmental, environmental, and metabolic signals (Rodríguez-Concepción, 2006), recent reports have demonstrated that enzyme levels are regulated at posttranscriptional levels (Guevara-Garcia et al., 2005; Sauret-Güeto et al., 2006). The molecular mechanisms involved in such regulation, however, remain to be established.
Growth of Arabidopsis seedlings in the presence of fosmidomycin (FSM), a strong competitive inhibitor of DXR (Figure 1), results in a specific block in the biosynthesis of MEP-derived plastid isoprenoids such as chlorophylls and carotenoids (required for photosynthesis and photoprotection), eventually causing a bleached phenotype and a developmental arrest that can be rescued by upregulating DXS and/or DXR levels (Rodríguez-Concepción et al., 2004; Guevara-Garcia et al., 2005; Carretero-Paulet et al., 2006; Sauret-Güeto et al., 2006). Our screening for Arabidopsis rif (for resistant to inhibition by FSM) lines able to develop in the presence of this inhibitor resulted in the unexpected isolation of several pale mutants, including rif10 (Sauret-Güeto et al., 2006). Impaired plastid RNA processing in rif10 plants and reduced protein synthesis in chloroplasts resulted in a posttranscriptional accumulation of DXS and DXR proteins and FSM resistance (Sauret-Güeto et al., 2006). By contrast, other processes affecting plastid development and causing a pale phenotype did not result in FSM resistance (Sauret-Güeto et al., 2006). The link between an altered production of plastid proteins and a posttranscriptional accumulation of nucleus-encoded MEP pathway enzymes in plastids has not been explored yet.
A number of albino, pale, and variegated mutants with defects in chloroplast development have been identified that affect a variety of plastid functions, mostly related to import and processing of nucleus-encoded proteins, expression of the plastid genome (plastome), and photosynthesis (López-Juez, 2007). Additionally, results from mutant screens suggest that a large number of genes with unknown function or unsuspected plastid relevance still remain to be identified as essential for chloroplast development (Budziszewski et al., 2001). An arrest in plastid development is also observed when the MEP pathway is blocked by FSM treatment or in mutants with defective biosynthetic genes, resulting in proplastid-like organelles with rudimentary thylakoids and an accumulation of vesicle structures, very low levels of photosynthetic pigments, and little or no expression of nuclear and plastidial genes required for chloroplast function (Mandel et al., 1996; Nagata et al., 2002; Gutierrez-Nava et al., 2004). Our previous results (Sauret-Güeto et al., 2006) provided strong evidence that proteins encoded by plastidial genes might in turn modulate the accumulation of flux-controlling MEP pathway enzymes within plastids. Plastome-encoded proteins include components of the plastidial gene expression machinery and photosynthetic apparatus and a few other polypeptides, including one of the catalytic subunits of the stromal Clp protease complex (Wakasugi et al., 2001; Adam et al., 2006).
To gain a deeper insight into the mechanisms that regulate the levels of flux-controlling MEP pathway enzymes, we have characterized another FSM-resistant mutant with a pale phenotype, rif1. Mutant rif1 plants show a posttranscriptional upregulation of DXS and DXR caused by the loss of function of the At3g47450 gene, originally reported to encode a mitochondrial protein related to nitric oxide synthesis. However, we demonstrate here that RIF1 is a plastidial protein and that nitric oxide is not involved in the regulation of the MEP pathway. Our data indicate that the RIF1 protein is most likely required for plastid ribosome assembly, confirming that defective expression of the plastid genome eventually results in the upregulation of MEP pathway enzyme levels. We also show that the mechanism responsible for such upregulation involves the stromal Clp protease complex and protein degradation within the plastid.
RESULTS
Loss of Function of the At3g47450 Gene Causes a Posttranscriptional Upregulation of MEP Pathway Enzymes
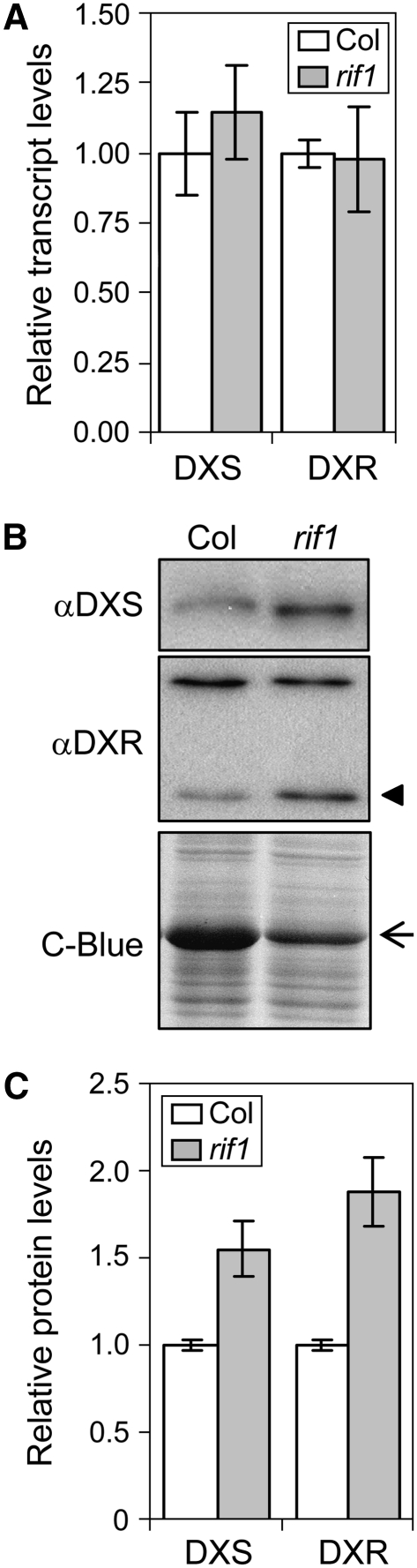
As reported for rif10 (Sauret-Güeto et al., 2006), mutant rif1 seedlings show pale cotyledons and a clearly delayed development and greening of true leaves compared with the Columbia (Col) wild type (Figures 2A to 2F), eventually resulting in smaller plants with a characteristic virescent phenotype of pale young leaves (those in the inner whorls of the rosette) but green mature leaves (Figures 2G and 2H). The rif1 mutant also showed a strong resistance to FSM (see Supplemental Figure 1 online). As a result, most rif1 seedlings remained virtually unaltered in the presence of FSM at concentrations that are lethal for the Col wild type (Figures 2A to 2D). Real-time quantitative RT-PCR experiments and immunoblot analyses showed that the FSM-resistant phenotype of rif1 seedlings likely results from the accumulation of increased levels of DXS (1.5-fold) and DXR (almost 2-fold) proteins in mutant plastids without changes in gene expression (Figure 3).
Figure 2.
Phenotypes of rif1 Plants and Complemented Lines.
Wild-type (Col) and transgenic lines were germinated on MS plates supplemented or not with 50 μM FSM and grown under long-day conditions. Plants grown in the absence of inhibitor for 15 d were then transferred to soil and grown under long days until completing their life cycle. Representative individuals at different developmental stages are shown. Panels in each row are to the same scale.
(A) to (D) Five-day-old Col ([A] and [B]) and rif1 ([C] and [D]) seedlings grown without FSM ([A] and [C]) or with FSM ([B] and [D]).
(E) and (F) Ten-day-old Col (E) and rif1 (F) plants grown on MS plates.
(G) to (I) One-month-old plants from Col (G), rif1 (H), and a transgenic rif1 line constitutively overexpressing a RIF1-GFP fusion protein (I) grown on soil.
(J) to (L) Young leaves from the complemented mutant line shown in (I) were used for confocal microscopy to detect the green fluorescence of RIF1-GFP (J) and the red autofluorescence of chlorophyll (K) in the same area of the leaf. Images were merged to show overlapping green and red fluorescence in yellow (L). Bars = 50 μm.
Figure 3.
Analysis of Gene Expression and Protein Levels of MEP Pathway Enzymes in rif1 Seedlings.
RNA and protein were extracted from 5-d-old Col and rif1 seedlings grown on MS plates under long-day conditions.
(A) Real-time quantitative RT-PCR analysis of transcript levels of the indicated genes in wild-type and mutant seedlings. Levels are represented relative to those in Col seedlings and correspond to the mean and sd from duplicate PCR analyses of two independent experiments.
(B) Immunoblot analysis with antibodies against DXS and DXR. The position of the DXR protein (Sauret-Güeto et al., 2006) is indicated with an arrowhead. The other major band recognized by the anti-DXR serum is shown as a protein-loading control. Coomassie blue (C-Blue) staining was also used to monitor total protein loading. The arrow marks the position of the RBCL protein.
(C) Quantification of DXS and DXR protein levels in Col and rif1 seedlings from immunoblot band intensity. Levels were normalized to those of the unspecific band recognized by the anti-DXR serum and are represented relative to the level in Col. Mean and se from duplicate blots of three independent experiments are represented.
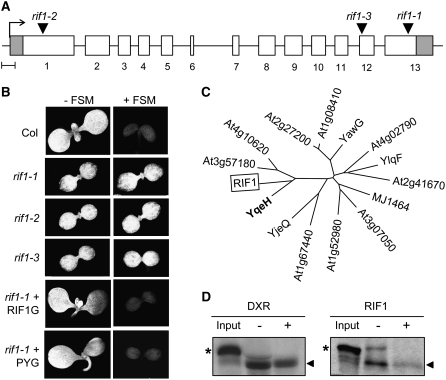
Backcrossing of homozygous rif1 plants with the Col wild type followed by analysis of the offspring showed that all of the phenotypes described above were recessive and linked to the presence of the T-DNA used to generate the activation-tagging lines (Weigel et al., 2000). A wild-type phenotype was observed in all F1 individuals resulting from the cross of homozygous rif1 and rif10 plants, indicating that they were not alleles but corresponded to different genes. Sequencing of the T-DNA–flanking sequences in the rif1 genome showed that the T-DNA was inserted in the last exon of the At3g47450 gene (Figure 4A). Other insertion alleles identified in the Salk collection (SALK_047882; herein named rif1-2) and the Cold Spring Harbor Laboratory Ds-GeneTrap lines (GT6235; herein named rif1-3) displayed the same phenotypes reported for the original rif1 mutant (renamed rif1-1), including a developmental delay, pale green cotyledons, and FSM resistance (Figure 4B). Furthermore, transformation of homozygous rif1-1 plants with a construct to constitutively express the full-length At3g47450 cDNA fused to green fluorescent protein (P35S:RIF1-GFP) completely restored the wild-type phenotype (Figures 2I and 4B), confirming that all of the distinctive phenotypes described here for rif1 are indeed due to the loss of function of this gene.
Figure 4.
Analysis of the Gene Mutated in rif1 and the Encoded Protein.
(A) Map of the coding region of the At3g47450 gene showing the transcription start (arrow), the exons (boxes), and the positions of the T-DNA in rif1-1, rif1-2, and rif1-3 mutants. Untranslated region sequences are represented in gray. Bar = 0.1 kb.
(B) Chlorophyll autofluorescence of 6-d-old seedlings of the indicated genotype grown on MS plates with (+) or without (−) 50 μM FSM. Representative individuals of transgenic rif1-1 plants constitutively expressing the fusion protein RIF1-GFP (RIF1G) or a plastid-targeted bacterial YqeH protein fused to GFP (PYG) are included. All panels are to the same scale.
(C) Phylogenetic tree of putative GTPases of the YlqF/YawG family identified in Arabidopsis. Representative members of each proposed subfamily (Leipe et al., 2002) are also included: B. subtilis YqeH and YlqF, Escherichia coli YjeQ, Methanococcus jannaschii MJ1464, and Schizosaccharomyces pombe YawG.
(D) In vitro import of DXR and RIF1 into wild-type chloroplasts. Import of in vitro-translated labeled DXR (used as a positive control) and RIF1 preproteins (positions marked with asterisks) was allowed to proceed for 10 min, and then the samples were either treated (+) or not (−) with thermolysin to remove nonimported proteins and analyzed by SDS-PAGE and fluorography. An aliquot of the input translation mixture was also included. The positions of the mature proteins (i.e., processed to remove the transit peptide after import into chloroplasts) are marked with arrowheads.
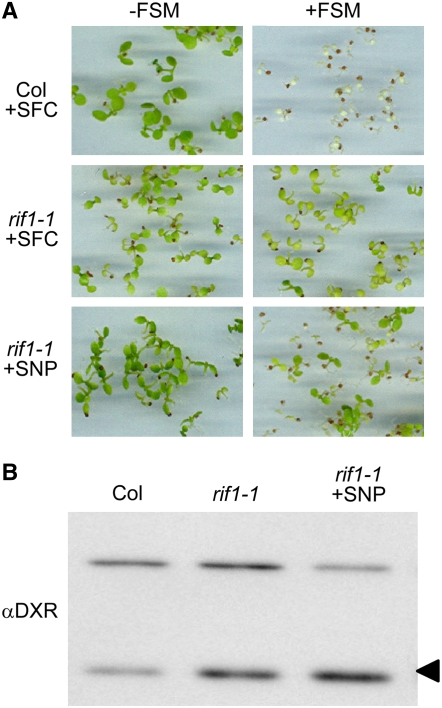
The protein encoded by the At3g47450 gene was originally reported to function as a nitric oxide (NO) synthase (NOS1) in Arabidopsis (Guo et al., 2003), but concerns about the proposed synthase activity of the protein led to its later being renamed NOA1 for NITRIC OXIDE–ASSOCIATED1 (Crawford et al., 2006; Zemojtel et al., 2006a). The pale phenotype and delayed growth of the loss-of-function nos1 mutant (our rif1-2 allele) was rescued by treating seedlings with sodium nitroprusside (SNP), a NO donor (Guo et al., 2003). If defective production of NO was also responsible for the enhanced accumulation of active DXS and DXR proteins detected in the mutant, it was expected that treatment of rif1 seedlings with SNP would result in wild-type levels of these enzymes and FSM sensitivity. Growth of rif1-1 seedlings on SNP-supplemented plates resulted in a partial rescue of the pale phenotype (Figure 5A). However, no significant changes were detected in the resistance to FSM (Figure 5A) or the levels of DXR protein (Figure 5B) compared with untreated controls or seedlings treated with the same concentration of sodium ferrocyanide (SFC; a SNP analog that does not produce NO). FSM resistance of the rif1-1 line was higher than that of Col seedlings at all of the concentrations of SNP tested up to 150 μM. Higher concentrations of SNP or SFC negatively influenced seedling growth. These results indicate that the posttranscriptional upregulation of MEP pathway enzymes in rif1 is not caused by a defect in the production of NO.
Figure 5.
Putative Role of NO in the Regulation of the MEP Pathway.
(A) Representative photographs of Col and rif1-1 seedlings germinated and grown for 5 d on MS plates containing a NO donor (50 μM SNP) or a NO donor analog as a control (50 μM SFC) and either supplemented (+) or not (−) with 50 μM FSM.
(B) Immunoblot analysis of DXR levels in samples grown without FSM as described for (A). The position of the DXR protein is indicated with an arrowhead. The unspecific band recognized by the antibody is also shown as a loading control.
The RIF1 Protein Is Likely Required for Ribosome Function in Plastids
RIF1 bears similarity to P-loop GTP binding proteins of the YlqF/YawG family containing a circularly permuted GTPase domain (Leipe et al., 2002). Comparison of representative members of the different subfamilies described for the YlqF/YawG family and homologous Arabidopsis proteins (Figure 4C; see Supplemental Figure 2 online) showed that RIF1 is most similar to the Bacillus subtilis YqeH protein, recently shown to be required for the correct formation of the bacterial 70S ribosome and the assembly or stability of the small (30S) ribosomal subunit (Uicker et al., 2007). Overall similarity and identity percentages are relatively low (23 and 33%, respectively), but they are higher when only conserved domains are considered (46% similar at the GTP binding domain and 39% similar at the putative Zn binding domain; see Supplemental Figure 2 online). The RIF1 protein contains an N-terminal domain that is absent from the bacterial YqeH protein and shows features of organellar targeting peptides (see Supplemental Figure 2 online). A GFP-tagged version of the full-length Arabidopsis RIF1/NOS1/NOA1 protein was previously found to be targeted to mitochondria in roots of stably transformed plants (Guo and Crawford, 2005). Using a similar P35S:RIF1-GFP construct, a wild-type phenotype (including FSM sensitivity) was fully restored in transgenic rif1-1 seedlings (Figure 4B) and adult plants (Figures 2G to 2I). However, we were unable to detect any green fluorescence from the biologically active RIF1-GFP fusion protein in mitochondria of roots of any of the transgenic lines generated. Analysis of photosynthetic tissues (cotyledons and leaves) also failed to reveal GFP fluorescence in mitochondria, but a clear signal was found in organelles identified as chloroplasts because of their size and chlorophyll autofluorescence (Figures 2J to 2L). Furthermore, untagged RIF1 was efficiently imported into isolated wild-type chloroplasts in vitro (Figure 4D), demonstrating that plastid targeting is an intrinsic feature of this protein and not an artifact caused by overexpression and/or GFP fusion.
To investigate whether RIF1 and YqeH were functional homologs and to confirm whether a plastidial localization of the protein was required to rescue the rif1 phenotype, a sequence encoding the bacterial protein fused to GFP was cloned in-frame with the plastid-targeting peptide of the MEP pathway HDS/GCPE protein (Querol et al., 2002) and constitutively expressed in mutant rif1-1 plants. As shown in Figure 4B and Supplemental Figure 3 online, the plastid-localized P-YqeH-GFP protein was able to complement the greening and growth defects of rif1-1 seedlings and to restore FSM sensitivity. These results confirm that the bacterial YqeH protein has a similar biochemical function as RIF1 and that the rif1 phenotype is due to a deficiency of this activity in the plastid.
Defective Development of Plastids in rif1 Seedlings Correlates with a General Decrease in Plastome-Encoded Protein Synthesis
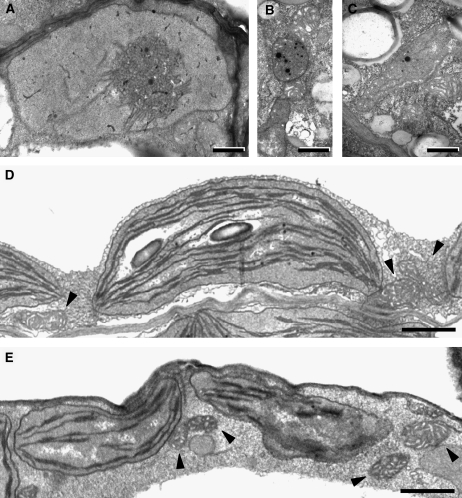
The results shown above indicate that the main biological function of RIF1 takes place in plastids, presumably in ribosome assembly. As a first approach to investigate whether plastid function was altered in the mutant, ultrathin sections of cotyledons from Col and rif1-1 seedlings grown in the dark (etiolated) or under long-day conditions for 3 d were examined using transmission electron microscopy (Figure 6). Etioplasts with a characteristic prolamellar body were observed in etiolated Col seedlings (Figure 6A) but not in rif1 seedlings, which only harbored small rounded plastids with plastoglobuli-like vesicles that in some cells developed into elongated plastids with rudimentary membranous structures (Figures 6B and 6C). When compared with the wild type, chloroplasts of light-grown rif1 seedlings were smaller, more irregularly shaped, and showed a general reduction in the development of thylakoid membranes and granal stacks (Figures 6D and 6E). Similar features were observed in leaf chloroplasts (see Supplemental Figure 4 online). By contrast, mitochondrial ultrastructure showed no apparent differences between Col and rif1 seedlings in any of the tissues or growing conditions analyzed (Figure 6; see Supplemental Figure 4 online). These results demonstrate that defects in the development and differentiation of different types of plastids (etioplasts and chloroplasts), but not mitochondria, result from the loss of RIF1 function.
Figure 6.
Ultrastructure of Plastids in Wild-Type and rif1-1 Cotyledons.
Seedlings were germinated and grown on MS plates in the dark (etiolated) or under long-day conditions for 3 d, and cotyledons were then examined by transmission electron microscopy. Mitochondria are indicated with arrowheads. Bars = 0.5 μm in (A) to (C) and 1 μm in (D) and (E).
(A) Etiolated Col seedling. The prolamellar body is clearly visible in the central-right area of the etioplast.
(B) and (C) Etiolated rif1 seedling. Plastids lack a prolamellar body and contain electrodense plastoglobuli-like vesicles.
(D) Light-grown Col seedling.
(E) Light-grown rif1 seedling.
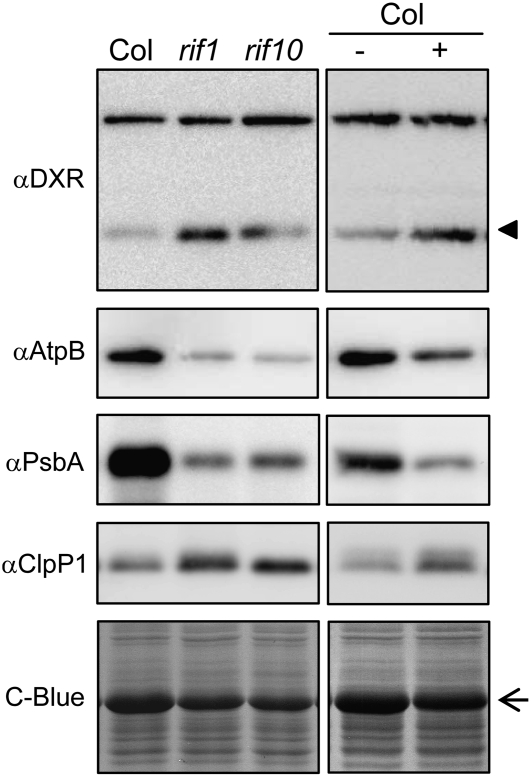
Assuming that the main role of RIF1 in Arabidopsis plastids is related to its ability to influence ribosome activity, as suggested by complementation analysis (Figure 4B; see Supplemental Figure 3 online), it is predicted that altered ribosome function in rif1 mutant plastids could result in defects in protein translation, eventually leading to decreased levels of plastome-encoded proteins. Consistently, the levels of the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (RBCL) detected by Coomassie blue staining after SDS-PAGE of protein extracts were much lower in rif1 than in Col seedlings (Figures 3 and 7). Immunoblot analysis of Col and rif1 seedling extracts with specific antibodies against the plastome-encoded proteins AtpB (for ATPase β chain) and PsbA (for photosystem II protein D1) confirmed a decreased production of plastome-encoded proteins in the absence of RIF1 function (Figure 7). As expected, similar reductions were observed in seedlings of the rif10 mutant, which is defective in plastidial RNA processing (Sauret-Güeto et al., 2006), and in Col seedlings grown in the presence of sublethal concentrations of chloramphenicol (CAP), an inhibitor of plastid protein synthesis. In all cases, partially impaired synthesis of plastome-encoded proteins correlated with concomitantly upregulated DXR levels (Figure 7), confirming our previous observations (Sauret-Güeto et al., 2006).
Figure 7.
Levels of Plastidial Proteins in Seedlings with Impaired Expression of the Plastome.
Proteins were extracted from Col, rif1-1, and rif10-1 seedlings grown under long-day conditions for 5 d on MS plates (left panels) and from Col seedlings grown under the same conditions on MS plates supplemented (+) or not (−) with 15 μM CAP (right panels). Immunoblot analyses were performed with antibodies raised against the indicated proteins. The position of the DXR protein is indicated with an arrowhead. The arrow marks the position of the RBCL protein detected by Coomassie blue (C-Blue) staining.
Homeostasis of DXS and DXR Levels Is Controlled by the Plastidial Clp Protease Complex
The discovery that the expression of the plastid genome is impaired in the rif1 mutant and causes similar phenotypes to those described for the rif10 mutant and CAP-treated wild-type seedlings (Sauret-Güeto et al., 2006) confirms a link between plastome-encoded proteins and the regulation of MEP pathway enzyme levels in plastids. Most of the ∼100 genes found in the plastid genome encode proteins of the photosynthetic apparatus and the transcription/translation system. The only plastome genes of known function in higher plants that fall out of these established groups encode enzymes (accD and ccsA) and ClpP1, a catalytic component of the plastidic ATP-dependent Clp protease (Wakasugi et al., 2001). Since the Clp protease has been proposed to target nucleus-encoded chloroplast-imported proteins (Sjögren et al., 2006) and decreased proteolytic degradation might be a plausible explanation for the increased levels of MEP pathway enzymes in rif1 plastids, we aimed to analyze the role of ClpP1 and the plastidic Clp protease complex in the posttranscriptional regulation of the MEP pathway.
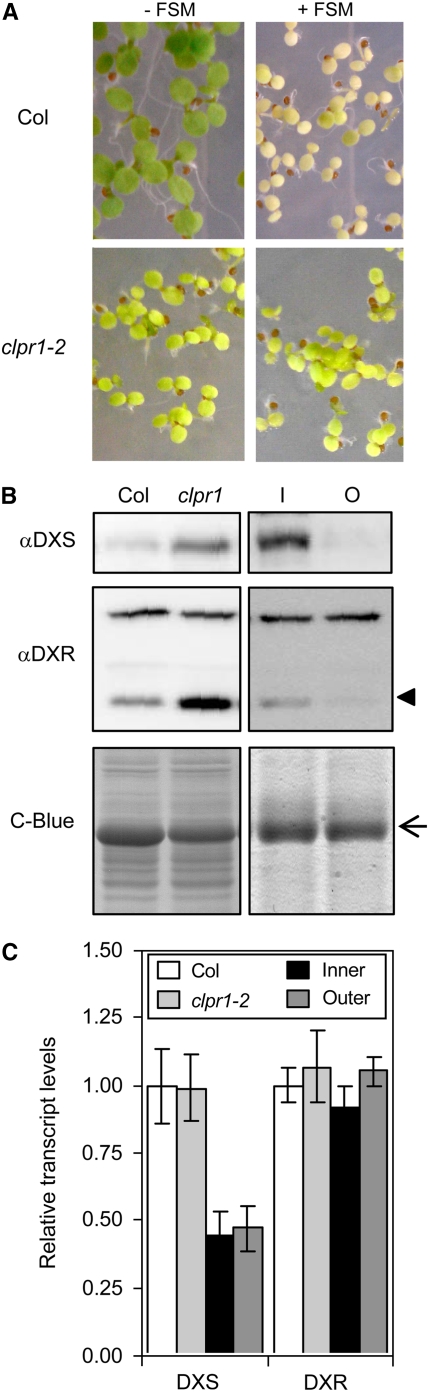
Unexpectedly, the levels of plastome-encoded ClpP1 protein were not reduced but increased in mutant rif1 and rif10 and in CAP-treated Col seedlings compared with untreated wild-type controls (Figure 7). ClpP1 together with four more Ser-type proteases (ClpP3 to ClpP6) and four related nonproteolytic proteins (ClpR1 to ClpR4) form the catalytic core of the Clp complex. Because the formation of a functionally active Clp protease relies on the correct stoichiometry of each of these subunits (Sjögren et al., 2006), the observed upregulation of ClpP1 in response to an impaired expression of the plastome might be a compensatory response to the inability to form sufficient active stromal Clp protease (Sjögren et al., 2004). To confirm whether a reduction in the activity of the plastidial Clp proteolytic complex had an effect on MEP pathway enzyme levels and FSM resistance, we used the publicly available clpr1-2 mutant (SALK_088407), in which the loss of function of nucleus-encoded ClpR1 causes a reduction of other ClpPR subunits, including ClpP1 (Koussevitzky et al., 2007). As shown in Figure 8, clpr1-2 seedlings showed a clear FSM resistance phenotype and a posttranscriptional increase in the levels of DXS and DXR compared with wild-type siblings, supporting a role of the plastidic Clp protease complex in the degradation of these MEP pathway enzymes. Because stromal substrates of the plastidic Clp machinery are reportedly more abundant in developing chloroplasts of young expanding leaves (Sjögren et al., 2006), we next compared the levels of DXS and DXR proteins in young leaves of the inner whorls of the rosettes of 3-week-old Col plants with those in fully expanded leaves of the outer whorl. Immunoblot analyses showed that DXS and DXR were much more abundant in younger leaves, whereas no differences were observed at the transcript level (Figure 8), providing additional evidence that the stromal Clp protease could be involved in the degradation of MEP pathway enzymes in functional wild-type chloroplasts.
Figure 8.
Role of the Plastidial Clp Protease Complex in the Degradation of DXS and DXR.
(A) Representative photographs of Col and mutant clpr1-2 seedlings germinated and grown under long-day conditions for 5 d on MS plates supplemented (+) or not (−) with 50 μM FSM.
(B) Immunoblot analysis of DXS and DXR levels in protein extracts from seedlings grown as described for (A) without FSM (left panels) and from leaves of the inner (I) or outer (O) whorls of the rosette of 3-week-old Col plants (right panels). The position of the DXR protein is indicated with an arrowhead. The arrow marks the position of the RBCL protein detected by Coomassie blue (C-Blue) staining.
(C) Real-time quantitative RT-PCR analysis of transcript levels of the indicated genes in the seedling and rosette leaf samples described for (B). Levels are represented relative to those in Col seedlings and correspond to the mean and se from duplicate PCR analyses of two independent experiments.
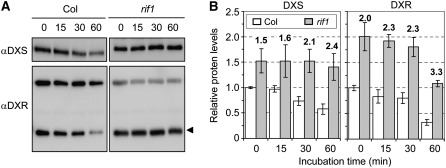
To further investigate whether DXS and DXR could be substrates for the plastidic Clp protease complex, we performed an in vivo degradation assay as described (Sjögren et al., 2006) and compared the stability of these stromal proteins in wild-type and mutant rif1 chloroplasts. Intact chloroplasts were incubated in the light with ATP, and samples were taken at different time points for immunoblot analysis. Figure 9 shows that both DXS and DXR proteins were degraded at a lower rate in mutant rif1 chloroplasts. As a result, the ratio of DXS levels in mutant versus wild-type chloroplasts was increased by 60% after 1 h of incubation in degradation buffer (from 1.5 to 2.4). A similar increase (65%, from 2.0 to 3.3) was observed for DXR (Figure 9). Our results are in agreement with these two MEP pathway enzymes being targets of the stromal Clp protease complex.
Figure 9.
Degradation of DXS and DXR in Isolated Chloroplasts.
(A) Chloroplasts isolated from Col and rif1-1 seedlings grown for 2 weeks on MS plates under long-day conditions were incubated for 1 h in the light at 25°C in the presence of 5 mM ATP. Aliquots were taken at the indicated times (minutes) and used for protein extraction and immunoblot analysis with antibodies against DXS and DXR. The amount of protein loaded was doubled in the case of Col samples, so bands of similar intensity were obtained in lanes corresponding to untreated Col and rif1 chloroplasts (time 0) in order to better compare degradation rates. The position of the DXR protein is indicated with an arrowhead. The other major band recognized by the anti-DXR serum is shown to monitor protein loading.
(B) Quantification of protein levels from two degradation experiments performed as described for (A). DXS and DXR levels (from immunoblot band intensity) were normalized to those of the unspecific band recognized by the anti-DXR serum and are represented relative to the level in untreated Col chloroplasts. Mean and se from duplicate blots of two experiments are represented. Boldface numbers above the columns indicate the ratio between DXS or DXR protein levels in rif1 versus Col chloroplasts.
DISCUSSION
A New Function for the RIF1 Protein in Chloroplasts
The RIF1/NOS1/NOA1 protein was first described to function as a NO synthase (Guo et al., 2003), but concerns were later raised about this activity (Crawford et al., 2006; Zemojtel et al., 2006a). Although our data do not rule out the participation of RIF1 in the production of NO in Arabidopsis, the fact that not all of the phenotypes of rif1 plants can be rescued by treatment with the NO donor SNP (Figure 5) suggests that the loss of RIF1 function affects other processes unrelated to NO synthesis. In particular, the increase in DXR levels eventually responsible for the FSM resistance phenotype of mutant rif1 seedlings was unaffected by treatment with SNP (Figure 5), indicating that the posttranscriptional accumulation of MEP pathway enzymes is not regulated by NO levels but most likely by an alternative function of RIF1. Although it has been reported that this protein is targeted to mitochondria in root cells (Guo and Crawford, 2005), here we provide several lines of evidence that RIF1 functions predominantly in plastids. RIF1 was imported into wild-type chloroplasts in vitro (Figure 4D), and a functional RIF1-GFP fusion protein was found to be localized in chloroplasts of rescued rif1 plants (Figure 2). The fact that the ultrastructure of etioplasts and chloroplasts but not mitochondria was clearly affected in rif1 seedlings (Figure 6) further supports a function of RIF1 in different types of plastids.
RIF1 is most similar to GTP binding proteins of the YlqF/YawG family, which are found in many prokaryotic and eukaryotic organisms (Leipe et al., 2002; Zemojtel et al., 2006a). Increasing evidence shows that prokaryotic members of this family are involved in the biogenesis, assembly, and/or stability of bacterial ribosomes by acting as chaperones or enzymes modifying rRNA or protein functions (Comartin and Brown, 2006; Uicker et al., 2007). The closest RIF1 homolog is YqeH (Figure 4C), a B. subtilis protein required for the formation of the bacterial 70S ribosome and the assembly or stability of the small (30S) ribosomal subunit (Uicker et al., 2007). Homologs from eukaryotic organisms lacking plastids, such as yeast and mammals, have been experimentally found in mitochondria, and the yeast YqeH protein (YOR205C) was shown to copurify in a complex with mitochondrial ribosomal proteins of the small subunit and to interact with mitochondrial translation elongation factor EF1α (Zemojtel et al., 2006b). Consistent with RIF1 having a similar activity in plastids, the mature protein contains an N-terminal Zn ribbon domain (see Supplemental Figure 2 online) that might be involved in binding to RNA (Zemojtel et al., 2006a). Also, a recombinant version of the bacterial YqeH protein targeted to plastids of rif1 plants was able to complement the mutant phenotype (Figure 4B; see Supplemental Figure 3 online). These results, together with the observed decrease in the levels of plastome-encoded proteins, support a main role for RIF1 in the expression of the plastid genome (plastome), most likely in a process required for normal plastid ribosome function. This conclusion is further supported by the striking phenotypic similarities between rif1 and rif10, a mutant known to be defective in the processing of all types of plastid RNA, including rRNA (Sauret-Güeto et al., 2006).
The Levels of Key MEP Pathway Enzymes Are Modulated by the Plastidic Clp Protease
The results we have shown here and in a previous report (Sauret-Güeto et al., 2006) confirm a strong and specific influence of plastid cues in the regulation of the MEP pathway for isoprenoid biosynthesis. Evidence provided in this work unveils a mechanism for such regulation involving the participation of the plastidic Clp protease complex. Impaired expression of the plastid genome in rif1, rif10, and CAP-treated Col seedlings unexpectedly resulted in increased levels of the plastome-encoded ClpP1 subunit of the catalytic ClpPR core of the complex (Figure 7). It is possible that ClpP1 levels are modulated not only by their biosynthetic rate but also by a regulatory feedback mechanism at the posttranslational level. As a result, a defective production of ClpP1 in the first stages of plastid development might result in an altered proportion of subunits within the ClpPR core and an insufficient Clp protease activity, which in turn might lead to the observed upregulation of ClpP1 levels as a compensatory mechanism (Sjögren et al., 2004). Most importantly, altered levels of ClpPR subunits in mutant clpr1-2 seedlings (Koussevitzky et al., 2007) also result in enhanced levels of both DXS and DXR and a concomitant phenotype of FSM resistance (Figure 8). The fact that these MEP pathway enzymes are more abundant in chloroplasts from young leaves of wild-type plants compared with fully expanded leaves of the outer whorl of the rosette (Figure 8), as expected for most substrates of the plastidic Clp complex (Sjögren et al., 2006), further implicates DXS and DXR as targets of the plastidial Clp protease. Consistently, a lower degradation rate of these two stromal enzymes was observed in rif1 compared with wild-type chloroplasts (Figure 9).
Proteolytic activities are required for the biogenesis and functioning of plastids and for their adaptation to changing environmental conditions. Plastid proteases of different classes remove and degrade targeting peptides, eliminate and recycle damaged, misfolded, or misassembled proteins and complexes, and help maintain the correct stoichiometry between different proteins and pathways (Adam et al., 2006; Sakamoto, 2006). The plastid Clp complex is the most prominent stromal protease, and it shows far more complexity in higher plants than in any other organism. The presence of similar Clp core complexes in all plastid types, their abundance, and the essential character of the catalytic subunits has led to the proposal that this protease plays a central role in plastid homeostasis and biogenesis, similar to that of the ubiquitin-proteasome system for plant development (Peltier et al., 2004). In contrast with the impressive progress in the structural characterization of this complex, little is currently known concerning the specific function of this stromal protease (Adam et al., 2006; Sakamoto, 2006). Genetic approaches have revealed that ClpP1 and other subunits of the catalytic ClpPR core are required for the correct differentiation of plastids and other aspects of plant development (Shikanai et al., 2001; Kuroda and Maliga, 2003; Rudella et al., 2006; Sjögren et al., 2006; Zheng et al., 2006; Koussevitzky et al., 2007), but the identification of protein substrates in higher plants has remained elusive. Several putative substrates, including a variety of chaperones and components required for the synthesis of plastome-encoded proteins, were recently identified by searching for stromal proteins differentially accumulated in chloroplasts from transgenic plants with reduced levels of Clp protease activity (Sjögren et al., 2004, 2006; Rudella et al., 2006). Our results (Figures 8 and 9) support the hypothesis that some of the MEP pathway enzymes, which are also localized in the stroma (Carretero-Paulet et al., 2002; Oudin et al., 2007), could be targets of the plastidic Clp protease machinery as well. However, current evidence does not allow us to conclude whether these proteins are direct targets (actual substrates) of this protease. It is possible that downregulation of Clp activity in plastids results in changes that are ultimately responsible for the observed accumulation of DXS, DXR, and/or the other proposed substrates. For example, the observed upregulation of a variety of stromal chaperones in plants with reduced levels of Clp protease (Sjögren et al., 2004, 2006; Rudella et al., 2006) might contribute to the observed phenotype of higher DXS and DXR levels and FSM resistance.
Although more experiments are needed to fully establish how the plastidic Clp protease and/or other plastid proteins modulate the accumulation of DXS and DXR enzymes, our work represents an important step forward to understanding the regulation of isoprenoid production in plastids. Changes in light conditions and photosynthetic activity can affect the synthesis of plastome-encoded proteins in wild-type chloroplasts (Pfannschmidt, 2002) but also the accumulation of plastidic Clp complex subunits (Zheng et al., 2006), which in turn could result in changes in the levels of DXS and DXR proteins. Because both DXS and DXR have been shown to control flux through the MEP pathway in Arabidopsis (Estévez et al., 2001; Carretero-Paulet et al., 2006) and other plants (Rodríguez-Concepción, 2006), the described posttranscriptional mechanism could allow individual plastids to rapidly optimize the supply of isoprenoid precursors when needed for the biosynthesis of end products.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana seeds from activation-tagging T-DNA collections (Weigel et al., 2000), Salk T-DNA insertion lines (Alonso et al., 2003), and the Ds-GeneTrap line GT6235 (Martienssen, 1998) were obtained from the Nottingham Arabidopsis Stock Centre. Wild-type and mutant genotypes used in this work were in the Col background with the exception of the GT6235 line, which was generated in Landsberg erecta. Seeds were surface-sterilized before plating on Petri dishes with solid Murashige and Skoog (MS) medium as described (Sauret-Güeto et al., 2006). Where indicated, the medium was supplemented with FSM (Gateway Chemical Technology), SNP (Sigma-Aldrich), SFC (Sigma-Aldrich), or CAP (Sigma-Aldrich). Plates were kept for at least 2 d at 4°C for stratification and then transferred to a growth chamber at 22°C under long-day conditions (8 h in the dark and 16 h under fluorescent white light at a photon fluence rate of 100 μmol·m−2·s−1). For seed production, plants were transferred from the plates to a 1:1:1 (v/v) perlite:vermiculite:sphagnum soil mixture irrigated with mineral nutrients and grown in the long-day chamber.
Identification of the T-DNA Insertion Site and Sequence Analysis
For the identification of the gene responsible for the FSM resistance phenotype in rif1, homozygous mutant plants were backcrossed with the Col wild type to test whether the corresponding mutations were linked to the presence of the T-DNA. After identifying the recessive nature of the rif1 mutation and its linkage to the resistance marker associated with the T-DNA, the insertion site was identified using an inverse PCR strategy as described (Sauret-Güeto et al., 2006).
Sequence analyses were performed with the Vector NTI Suite 6 (InforMax) software package and the available web resources. RIF1 putative homologs were searched on the National Center for Biotechnology Information and The Arabidopsis Information Resource databases (www.ncbi.nlm.nih.gov and www.Arabidopsis.org, respectively). Multiple alignments were performed with the ClustalW program (www2.ebi.ac.uk/clustalw), edited with GeneDoc (www.psc.edu/biomed/genedoc), and used to construct a phylogenetic tree with MEGA 2.1 (Molecular Evolutionary Genetics Analysis; www.megasoftware.net).
Generation of Transgenic Plants
Constructs for the constitutive expression of the fusion proteins RIF1-GFP and P-YqeH-GFP in transgenic plants were made as described in the Supplemental Methods online and used for Agrobacterium tumefaciens–mediated transformation of rif1-1 plants as described (Sauret-Güeto et al., 2006). Plants expected to harbor the transgenes were selected based on their ability to survive when grown on MS plates supplemented with 50 μg/mL hygromycin. The resulting positive plants were transferred to soil and allowed to set seed. Lines harboring a single T-DNA insertion were identified by segregation of the hygromycin selection marker and used for analysis.
Analysis of Transcript and Protein Levels
Real-time quantitative RT-PCR was performed as described (Carretero-Paulet et al., 2006). Crude total protein extracts from Arabidopsis tissues were obtained by grinding samples in liquid nitrogen and resuspending in ice-cold TKMES homogenization buffer (100 mM Tricine-KOH, pH 7.5, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA, and 10% [w/v] sucrose) supplemented with 0.2% (v/v) Triton X-100, 1 mM DTT, 100 μg/mL phenylmethylsulfonyl fluoride, 3 μg/mL E64, and 20 μL/mL protease inhibitor cocktail (Sigma-Aldrich). Protein concentration was determined as described (Carretero-Paulet et al., 2002). Separation by SDS-PAGE and immunoblot analyses were performed as described (Rodríguez-Concepción et al., 2004). Antibodies against DXS, DXR, and ClpP1 were kind gifts of Patricia León (Instituto de Biotecnología-UNAM), Michael H. Walter (Leibniz Institute of Plant Biochemistry), and Zach Adam (Hebrew University), respectively. Sera against AtpB and PsbA were purchased from Antisera. In the case of mutant or inhibitor-treated Col seedlings with decreased levels of RBCL (the major protein in the extracts), protein loading was normalized to the levels of other proteins detected by Coomassie Brilliant Blue staining of the gels. Bands corresponding to DXS and DXR were identified by size and by comparison with samples from transgenic overexpression lines (Carretero-Paulet et al., 2006; Sauret-Güeto et al., 2006). Chemiluminescent signals of the selected bands were visualized using a LAS-3000 (Fujifilm) image analyzer and quantified with the Multigauge Fujifilm 3.0 software. The levels of the unspecific band recognized by the anti-DXR serum were also quantified for normalization and used as an additional control of equal loading. Similar results were obtained when different proteins detected by Coomassie Brilliant Blue staining were used for normalization.
Chloroplast Isolation, Import Assays, and Degradation Experiments
For chloroplast import experiments, a cDNA sequence encoding the full-length RIF1 protein was cloned into the SmaI site of pBluescript SK+ (Stratagene). A construct with the right orientation, pBS-RIF1, was used as a template for in vitro transcription and translation using the TNT Quick T7 system (Promega) with 35S-labeled Met and T7 RNA polymerase. Plasmid pDXR-At (Carretero-Paulet et al., 2002) was used to obtain labeled DXR protein as a positive control. Import experiments were performed with chloroplasts isolated from 10-d-old Arabidopsis seedlings as described (Kubis et al., 2007) (see also Supplemental Methods online). Import was performed in white light at 25°C for 10 min. Thermolysin treatment and detection of imported polypeptides were performed as described (Aronsson and Jarvis, 2002).
For degradation experiments, chloroplasts isolated from 14-d-old seedlings were resuspended in HMS buffer (see Supplemental Methods online) supplemented with 20 mM gluconic acid, 10 mM NaHCO3, 0.2% (w/v) BSA, and 5 mM Mg-ATP and incubated at 25°C under white light (100 μmol·m−2·s−1). Aliquots of ∼107 chloroplasts were taken at 0, 15, 30, and 60 min, immediately pelleted, and frozen in liquid nitrogen to stop all protein degradation, as described (Sjögren et al., 2004). Pelleted samples were resuspended in ice-cold TKMES buffer (see Supplemental Methods online) and incubated for 15 min on ice for chloroplast lysis and protein solubilization. Determination of protein concentration, separation by SDS-PAGE, and immunoblot analysis were performed as described above.
Microscopy
Methods for the observation and recording of chlorophyll autofluorescence in whole seedlings, confocal laser scanning microscopy, and transmission electron microscopy are described in the Supplemental Methods online.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AtpB, AtCg00480; ClpP1, AtCg00670; ClpR1, At1g49970; DXR, At5g62790; DXS, At4g15560; HDR, At4g34350; HDS, At5g60600; MJ1464, Q58859; PsbA, AtCg00020; RBCL, AtCg00490; RIF1/NOS1/NOA1, At3g47450; RIF10, At3g03710; YawG, Q10190; YjeQ, P39286; YlqF, F69880; and YqeH, P54453.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Quantification of FSM Resistance of rif1 Seedlings.
Supplemental Figure 2. Multiple Alignment of the B. subtilis YqeH Protein with the Arabidopsis Closest Homologs.
Supplemental Figure 3. Phenotype of rif1 Plants Constitutively Expressing a Plastid-Targeted Bacterial YqeH Protein Fused to GFP (PYG).
Supplemental Figure 4. Ultrastructure of Chloroplasts in Wild-Type and rif1 Leaves.
Supplemental Methods. Constructs for the Production of Transgenic Lines; Chloroplast Isolation; Microscopy.
Supplemental Data Set 1. Text File of the Alignment Corresponding to Figure 4C and Supplemental Figure 2 online.
Supplementary Material
Acknowledgments
We thank J. Martínez-García and M.A. Phillips for critical reading of the manuscript, S. Kubis and J. Bédard for help and advice with the import experiments, and P. León, M. Walter, and Z. Adam for the gift of antibodies. We are also grateful to the Nottingham Arabidopsis Stock Centre, the Salk Institute Genomic Analysis Laboratory, and the Cold Spring Harbor Laboratory for valuable seed and information resources. The excellent technical support from A. Orozco and the staffs of the Serveis Cientificotècnics and the Serveis de Camps Experimentals of the Universitat de Barcelona and the Greenhouse and Microscopy facilities at the Centre for Research on Agricultural Genomics is greatly appreciated. This work was supported by grants from the Generalitat de Catalunya (Distinció) and the Spanish Ministerio de Ciencia y Tecnología and FEDER to M.R.-C. (Grant BIO2005-00357). U.F.-P. and S.S.-G. received PhD fellowships from the Spanish Ministerio de Educación y Ciencia (FPU program) and the Generalitat de Catalunya, respectively.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Manuel Rodríguez-Concepción (mrcgmp@ibmb.csic.es).
Online version contains Web-only data.
References
- Adam, Z., Rudella, A., and van Wijk, K.J. (2006). Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9 234–240. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Aronsson, H., and Jarvis, P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529 215–220. [DOI] [PubMed] [Google Scholar]
- Botella-Pavía, P., Besumbes, O., Phillips, M.A., Carretero-Paulet, L., Boronat, A., and Rodríguez-Concepción, M. (2004). Regulation of carotenoid biosynthesis in plants: Evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. Plant J. 40 188–199. [DOI] [PubMed] [Google Scholar]
- Budziszewski, G.J., et al. (2001). Arabidopsis genes essential for seedling viability. Isolation of insertional mutants and molecular cloning. Genetics 159 1765–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet, L., Ahumada, I., Cunillera, N., Rodríguez-Concepción, M., Ferrer, A., Boronat, A., and Campos, N. (2002). Expression and molecular analysis of the Arabidopsis DXR gene encoding 1-deoxy-D-xylulose 5-phosphate reductoisomerase, the first committed enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway. Plant Physiol. 129 1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet, L., Cairo, A., Botella-Pavia, P., Besumbes, O., Campos, N., Boronat, A., and Rodriguez-Concepcion, M. (2006). Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol. Biol. 62 683–695. [DOI] [PubMed] [Google Scholar]
- Comartin, D.J., and Brown, E.D. (2006). Non-ribosomal factors in ribosome subunit assembly are emerging targets for new antibacterial drugs. Curr. Opin. Pharmacol. 6 453–458. [DOI] [PubMed] [Google Scholar]
- Crawford, N.M., Gally, M., Tischner, R., Heimer, Y.M., Okamoto, M., and Mack, A. (2006). Plant nitric oxide synthase: Back to square one. Trends Plant Sci. 11 526–527. [Google Scholar]
- Eisenreich, W., Bacher, A., Arigoni, D., and Rohdich, F. (2004). Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell. Mol. Life Sci. 61 1401–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enfissi, E.M.A., Fraser, P.D., Lois, L.M., Boronat, A., Schuch, W., and Bramley, P.M. (2005). Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotechnol. J. 3 17–27. [DOI] [PubMed] [Google Scholar]
- Estévez, J.M., Cantero, A., Reindl, A., Reichler, S., and León, P. (2001). 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 276 22901–22909. [DOI] [PubMed] [Google Scholar]
- Guevara-Garcia, A., San Roman, C., Arroyo, A., Cortes, M.E., Gutierrez-Nava, M.L., and Leon, P. (2005). Characterization of the Arabidopsis clb6 mutant illustrates the importance of posttranscriptional regulation of the methyl-D-erythritol 4-phosphate pathway. Plant Cell 17 628–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, F.Q., and Crawford, N.M. (2005). Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell 17 3436–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, F.Q., Okamoto, M., and Crawford, N.M. (2003). Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302 100–103. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Nava, M.L., Gillmor, C.S., Jimenez, L.F., Guevara-Garcia, A., and Leon, P. (2004). CHLOROPLAST BIOGENESIS genes act cell and noncell autonomously in early chloroplast development. Plant Physiol. 135 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara, H., Hanada, A., Kuzuyama, T., Takagi, M., Kamiya, Y., and Yamaguchi, S. (2002). Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. J. Biol. Chem. 277 45188–45194. [DOI] [PubMed] [Google Scholar]
- Koussevitzky, S., Stanne, T.M., Peto, C.A., Giap, T., Sjogren, L.L., Zhao, Y., Clarke, A.K., and Chory, J. (2007). An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 63 85–96. [DOI] [PubMed] [Google Scholar]
- Kubis, S.E., Lilley, K.S., and Jarvis, P. (2007). Isolation and preparation of chloroplast from Arabidopsis thaliana plants. In Methods in Molecular Biology, A. Posch, ed (Totowa, NJ: Humana Press), pp. 171–186. [DOI] [PubMed]
- Kuroda, H., and Maliga, P. (2003). The plastid clpP1 protease gene is essential for plant development. Nature 425 86–89. [DOI] [PubMed] [Google Scholar]
- Laule, O., Furholz, A., Chang, H.S., Zhu, T., Wang, X., Heifetz, P.B., Gruissem, W., and Lange, M. (2003). Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100 6866–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe, D.D., Wolf, Y.I., Koonin, E.V., and Aravind, L. (2002). Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317 41–72. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1999). The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 47–65. [DOI] [PubMed] [Google Scholar]
- López-Juez, E. (2007). Plastid biogenesis, between light and shadows. J. Exp. Bot. 58 11–26. [DOI] [PubMed] [Google Scholar]
- Mahmoud, S.S., and Croteau, R.B. (2001). Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc. Natl. Acad. Sci. USA 98 8915–8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel, M.A., Feldmann, K.A., Herrera-Estrella, L., Rocha-Sosa, M., and Leon, P. (1996). CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 9 649–658. [DOI] [PubMed] [Google Scholar]
- Martienssen, R. (1998). Functional genomics: Probing plant gene function and expression with transposons. Proc. Natl. Acad. Sci. USA 95 2021–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, N., Suzuki, M., Yoshida, S., and Muranaka, T. (2002). Mevalonic acid partially restores chloroplast and etioplast development in Arabidopsis lacking the non-mevalonate pathway. Planta 216 345–350. [DOI] [PubMed] [Google Scholar]
- Oudin, A., Mahroug, S., Courdavault, V., Hervouet, N., Zelwer, C., Rodriguez-Concepcion, M., St-Pierre, B., and Burlat, V. (2007). Spatial distribution and hormonal regulation of gene products from methyl erythritol phosphate and monoterpene-secoiridoid pathways in Catharanthus roseus. Plant Mol. Biol. 65 13–30. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Ripoll, D.R., Friso, G., Rudella, A., Cai, Y., Ytterberg, J., Giacomelli, L., Pillardy, J., and van Wijk, K.J. (2004). Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 279 4768–4781. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt, T. (2002). Chloroplast redox signals: How photosynthesis controls its own genes. Trends Plant Sci. 8 33–41. [DOI] [PubMed] [Google Scholar]
- Querol, J., Campos, N., Imperial, S., Boronat, A., and Rodriguez-Concepcion, M. (2002). Functional analysis of the Arabidopsis thaliana GCPE protein involved in plastid isoprenoid biosynthesis. FEBS Lett. 514 343–346. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción, M. (2006). Early steps in isoprenoid biosynthesis: Multilevel regulation of the supply of common precursors in plant cells. Phytochem. Rev. 5 1–15. [Google Scholar]
- Rodríguez-Concepción, M., and Boronat, A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción, M., Forés, O., Martínez-García, J.F., González, V., Phillips, M.A., Ferrer, A., and Boronat, A. (2004). Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. Plant Cell 16 144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudella, A., Friso, G., Alonso, J.M., Ecker, J.R., and van Wijk, K.J. (2006). Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18 1704–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, W. (2006). Protein degradation machineries in plastids. Annu. Rev. Plant Biol. 57 599–621. [DOI] [PubMed] [Google Scholar]
- Sauret-Güeto, S., Botella-Pavia, P., Flores-Perez, U., Martinez-Garcia, J.F., San Roman, C., Leon, P., Boronat, A., and Rodriguez-Concepcion, M. (2006). Plastid cues posttranscriptionally regulate the accumulation of key enzymes of the methylerythritol phosphate pathway in Arabidopsis. Plant Physiol. 141 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai, T., Shimizu, K., Ueda, K., Nishimura, Y., Kuroiwa, T., and Hashimoto, T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42 264–273. [DOI] [PubMed] [Google Scholar]
- Sjögren, L.L., MacDonald, T.M., Sutinen, S., and Clarke, A.K. (2004). Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 136 4114–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren, L.L., Stanne, T.M., Zheng, B., Sutinen, S., and Clarke, A.K. (2006). Structural and functional insights into the chloroplast ATP-dependent Clp protease in Arabidopsis. Plant Cell 18 2635–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uicker, W.C., Schaefer, L., Koenigsknecht, M., and Britton, R.A. (2007). The essential GTPase YqeH is required for proper ribosome assembly in Bacillus subtilis. J. Bacteriol. 189 2926–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi, T., Tsudzuki, T., and Sugiura, M. (2001). The genomics of land plant chloroplasts: Gene content and alteration of genomic information by RNA editing. Photosynth. Res. 70 107–118. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel, T., Frohlich, A., Palmieri, M.C., Kolanczyk, M., Mikula, I., Wyrwicz, L.S., Wanker, E.E., Mundlos, S., Vingron, M., Martasek, P., and Durner, J. (2006. a). Plant nitric oxide synthase: A never-ending story? Trends Plant Sci. 11 524–525. [DOI] [PubMed] [Google Scholar]
- Zemojtel, T., Kolanczyk, M., Kossler, N., Stricker, S., Lurz, R., Mikula, I., Duchniewicz, M., Schuelke, M., Ghafourifar, P., Martasek, P., Vingron, M., and Mundlos, S. (2006. b). Mammalian mitochondrial nitric oxide synthase: Characterization of a novel candidate. FEBS Lett. 580 455–462. [DOI] [PubMed] [Google Scholar]
- Zheng, B., MacDonald, T.M., Sutinen, S., Hurry, V., and Clarke, A.K. (2006). A nuclear-encoded ClpP subunit of the chloroplast ATP-dependent Clp protease is essential for early development in Arabidopsis thaliana. Planta 224 1103–1115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.