Abstract
Rice (Oryza sativa) accumulates very high concentrations of silicon (Si) in the shoots, and the deposition of Si as amorphous silica helps plants to overcome biotic and abiotic stresses. Here, we describe a transporter, Lsi6, which is involved in the distribution of Si in the shoots. Lsi6 belongs to the nodulin-26 intrinsic protein III subgroup of aquaporins and is permeable to silicic acid. Lsi6 is expressed in the leaf sheath and leaf blades as well as in the root tips. Cellular localization studies revealed that Lsi6 is found in the xylem parenchyma cells of the leaf sheath and leaf blades. Moreover, Lsi6 showed polar localization at the side facing toward the vessel. Knockdown of Lsi6 did not affect the uptake of Si by the roots but resulted in disordered deposition of silica in the shoots and increased excretion of Si in the guttation fluid. These results indicate that Lsi6 is a transporter responsible for the transport of Si out of the xylem and subsequently affects the distribution of Si in the leaf.
INTRODUCTION
Silicon (Si) is the second most abundant element in the Earth's crust and soil. It has been considered to be a “quasi-essential element for plant growth” (Epstein and Bloom, 2005). Si functions to protect plants from various biotic and abiotic stresses (Epstein, 1999; Ma, 2004; Ma and Yamaji, 2006). For example, Si increases rice (Oryza sativa) resistance to leaf and neck blast, sheath blight, brown spot, leaf scald, and stem rot (Datnoff and Rodrigues, 2005) and decreases the incidence of powdery mildew in several plants (Fauteux et al., 2005, 2006). Si also reduces lodging of rice and alleviates the adverse impacts of drought and nutrient imbalance (Ma, 2004).
Although all plants contain Si, there is a wide variation in Si accumulation between species, ranging from 0.1% to 10% of aboveground dry weight (Epstein, 1999; Ma and Takahashi, 2002). This range is attributed to differences in the abilities of roots to take up Si (Takahashi et al., 1990). Rice is able to accumulate Si to up to 10% of shoot dry weight, which is often in the range of or even higher than the levels of essential macronutrients such as nitrogen, phosphate, and potassium (Ma and Takahashi, 2002). A large amount of Si is required for high and sustainable rice production (Savant et al., 1997). Recently, two transporters (Lsi1 and Lsi2) responsible for the high capacity of rice for Si uptake have been identified (Ma et al., 2006, 2007a). Lsi1 belongs to the nodulin-26 intrinsic protein III (NIP III) subgroup of aquaporins and is an influx transporter of silicic acid, while Lsi2 is an active efflux transporter of silicic acid. Both Lsi1 and Lsi2 are localized in the root exodermis and endodermis, but Lsi1 is on the distal side and Lsi2 is on the proximal side (Ma et al., 2006, 2007a). Lsi2 actively transports Si into the stele and thereby maintains a low Si concentration in exodermis and endodermis cells. The resulting concentration gradient between endodermis and exodermis on the one hand and cortex and soil solution on the other hand drives Si influx through Lsi1.
Si transported via Lsi1 and Lsi2 into the stele is then translocated to the shoot by transpirational flow through the xylem. Si is present in the xylem at a high concentration but in the form of monosilicic acid (Casey et al., 2003; Mitani et al., 2005), and >90% of Si taken up by the roots is translocated to the shoots (Ma and Takahashi, 2002). Finally, Si is deposited into the cell wall materials as a polymer of hydrated amorphous silica, forming silica-cuticle double layers, and also deposited in specific shoot cells as silica bodies (Ma and Takahashi, 2002; Prychid et al., 2004). Si must be transported out of the xylem for the deposition; however, the transporters involved in this process are unknown. Here, we describe a transporter, Lsi6, which is responsible for the transport of silicic acid from the xylem into xylem parenchyma cells, thereby having influence on the subsequent Si distribution in rice shoots.
RESULTS
Expression Pattern of Lsi6
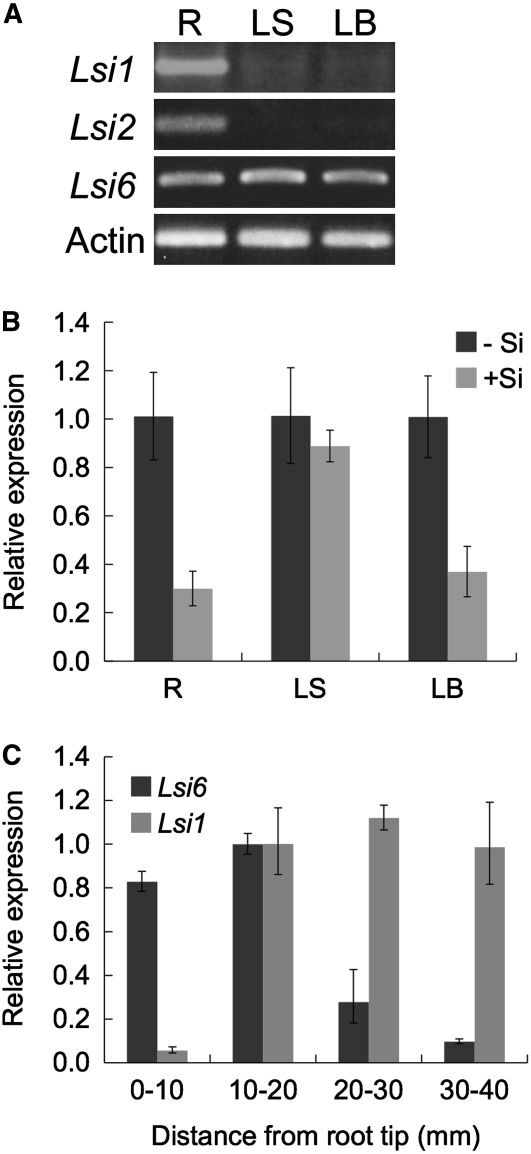
Based on homology research, there was a close homolog (Os NIP2;2; named Lsi6 in this article) of Si influx transporter Lsi1 in the rice genome. We isolated this gene from cDNA of rice roots by PCR. Lsi6 consisted of five exons and four introns with an 894-bp open reading frame that encoded a protein of 298 amino acids. The protein shared 77% identity with Lsi1 (see Supplemental Figure 1 online). Like Lsi1, Lsi6 contained two conserved NPA motifs and four residues for ar/R selectivity filter. However, when we examined the expression of Lsi6 in different tissues, we found that, in contrast with Lsi1 and Lsi2, which were expressed only in the roots, Lsi6 was also expressed in the leaf sheaths and leaf blades (Figure 1A). Interestingly, the expression level was decreased by the presence of a supply of Si in the roots and leaf blades but not in the leaf sheath (Figure 1B). Furthermore, in the roots, more expression was found in the immature region (0 to 20 mm) of the root tip, containing the root apical meristem and elongation zone (Figure 1C). This expression pattern was different from Lsi1 and Lsi2, which are expressed in the mature region above 10 mm from the root tip (Figure 1C; Ma et al., 2007a; Yamaji and Ma, 2007). The difference in the expression patterns suggests that Lsi6 plays a role different from Lsi1 in Si transport.
Figure 1.
Lsi6 Gene Expression.
(A) Transcripts of Lsi1, Lsi2, and Lsi6 were detected by RT-PCR in wild-type rice cv Nipponbare. Total RNA was isolated from the root (R), leaf sheath (LS), and leaf blade (LB) of 4-week-old seedlings grown hydroponically. Actin was used as an internal standard.
(B) Effect of Si supply on Lsi6 expression. Four-week-old seedlings (cv Nipponbare) grown hydroponically were treated with or without 0.5 mM Si for 6 d, and then relative expression levels of Lsi6 in the root, leaf sheath, and leaf blade were compared by quantitative RT-PCR. Means ± sd of biological replicates (n = 3) are shown.
(C) Lsi6 and Lsi1 expression in the root. The primary roots of hydroponically grown 5-d-old rice seedlings (cv Nipponbare) were used. The region 0 to 40 mm from the root tips was separated into four segments as indicated in the figure, and the transcript levels were quantified by RT-PCR. Means ± sd of biological replicates (n = 3) are shown.
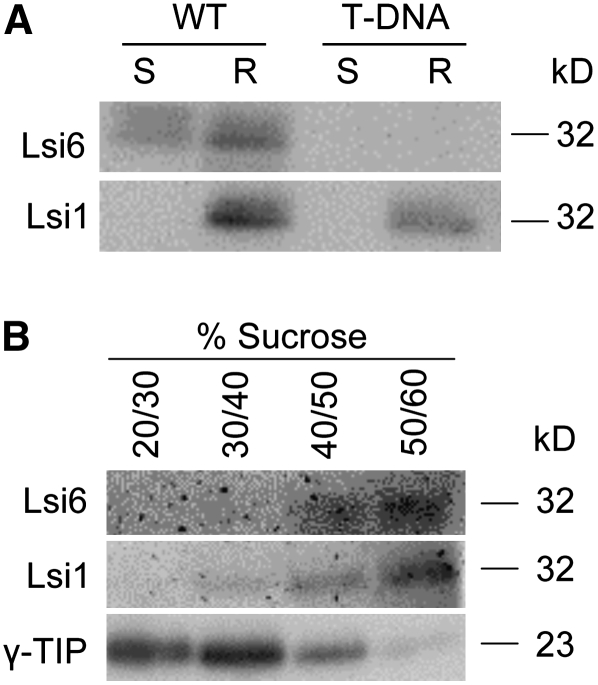
Protein gel blot analysis of membrane fractions also showed that Lsi6 protein was expressed in both the shoots and roots, in contrast with Lsi1, which was expressed only in the roots (Figure 2A).
Figure 2.
Protein Gel Blot Analysis.
(A) Expression of Lsi6 protein in the roots and shoots. Membrane fractions were isolated from the shoots (S) and roots (R) of wild-type rice (cv Dongjin) and the Lsi6 T-DNA knockdown line. SDS-PAGE and protein gel blot analyses were conducted using anti-Lsi6 and anti-Lsi1 antibodies.
(B) Subcellular localization of Lsi6 protein. Membrane fractions of the wild-type root (cv Dongjin) were separated by sucrose gradient centrifugation as indicated, and each fraction was subjected to SDS-PAGE and protein gel blot analysis. Anti-Lsi1 and anti-γ-TIP antibodies were used as markers of the plasma membrane and tonoplast, respectively.
Subcellular Localization and Tissue Specificity
To investigate the localization of Lsi6, we raised an antibody against it. The specificity of the antibody was tested in an Lsi6 knockdown T-DNA insertion line. Since T-DNA was inserted into the second intron of Lsi6 in this line (see Supplemental Figure 2A online), full-length mRNA was still detected by RT-PCR, but the expression level was reduced to a few percent of that in wild-type rice (see Supplemental Figure 2B online). Our antibody generated a signal only in the wild type but not in the knockdown line (Figure 2A), indicating that this antibody was specific for Lsi6. In addition, there was no signal detected for Lsi1 in the shoots using either that antibody or an antibody against Lsi1 (Figure 2A), showing that there was no cross-reactivity between Lsi1 and Lsi6.
Fractionation of the root microsomal membranes with sucrose density gradients showed that Lsi6 was detected in the lower fraction (mainly at the 50/60% sucrose boundary) as was Lsi1 (Figure 2B), whereas tonoplast intrinsic protein (γ-TIP) was detected in upper fractions. Lsi1 is a plasma membrane protein (Ma et al., 2006), while γ-TIP is a tonoplast intrinsic protein (Maeshima, 1992). These results suggest that Lsi6 protein was localized at the plasma membrane like Lsi1 but not in the tonoplast.
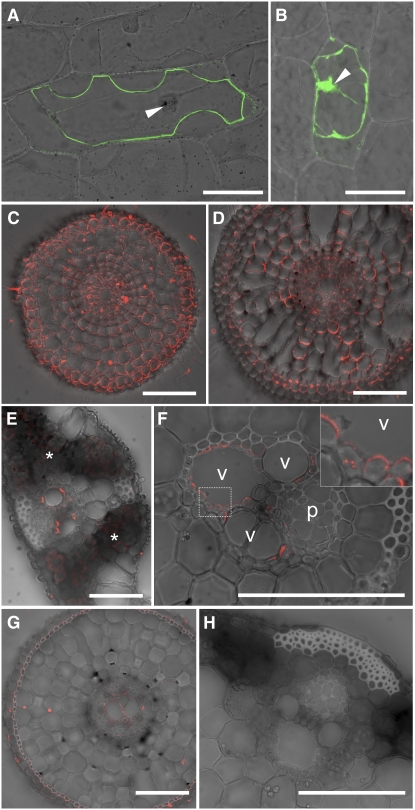
We further investigated the subcellular localization of Lsi6 by delivering a translational fusion between green fluorescent protein (GFP) and Lsi6 into onion epidermal cells by particle bombardment. Cells expressing the GFP-Lsi6 fusion showed GFP fluorescence only in the plasma membrane (outside of the nucleus), whereas the signal for cells expressing GFP alone was found in the nucleus and cytosol (Figures 3A and 3B).
Figure 3.
Subcellular and Tissue-Specific Localization of Lsi6 Protein.
(A) and (B) Transient expression of a GFP-Lsi6 fusion (A) and GFP alone as control (B) introduced by particle bombardment into onion epidermal cells. The fluorescence was observed after plasmolysis of cells with 1 M mannitol. Merged images of confocal GFP fluorescence and bright field are shown. Arrowheads indicate the nuclei.
(C) to (F) Immunostaining of Lsi6 protein in wild-type rice. Seminal root cross sections 5 and 30 mm from the tip ([C] and [D], respectively), leaf blade (E), and leaf sheath (F) with an enlarged image of xylem parenchyma (inset, corresponding to the frame in [F]) are shown. Fluorescence from secondary antibodies labeled with Alexa Fluor 555 is shown in red. (C) and (D) were stained and observed under identical conditions on the same slide. Asterisks in (E) indicate weak autofluorescence from chloroplasts in mesophyll cells. Vessel and phloem are indicated as v and p, respectively, in (F).
(G) and (H) Anti-Lsi6 antibody immunostaining in the Lsi6 T-DNA insertion line. A root cross section at 30 mm from the tip (G) and a leaf sheath (H) are shown.
Bars = 100 μm.
To investigate the localization of Lsi6 in rice tissues, we probed the roots, leaf sheaths, and leaf blades with the anti-Lsi6 antibody. In the roots, Lsi6 was localized in the plasma membrane of all cells of the young region (Figure 3C); however, the expression was markedly decreased in the mature region (Figure 3D), which was consistent with the gene expression levels measured earlier (Figure 1C). Furthermore, Lsi6 showed higher abundance at the distal side of each cell than the proximal side, indicating a polar localization similar to that of Lsi1 (Figures 3C and 3D). In the shoot, Lsi6 protein was observed only in the xylem parenchyma cells that were adjacent to vessels in both leaf sheaths and leaf blades (Figures 3E and 3F). Lsi6 in the shoots also exhibited polar localization on the side facing toward the vessel (Figures 3E and 3F). No signal was observed in the T-DNA insertion line (Figures 3G and 3H), further confirming the specificity of the anti-Lsi6 antibody.
Role of Lsi6 in Si Uptake
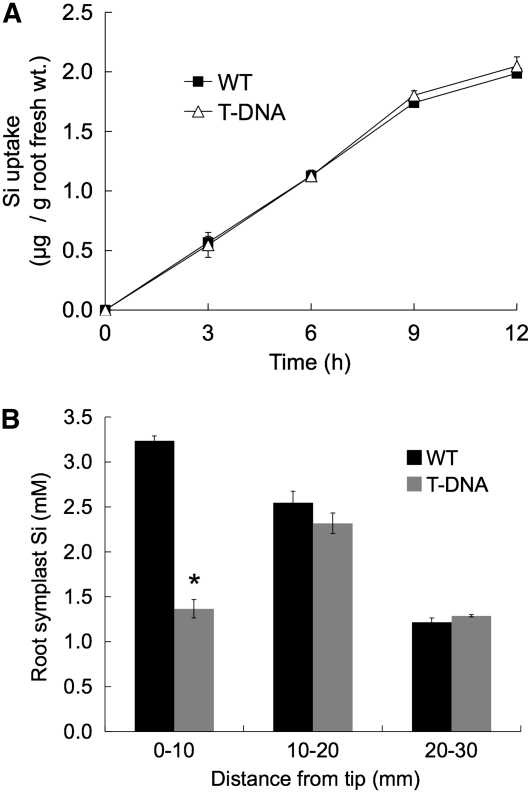
To understand the role of Lsi6 in the rice plant, we compared the Si uptake by the roots of wild-type rice to that of the T-DNA insertion line. A time-course experiment up to 12 h showed that Si uptake increased linearly with time, but there was no difference between the Si uptake levels of the wild-type and the T-DNA line (Figure 4A). Because Lsi6 was mainly expressed in the root tips, we then compared the Si concentrations in the root symplastic solution of different root segments. The concentration of Si in the symplast of root segments at 0 to 10 mm from the tip was significantly (P < 0.01) higher in the wild-type rice than in the T-DNA line (Figure 4B), but there was no difference in the Si concentration of mature segments (10 to 20 mm and 20 to 30 mm) between the two lines. Although Lsi6 was also expressed in the 10- to 20-mm segments, the expression of Lsi1 was much higher, and this probably resulted in no difference in the symplastic Si concentration. These data indicate that although Lsi6 functions as a transporter for Si uptake, its contribution to the whole root uptake is small.
Figure 4.
Effect of the lsi6 Mutation on Rice Si Uptake and Accumulation.
The T-DNA insertion mutant of lsi6 (T-DNA) and wild-type rice (cv Dongjin) were compared.
(A) Si uptake by individual plants. Three-week-old rice plants grown hydroponically were used. Si uptake from medium containing 0.5 mM Si was measured for 12 h. Means ± sd of biological replicates (n = 3) are shown.
(B) Root symplastic Si concentration. Cell sap of primary root segments was collected from 4-d-old seedlings treated with 0.5 mM Si for 6 h, and Si concentrations in the symplastic solution were measured. Means ± sd of biological replicates (n = 3) are shown. An asterisk above a bar indicates a significant difference (P < 0.01) between the wild type and the T-DNA by the Student's t test.
Role of Lsi6 in Regulating Si Distribution in Rice Shoots
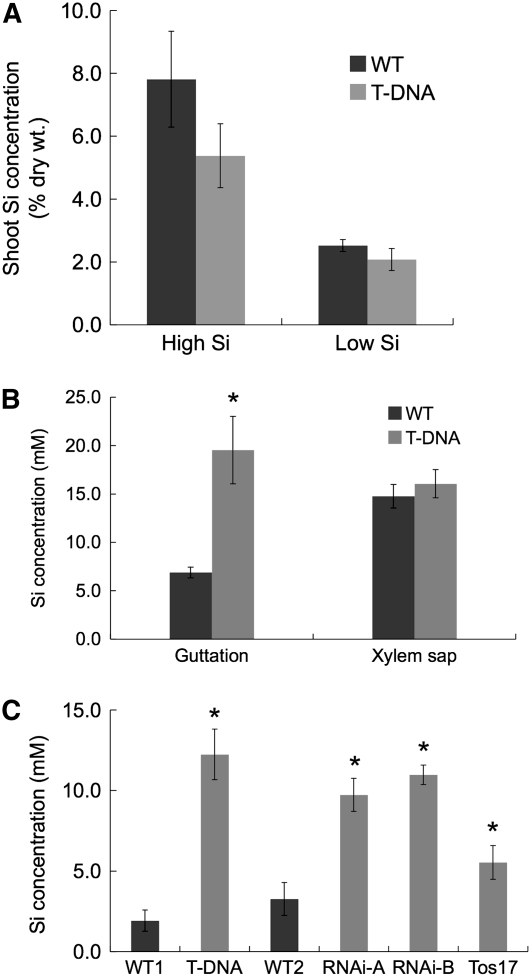
To further understand the role of Lsi6 in rice, we compared the shoot Si concentration between the wild-type rice and the T-DNA line. Unexpectedly, the shoot Si concentration in the wild type was slightly higher than that in the T-DNA line after both lines were grown in a soil amended with or without silica fertilizer (High Si and Low Si, Figure 5A). Since there was no difference in the Si uptake by the roots between these two lines (Figure 4A), the lower shoot concentration in the T-DNA line puzzled us with regard to the fate of Si translocated from the roots. Rice is known to produce guttation fluid during the night, a phenomenon in which positive pressure causes xylem sap exudation through hydathodes, which are located near terminal tracheids of the bundle ends around the margins of leaves (see Supplemental Figure 3 online; Taiz and Zeiger, 2006). This led us to suppose that Si from the xylem might be lost through exudation to guttation fluid in the T-DNA line. We therefore determined the Si concentration in the guttation fluid. Although there was no difference in the Si concentration of xylem sap between the two lines, the Si concentration in the guttation fluid was more than three times higher in the T-DNA line than that in the wild-type rice (Figure 5B). To confirm this result, we produced knockdown lines using RNA interference (RNAi) and obtained a Tos17 insertion line (see Supplemental Figure 2A online). In the T2 homozygous plants of two independent RNAi lines and the Tos17 insertion line, expression of Lsi6 was suppressed to ∼10 to 15%, and 35%, respectively (see Supplemental Figure 2B online), of that of the wild-type rice (cv Nipponbare). Similar to the T-DNA line, the RNAi and Tos17 lines exhibited a significant (P < 0.05) increase of Si concentration in the guttation fluid (Figure 5C). These results indicated that suppression of Lsi6 in these lines changed the pathways of Si from the xylem sap to the leaf cells, resulting in enhanced exudation of Si into the guttation fluid.
Figure 5.
Shoot Si Concentration and Si Concentration in the Leaf Guttation Fluid and Xylem Sap.
(A) Shoot Si concentration. Two-week-old seedlings of wild-type rice (cv Dongjin) and the Lsi6 T-DNA line grown hydroponically without Si were transferred to 1.2-liter plastic pots containing 1 kg of soil with or without silica fertilizer (high and low Si, respectively). Three weeks later, the aboveground parts were harvested to measure Si concentration. Means ± sd of biological replicates (n = 3) are shown.
(B) and (C) Si concentration in the leaf guttation fluid and xylem sap.
(B) Four-week-old seedlings of wild-type rice (cv Dongjin) and the T-DNA insertion mutant (T-DNA) were cultured on hydroponic medium with 0.5 mM Si in the greenhouse under natural right. Guttation fluid was collected just after sunset and then xylem sap was collected from the cut end of the shoot basal region for Si measurement. Means ± sd of biological replicates (n = 4) are shown.
(C) Four-week-old seedlings of the wild type (WT1, cv Dongjin; WT2, cv Nipponbare), the Lsi6 T-DNA line (T-DNA), two independent lines of Lsi6 RNAi (RNAi-A and B), and a Tos17 insertion mutant (Tos17) were cultured on soil in pots in the greenhouse under natural right. Guttation fluid was collected just after sunset, and Si concentrations were measured. Means ± sd of three biological replicates are shown. Asterisks above a bar indicate a significant difference (P < 0.05) between the knockdown lines and the corresponding wild types by the Student's t test.
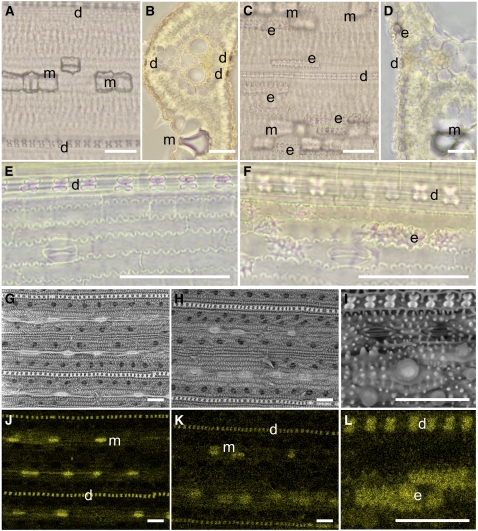
Si from the xylem is ultimately deposited in leaf blades and sheaths. There are two types of silicified cells in rice leaf blades; silica cells, and silica bodies or silicified motor cells (Ma and Takahashi, 2002). Silica cells are located in the epidermis above the veins and are dumbbell-like in shape, while silica bodies, known as plant opal or phytoliths, are in bulliform motor cells of rice leaves. We compared the leaf cell silicification patterns between the wild-type rice and the T-DNA line using a phenol-safranin staining method. In wild-type rice leaf blades, silicified dumbbell shape and motor cells were observed to be arrayed in an orderly fashion parallel to leaf veins (Figures 6A and 6B). However, in the T-DNA line, although silicified dumbbell shape and motor cells were observed, a fraction of abaxial epidermal cells were also silicified, an infrequent occurrence in the wild-type leaf blades (Figures 6C and 6D). Silicified epidermal cells were also frequently observed in leaf sheaths from the T-DNA line but not from the wild-type rice (Figures 6E and 6F). Similar results were obtained using energy-dispersive x-ray (EDX) elemental analysis combined with scanning electron microscopy (Figures 6G to 6L). Abaxial epidermis containing a high density of Si was frequently observed in the T-DNA line (Figures 6K and 6L). Furthermore, the signal density from Si in the dumbbell shape and the motor cells in the T-DNA line was evidently weaker than that of the wild-type rice.
Figure 6.
Silica Distribution in the Leaf Blade.
(A) to (F) Phenol-safranin staining of leaf blade ([A] to [D]) and leaf sheath ([E] and [F]) of wild-type cv Dongjin ([A], [B], and [E]) and the Lsi6 T-DNA insertion mutant ([C], [D], and [F]) after 0.5 mM Si treatment for 6 d. Longitudinal ([A], [C], [E], and [F]) and cross sections ([B] and [D]) were observed by optical microscopy.
(G) to (L) Images from scanning electron microscopy coupled with EDX spectroscopy. Leaf blade of cv Dongjin ([G] and [J]) and T-DNA line ([H], [I], [K], and [L]) treated with 0.5 mM Si for 6 d were observed by scanning electron microscopy ([G] to [I]) and simultaneously detected with EDX of Si ([J] to [L]). Yellow color in (J) to (L) indicates the characteristic x-rays from Si.
d, silicified dumbbell-shaped cell; m, silicified motor cell; e, silicified abaxial epidermis. Bars = 50 μm.
DISCUSSION
Lsi6 Is a Si Transporter Localized in the Shoot
Lsi6, which was identified in this study, is permeable to silicic acid like the Si influx transporter Lsi1 (Mitani et al., 2008). Moreover, both Lsi1 and Lsi6 are localized in the plasma membrane (Figures 2B and 3A; Ma et al., 2006), indicating that, similar to Lsi1, Lsi6 functions as a plasma membrane Si transporter. However, the expression pattern and localization of Lsi6 is different from that of Lsi1. Lsi1 is expressed in the roots but not in the shoots (Figures 1A and 2A). By contrast, Lsi6 is expressed both in the roots and shoots (Figures 1A, 1B, and 2A). Furthermore, Lsi1 expression is higher in the mature region of the roots, but higher expression of Lsi6 was found in the immature region near the root tip (Figure 1C). Immunostaining showed that Lsi1 is localized in the exodermis and endodermis cells (Ma et al., 2006; Yamaji and Ma, 2007), where Casparian strips have been developed, but that Lsi6 is present in all cells in the root immature regions where Casparian strips have not yet developed (Figure 3C). These spatial differences in the localization suggest that Lsi6 and Lsi1 play different roles in Si uptake. A significant decrease in the symplast Si concentration of the 0- to 10-mm root tip region in the Lsi6 knockdown line indicates that Lsi6 contributed to the Si uptake in that region (Figure 4B). However, the contribution of Lsi6 to overall uptake is trivial, as demonstrated by the fact that there was no difference in the Si uptake between the wild-type rice and the T-DNA line (Figure 4A). This result is consistent with a previous study showing that the uptake site of Si is located in the root mature region (Yamaji and Ma, 2007). The noncontribution of Lsi6 to the Si uptake might be attributed to the lack of the efflux transporter Lsi2 in the root tip regions. Like Lsi1, Lsi2 is highly expressed in the exodermis and endodermis cells of the root mature regions. Different from Lsi1 and Lsi6, Lsi2 is an energy-dependent active transporter (Ma et al., 2007a). Therefore, Si transported into the cells by Lsi1 is exported by Lsi2, and the efficient coupling of Lsi1 and Lsi2 plays a major role in Si uptake in rice (Ma et al., 2006, 2007a; Yamaji and Ma, 2007). In fact, the loss of either Lsi1 or Lsi2 results in a significant decrease of Si uptake by rice roots (Ma et al., 2002). It is possible that Si transported by Lsi6 might not be translocated to the shoots and might remain in the cells of root tip region for protecting the roots from various stress, although the exact mechanisms remain to be examined in future.
Lsi6 Is Involved in the Si Distribution from Xylem to Leaf Tissues
The outstanding feature of Lsi6 is its expression in the leaf sheaths and leaf blades (Figures 1A and 1B). Immnuostaining revealed that Lsi6 is localized in the xylem parenchyma cells in the leaf blades and sheaths (Figures 3E and 3F). Furthermore, Lsi6 exhibits polar localization on the side facing toward the vessel (Figures 3E and 3F). This result suggests that Lsi6 plays a role in the unloading of Si from xylem to leaf tissues. Si in the xylem sap is present in the form of monosilicic acid (Mitani et al., 2005). Because silicic acid is a noncharged small molecule, it is assumed that silicic acid can diffuse across the plasma membrane. However, to avoid undesirable water loss, leaf xylem parenchyma cells are armed with an apoplastic barrier (Chonan et al., 1984), similar to the Casparian strip in the roots. The polar localization of Lsi6 in the xylem parenchyma cells indicates that Lsi6 is responsible for the delivery of Si from the xylem to the leaf cells. This is supported by the fact that more Si is excreted into the guttation fluid in the T-DNA insertion line, RNAi knockdown lines, and Tos17 insertion line (Figure 5).
The Leaf Si Deposition Pattern Is Altered by Lsi6
In the leaf, silicic acid is concentrated by transpirational water loss and deposited as amorphous silica in the specific cells after polymerization (Ma and Takahashi, 2002; Figure 6). The deposition of Si enhances the strength and rigidity of the leaves and thus increases the resistance of plants to various stresses. In this study, we found that Lsi6 affects the silica deposition pattern in the leaf blades and sheaths (Figure 6). Safranin staining and EDX analysis combined with scanning electron microscopy of the mutant leaves indicated that the density of silicified dumbbell shape and motor cells was decreased compared with the wild-type rice. Furthermore, abaxial epidermis cells were observed to be silicified in the mutant but infrequently in the wild-type rice (Figure 6). These results suggest that suppression of Lsi6 resulted in alteration of the Si transport to specific cells. Therefore, cell type–specific silicification depends on the symplastic pathway of Si delivered by Lsi6 rather than on the apoplastic pathway. The subsequent mechanisms of silicification and whether other transporters are involved remain to be examined in future.
In conclusion, Lsi6 is identified as a Si transporter and is shown to be mainly responsible for the transport of Si out of the xylem into the leaf tissues. Lsi6 is also required for wild-type patterns of deposition of Si in the leaves.
METHODS
Plant Materials and Growth Conditions
Wild-type rice (Oryza sativa cv Dongjin and cv Nipponbare), the T2 homozygous progeny of T-DNA insertion line 4A-01373 (POSTECH RISD) (Jeong et al., 2006), the T2 homozygous progeny of Tos17 insertion line NG0012 (Miyao et al., 2003), and the RNAi line described below were cultured hydroponically as described previously (Yamaji and Ma, 2007). Seeds were soaked in water overnight at 25°C in the dark and then transferred to a 0.5 mM CaCl2 solution. On day 7, seedlings were transferred to a 3.5-liter plastic pot containing half-strength Kimura B solution and grown in a closed greenhouse at 22 to 25°C. The nutrient solution was renewed every 2 d. For soil culture, 3- to 4-week-old seedlings grown hydroponically were transplanted to a 3.5-liter plastic pot filled with paddy soil amended with or without silica fertilizer (50 g per pot) as previously described (Ma et al., 2007b). Tap water was supplied daily.
To generate the RNAi suppression line of Lsi6, we amplified a 355-bp fragment (1168 to 1522 bases from transcriptional start) of the 3′ untranslated region of Lsi6 cDNA by PCR using a pair of primers (forward, 5′-AAAAAGCAGGCTTTGGGAAGCCATTGTTTGTACG-3′, and reverse, 5′-AGAAAGCTGGGTTCAAGCAAAGCTCAGCTCCTT-3′). We then cloned two copies of the fragment into the pANDA vector as inverted repeats according to Miki and Shimamoto (2004) and subsequently introduced that construct into rice (cv Nipponbare) using an Agrobacterium tumefaciens–mediated transformation system as described previously (Ma et al., 2006).
Cloning of Lsi6
Rice roots grown hydroponically (6 d old) were harvested and ground in liquid nitrogen and immediately subjected to RNA extraction with the RNeasy plant mini kit (Qiagen). Total RNA (1 μg) was reverse transcribed into cDNA using the SuperScript first-strand synthesis system (Invitrogen). The open reading frame of Lsi6 was amplified by PCR using high-fidelity KOD plus DNA polymerase (Toyobo) from the cDNA. A gene-specific primer pair (forward, 5′-ATGTACAAGGGGGCCATGGCATCGACG-3′, and reverse, 5′-AGGATCCATGGCGTCAGGGCGATTAG-3′) was used. Sequence of the amplified gene was confirmed by the ABI PRISM 310 genetic analyzer (Applied Biosystems) using the BigDye Terminators V3.1 cycle sequencing kit.
Quantitative Real-Time RT-PCR
Total RNA was extracted from the roots, leaf blades, and leaf sheaths and relative transcript levels of Ls6, Lsi1, Lsi2, and Actin (internal control) were determined by quantitative real-time RT-PCR as described previously (Yamaji and Ma, 2007). Quantitative real-time PCR was performed in a 20-μL reaction volume containing 2 μL of 1:5 diluted cDNA, 200 nM each gene-specific primers, and SYBR Premix Ex Taq (Takara Bio) using Applied Biosystems 7500. Primer sequences used are as follows: Lsi6, 5′-GAGTTCGACAACGTCTAATCGC-3′ (forward) and 5′-AGTACACGGTACATGTATACACG-3′ (reverse); Lsi1, 5′-CGGTGGATGTGATCGGAACCA-3′ (forward) and 5′-CGTCGAACTTGTTGCTCGCCA-3′ (reverse); Lsi2, 5′-ATCTGGGACTTCATGGCCC-3′ (forward) and 5′-ACGTTTGATGCGAGGTTGG-3′ (reverse); Actin, 5′-GACTCTGGTGATGGTGTCAGC-3′ (forward) and 5′-GGCTGGAAGAGGACCTCAGG-3′ (reverse).
Preparation of Membrane Proteins
Membrane proteins were prepared from rice roots and shoots basically according to the methods of Sugiyama et al. (2007). The seedlings (20 d old) of both wild-type rice (cv Dongjin) and the T-DNA insertion line were harvested and then homogenized in 30 mL of ice-cold homogenizing buffer comprising 100 mM Tris-HCl, pH 8.0, 150 mM KCl, 0.5% (w/v) polyvinylpolypyrrolidone, 5 mM EDTA, 3.3 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and 10% (v/v) glycerol. After filtration, the homogenates were centrifuged at 8000g for 10 min to yield the supernatant and centrifuged again under the same conditions. The supernatants were then ultracentrifuged at 100,000g for 30 min. The pellets were resuspended in a small volume of resuspension buffer containing 10 mM Tris-HCl, pH 7.6, 10% (v/v) glycerol, and 1 mM EDTA and fractionated thorough a 20, 30, 40, 50, and 60% (w/v) discontinuous sucrose gradient with 10 mM Tris-HCl, pH 7.6, 1 mM EDTA, and 1 mM DTT by ultracentrifugation at 100,000g for 120 min. The fractionated membranes were recovered by ultracentrifugation at 100,000g for 30 min. Each pellet was resuspended in resuspension buffer supplemented with 1/100 volume of Protease Inhibitor Cocktail for plant cell and tissue extracts (Sigma-Aldrich) and 1 mM DTT.
Lsi6 Antibody Generation and Protein Gel Blot Analysis
The synthetic peptide C-SMAADEFDNV (positions 289 to 298 of Lsi6) was used to immunize rabbits to obtain antibodies against Lsi6. Antiserum obtained was purified through the peptide affinity column before use.
Protein concentrations were measured by the Bradford method (Bio-Rad), and equal amounts of samples were mixed with same volume of sample buffer containing 250 mM Tris-HCl, pH 6.8, 8% (w/v) SDS, 40% (w/v) glycerol, 0.01% (w/v) bromophenol blue, and 200 mM β-mercaptoethanol. The mixture was allowed to incubate at 37°C for 10 min (65°C for 10 min for γ-TIP), and SDS-PAGE was performed using 11% polyacrylamide gels containing 0.1% SDS. The transfer to polyvinylidene difluoride membrane was performed with a semidry blotting system, and the membrane was treated with the purified primary rabbit anti-Lsi6, anti-Lsi1 (Ma et al., 2006), and anti-γ-TIP (Maeshima, 1992) polyclonal antibodies diluted at 1:100, 1:100, and 1:1000, respectively. Anti rabbit IgG horseradish peroxidase conjugate (1:10,000 dilution; GE Healthcare) was used as a secondary antibody, and ECL plus (GE Healthcare) was used for detection via chemiluminescence.
Transient Expression of GFP Fusion
The open reading frame of Lsi6 was amplified by gene-specific primers 5′-ATGTACAAGGGGGCCATGGCATCGACG-3′ and 5′-AGGATCCATGGCGTCAGGGCGATTAG-3′ using high-fidelity KOD plus DNA polymerase (Toyobo) to create BsrGI and BamHI sites on the ends. The fragment was treated with restriction enzymes and inserted in frame between the GFP coding sequence and NOS terminator under the control of the 35S promoter in pBluescript SK− vector. The fused gene was transiently introduced into the onion epidermal cells by particle bombardment as described previously (Ma et al., 2007a).
Immunostaining of Lsi6 Protein
The root, leaf sheath, and leaf blade of both wild-type rice and the T-DNA line were used for immunostaining of Lsi6 protein with 1:300 diluted anti-Lsi6 antibody as described previously (Yamaji and Ma, 2007). Fluorescence from the secondary antibody (Alexa Fluor 555 goat anti-rabbit IgG; Molecular Probes) was observed with a fluorescence microscope (Axio Imager with Apotome; Carl Zeiss).
Uptake Experiments
Si uptake by wild-type rice (cv Dongjin) and the T2 homozygous plants of the T-DNA insertion mutant of lsi6 were examined during short-term (up to 12 h) experiments. Two seedlings each (20 d old) were placed in 50-mL black bottles containing half-strength Kimura B solution, pH 5.6, with 0.5 mM Si as silicic acid. At various time points, a 1-mL aliquot of uptake solution was taken for determination of Si concentration (see below). Transpiration (water loss) was also measured by recording the total weight change at each sampling time. At the designated time point, the roots were harvested and their fresh weights were recorded.
Determination of Si in Root Symplastic Solution
Root symplastic solutions were collected as described previously (Mitani and Ma, 2005). Briefly, the root segments (0 to 10, 10 to 20, and 20 to 30 mm from the tip) were excised from 5-d-old seedlings of wild-type rice (cv Dongjin) and the T-DNA insertion line treated with 0.5 mM Si at 25°C for 6 h. For each sample, 40 roots were used. The cut segments were placed in a 0.22-μm filter unit (ULTRAFREER-MC; Millipore) with the cut ends facing down and centrifuged at 2000g for 15 min at 4°C to eliminate the apoplastic solution. After centrifugation, root segments were frozen at −80°C for 2 h and then thawed at room temperature. The symplastic solution was prepared from the frozen-thawed segments by centrifugation at 2000g for 15 min at 4°C and used for Si determination as described below.
Collection of Xylem Sap and Guttation Fluid
Four-week-old seedlings of wild-type rice (cv Dongjin and Nipponbare), T2 homozygous progeny of T-DNA and Tos17 insertion mutants and T2 progenies of two independent lines of Lsi6 RNAi were cultured on hydroponic medium with 0.5 mM Si or in 3-liter soil pots in the greenhouse under natural right. Leaf guttation fluid was collected with a micropipet just after sunset in the greenhouse. Xylem sap was collected from the cut end for 30 min with a micropipet after decapitating the plant 5 cm above the roots. Collected leaf guttation fluid and xylem sap were immediately diluted with distilled water and used for Si measurements.
Determination of Si
Plant samples harvested were dried at 70°C in an oven for at least 2 d and then ground to a powder. The samples were then digested in a mixture of 3 mL of HNO3 (62%), 3 mL of hydrogen peroxide (30%), and 2 mL of hydrofluoric acid (46%) by microwaving, and the digested samples were diluted to 100 mL with 4% boric acid. The Si concentration in the digest solution, uptake medium, xylem sap, and guttation fluid was determined by the colorimetric molybdenum blue method at 600 nm as described earlier (Ma et al., 2007b).
Detection of Silicified Cells
Rice leaves were decolorized with 70% ethanol at room temperature for 3 d and subsequently stained in boiled phenol containing 0.001% safranin for 5 min (Ma et al., 2001). Photographs of stained leafs were taken under optical microscopy.
Scanning electron microscopy and Si elementary analysis of the leaf blades were performed using scanning electron microscopy coupled with EDX spectroscopy (S-3400NX; Hitachi-hitec) (Takahashi et al., 2006).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL/DDBJ data libraries under accession numbers AB253627 (Lsi6), AB222272 (Lsi1), AB222273 (Lsi2), and X16280 (actin).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of the Amino Acid Sequences of Lsi1 and Lsi6.
Supplemental Figure 2. Lsi6 Gene Structure and Expression in Suppression Lines.
Supplemental Figure 3. Rice Leaf Guttation.
Supplementary Material
Acknowledgments
This research was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation IPG-0006 to J.F.M.) and a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (No. 17078008 to J.F.M.). We thank the Rice Genome Resource Center for providing Tos17 seeds. We also thank M. Maeshima for providing the anti-γ-TIP antibody.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jian Feng Ma (maj@rib.okayama-u.ac.jp).
Online version contains Web-only data.
References
- Casey, W.H., Kinrade, S.D., Knight, C.T.G., Rains, D.W., and Epstein, E. (2003). Aqueous silicate complexes in wheat, Triticum aestivum L. Plant Cell Environ. 27 51–54. [Google Scholar]
- Chonan, N., Kawahara, H., and Matsuda, T. (1984). Ultrastructure of vascular bundles and fundamental parenchyma in relation to movement of photosynthate in leaf sheath of rice. Jpn. J. Crop. Sci. 53 435–444. [Google Scholar]
- Datnoff, L.E., and Rodrigues, F.A. (2005). The role of silicon in suppressing rice diseases. APSnet, http://apsnet.org/online/feature/silicon/.
- Epstein, E. (1999). Silicon. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 641–664. [DOI] [PubMed] [Google Scholar]
- Epstein, E., and Bloom, A.J. (2005). Mineral Nutrition of Plants: Principles and Perspectives, 2nd ed. (Sunderland, MA: Sinauer Associates).
- Fauteux, F., Chain, F., Belzile, F., Menzies, J.G., and Belanger, R.R. (2006). The protective role of silicon in the Arabidopsis-powdery mildew pathosystem. Proc Natl Acad Sci U S A. 103 17554–17559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauteux, F., Remus-Borel, W., Menzies, J.G., and Belanger, R.R. (2005). Silicon and plant disease resistance against pathogenic fungi. FEMS Microbiol. Lett. 249 1–6. [DOI] [PubMed] [Google Scholar]
- Jeong, D.H., et al. (2006). Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 45 123–132. [DOI] [PubMed] [Google Scholar]
- Ma, J.F. (2004). Role of silicon in enhancing the resistance of plants to biotic and abiotic stresses. Soil Sci. Plant Nutr. 50 11–18. [Google Scholar]
- Ma, J.F., Goto, S., Tamai, K., and Ichii, M. (2001). Role of root hairs and lateral roots in silicon uptake by rice. Plant Physiol. 127 1773–1780. [PMC free article] [PubMed] [Google Scholar]
- Ma, J.F., and Takahashi, E. (2002). Soil, Fertilizer, and Plant Silicon Research in Japan. (Amsterdam: Elsevier).
- Ma, J.F., Tamai, K., Ichii, M., and Wu, G.F. (2002). A rice mutant defective in Si uptake. Plant Physiol. 130 2111–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.F., Tamai, K., Yamaji, N., Mitani, N., Konishi, S., Katsuhara, M., Ishiguro, M., Murata, Y., and Yano, M. (2006). A silicon transporter in rice. Nature 440 688–691. [DOI] [PubMed] [Google Scholar]
- Ma, J.F., and Yamaji, N. (2006). Silicon uptake and accumulation in higher plants. Trends Plant Sci. 11 392–397. [DOI] [PubMed] [Google Scholar]
- Ma, J.F., Yamaji, N., Mitani, N., Tamai, K., Konishi, S., Fujiwara, T., Katsuhara, M., and Yano, M. (2007. a). An efflux transporter of silicon in rice. Nature 448 209–212. [DOI] [PubMed] [Google Scholar]
- Ma, J.F., Yamaji, N., Tamai, K., and Mitani, N. (2007. b). Genotypic difference in silicon uptake and expression of silicon transporter genes in rice. Plant Physiol. 145 919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima, M. (1992). Characterization of the major integral protein of vacuolar membrane. Plant Physiol. 98 1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, D., and Shimamoto, K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45 490–495. [DOI] [PubMed] [Google Scholar]
- Mitani, N., and Ma, J.F. (2005). Uptake system of silicon in different plant species. J. Exp. Bot. 56 1255–1261. [DOI] [PubMed] [Google Scholar]
- Mitani, N., Ma, J.F., and Iwashita, T. (2005). Identification of the silicon form in xylem sap of rice (Oryza sativa L.). Plant Cell Physiol. 46 279–283. [DOI] [PubMed] [Google Scholar]
- Mitani, N., Yamaji, N., and Ma, J.F. (2008). Characterization of substrate specificity of a rice silicon transporter, Lsi1. Pflugers Arch. (in press). [DOI] [PubMed]
- Miyao, A., Tanaka, K., Murata, K., Sawaki, H., Takeda, S., Abe, K., Shinozuka, Y., Onosato, K., and Hirochika, H. (2003). Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prychid, C.J., Rudall, P.J., and Gregory, M. (2004). Systematics and biology of silica bodies in monocotyledons. Bot. Rev. 69 377–440. [Google Scholar]
- Savant, N.K., Snyder, G.H., and Datnoff, L.E. (1997). Silicon management and sustainable rice production. Adv. Agron. 58 151–199. [Google Scholar]
- Sugiyama, A., Shitan, N., and Yazaki, K. (2007). Involvement of a soybean ATP-binding cassette-type transporter in the secretion of genistein, a signal flavonoid in legume-Rhizobium symbiosis. Plant Physiol. 144 2000–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz, L., and Zeiger, E. (2006). Plant Physiology, 4th ed. (Sunderland, MA: Sinauer Associates).
- Takahashi, N., Kato, Y., Isogai, A., and Kurata, K. (2006). Silica distribution on the husk epidermis at different parts of the panicle in rice (Oryza sativa L.) determined by X-ray microanalysis. Plant Prod. Sci. 9 168–171. [Google Scholar]
- Takahashi, E., Ma, J.F., and Miyake, Y. (1990). The possibility of silicon as an essential element for higher plants. Comments Agric. Food Chem. 2 99–122. [Google Scholar]
- Yamaji, N., and Ma, J.F. (2007). Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol. 143 1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.