Abstract
Light absorbed by seed phytochromes of Arabidopsis thaliana modulates abscisic acid (ABA) and gibberellic acid (GA) signaling pathways at least partly via PHYTOCHROME-INTERACTING FACTOR3-LIKE5 (PIL5), a phytochrome-interacting basic helix-loop-helix transcription factor. Here, we report a new mutant, somnus (som), that germinates in darkness, independently of various light regimens. SOM encodes a nucleus-localized CCCH-type zinc finger protein. The som mutant has lower levels of ABA and elevated levels of GA due to expressional changes in ABA and GA metabolic genes. Unlike PIL5, however, SOM does not regulate the expression of GA-INSENSITIVE and REPRESSOR OF GA1 (RGA/RGA1), two DELLA genes encoding GA negative signaling components. Our in vivo analysis shows that PIL5 activates the expression of SOM by binding directly to its promoter, suggesting that PIL5 regulates ABA and GA metabolic genes partly through SOM. In agreement with these results, we also observed that the reduced germination frequency of a PIL5 overexpression line is rescued by the som mutation and that this rescue is accompanied by expressional changes in ABA and GA metabolic genes. Taken together, our results indicate that SOM is a component in the phytochrome signal transduction pathway that regulates hormone metabolic genes downstream of PIL5 during seed germination.
INTRODUCTION
The pioneering work by Borthwick et al. (1952) led to the discovery of phytochrome as a photoreceptor that promotes germination in many plant species. Arabidopsis thaliana has five phytochrome genes (PHYA to PHYE) that have shared but distinct roles in regulating seed germination, hypocotyl elongation, shade avoidance, flowering, and other light responses (Mathews, 2006). PHYA and PHYB are the major phytochromes for the regulation of seed germination (Reed et al., 1994). In particular, PHYA mediates the very low fluence response and the far-red high irradiance response of seeds (Shinomura et al., 1994, 1996; Botto et al., 1996). By contrast, PHYB mediates the red light low fluence response, which can be reversed by subsequent far-red light (Reed et al., 1993). Due to the presence of PHYB and the absence of PHYA in dry seeds, only the low fluence response promotes germination early in seed imbibition. In hydrated seeds, however, PHYA accumulates and the very low fluence response and far-red high irradiance response can regulate germination (Shinomura et al., 1994).
Phytochromes promote seed germination by converting light signals into hormonal signals. The activation of phytochromes decreases the level of abscisic acid (ABA) and increases the level of gibberellic acid (GA) (Yamaguchi et al., 1998; Ogawa et al., 2003; Seo et al., 2006; Oh et al., 2007). The light-promoted decrease in ABA is largely due to the transcriptional repression of ABA anabolic genes (ABA-DEFICIENT1 [ABA1], NINE-CIS-EPOXYCAROTENOID DEOXYGENASE6 [NCED6], and NCED9) and the transcriptional activation of an ABA catabolic gene (CYP707A2) (Seo et al., 2006; Oh et al., 2007). The significance of ABA biosynthesis in seed germination can be seen in the altered germination frequencies of ABA metabolic mutants, such as aba1, aba2, nced6, nced9, and cyp707a2 (Koornneef et al., 1982; Karssen et al., 1983; Kushiro et al., 2004; Lefebvre et al., 2006; Okamoto et al., 2006). ABA establishes and maintains seed dormancy and inhibits seed germination through various signaling components, including three positive components, ABA-INSENSITIVE3 (ABI3), ABI4, and ABI5 (Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000; Soderman et al., 2000). The light-induced decrease in ABA leads to decreased activities of these factors, causing decreased expression of ABA-inducible genes and promotion of germination.
Light also regulates the level of GA by transcriptional regulation of GA anabolic genes and a GA catabolic gene (Yamaguchi, 2008). Light increases the expression of two GA anabolic genes (GIBBERELLIN 3 β-HYDROXYLASE1 [GA3ox1] and GA3ox2) and inhibits the expression of a GA catabolic gene (GIBBERELLIN 2-OXIDASE2 [GA2ox2]), and this leads to an increase in the level of bioactive GA (Ogawa et al., 2003; Oh et al., 2006). The significance of GA metabolic genes in germination can be seen in the altered germination frequencies of GA metabolic mutants, such as ga requiring1 (ga1), ga2, ga3, ga4, ga5, and ga2ox2 (Koornneef and Van der Veen, 1980; Talon et al., 1990; Sun et al., 1992; Sun and Kamiya, 1994; Xu et al., 1995; Williams et al., 1998; Yamauchi et al., 2007). Light-induced increase in GA leads to the degradation of DELLA proteins via SCFSLY1 ubiquitin E3 ligase in conjunction with GA receptors (GA INSENSITIVE DWARF 1a [GID1a], -1b, and -1c) (Itoh et al., 2003; McGinnis et al., 2003; Dill et al., 2004; Ueguchi-Tanaka et al., 2005, 2007; Griffiths et al., 2006; Iuchi et al., 2007; Jiang and Fu, 2007). DELLA proteins are nucleus-localized negative GA signaling components that inhibit various GA responses, including seed germination, stem elongation, and floral development (Peng et al., 1997, 1999; Silverstone et al., 1998; Ikeda et al., 2001; Sun and Gubler, 2004; Tyler et al., 2004). The molecular functions of DELLA proteins are not clearly defined, but a recent report shows that one DELLA protein binds directly to DNA and regulates the expression of various genes, including GA receptor genes (GID1a and -1b), GA anabolic genes (GA20ox2 and GA3ox1), and XERICO, which encodes a ring finger ubiquitin E3 ligase that activates ABA biosynthesis (Ko et al., 2006; Zentella et al., 2007). There are five DELLA proteins in Arabidopsis (GA-INSENSITIVE [GAI], RGA, RGA LIKE1 [ RGL1], RGL2, and RGL3), which have shared but distinct roles in mediating different GA responses. Among them, GAI, RGA, and RGL2 play important roles in inhibiting seed germination (Lee et al., 2002; Cao et al., 2005; Oh et al., 2007). A triple mutant of these three DELLA genes (gai rga rgl2) germinates irrespective of light conditions, even in the absence of GA synthesis (ga1 mutant background) (Cao et al., 2005). This suggests that PHY-mediated light signaling promotes seed germination at least partly by increasing GA biosynthesis and the subsequent degradation of these three DELLA proteins.
In addition to ABA and GA metabolic genes, light also regulates ABA and GA signaling genes. The light activation of phytochromes inhibits the expression of two DELLA genes (GAI and RGA) in seeds (Oh et al., 2007). This indicates that PHY-mediated light signaling removes DELLA proteins by decreasing the expression of GAI and RGA at the transcriptional level and by activating the degradation of all DELLA proteins, including RGL2, at the protein level. PHY-mediated regulation of ABA signaling genes in seeds is less clearly understood. However, a recent microarray experiment showed that expression of ABI3 is significantly increased in a phyB mutant, suggesting that light represses ABA signaling by decreasing the expression of an ABI-positive component (Nakabayashi et al., 2005). These results suggest that light promotes seed germination by shifting the balance of the ABA and GA pathways via transcriptional regulation of hormone metabolic and signaling genes.
In seeds, PHYTOCHROME-INTERACTING FACTOR3-LIKE5 (PIL5, also known as PIF1 and bHLH015) plays a key role in converting light signals to alterations in the ABA and GA pathways. PIL5 encodes a phytochrome-interacting basic helix-loop-helix (bHLH) transcription factor that is a member of bHLH subgroup 15, which includes 14 other bHLH transcription factors, including PIF3, PIF4, and PIL6 (Toledo-Ortiz et al., 2003; Huq et al., 2004; Oh et al., 2004). Phenotypic analysis of a pil5 mutant and PIL5 overexpression lines shows that PIL5 is a negative regulator of seed germination in PHYA- and PHYB-dependent seed germination (Oh et al., 2004). The negative role of PIL5 on germination is partly due to its regulation of GA and ABA metabolism. In particular, PIL5 represses the expression of two GA anabolic genes (GA3ox1 and GA3ox2) and activates the expression of a GA catabolic gene (GA2ox2), causing a decrease in the level of bioactive GA in seeds (Oh et al., 2006). By contrast, PIL5 also activates the expression of ABA anabolic genes (ABA1, NCED6, and NCED9) and represses the expression of an ABA catabolic gene (CYP707A2), resulting in an increase in the level of ABA in seeds (Oh et al., 2007). Expression analyses show that PIL5 also activates the expression of GAI and RGA, indicating that it regulates both GA levels and GA responsiveness (Oh et al., 2007). Consistent with this interpretation, pil5 mutant seeds have an elevated level of GA, a decreased level of ABA, and an increased responsiveness to GA irrespective of light conditions. These expression patterns and genetic analyses indicate that phytochromes regulate the expression of these genes by inhibiting the function of PIL5. Inhibition of PIL5 function by phytochromes occurs at the protein level, as shown by the phytochrome-induced rapid degradation of PIL5 proteins in seeds and seedlings (Shen et al., 2005; Oh et al., 2006). Taken together, these previous results indicate that in Arabidopsis seeds, phytochromes activate the expression of hormone metabolic genes and GA signaling genes by promoting the degradation of PIL5.
Chromatin immunoprecipitation (ChIP) analyses indicate that PIL5 directly regulates the expression of GAI and RGA by binding to G-box elements on their promoters (Oh et al., 2007). However, PIL5 does not bind to the promoters of hormone metabolic genes, even though some of them contain G-box elements (Oh et al., 2007). This indicates that PIL5 regulates the expression of ABA and GA metabolic genes indirectly. To identify components involved in PHY- and PIL5-mediated light signaling processes, we isolated a novel mutant called somnus (som) that germinates in complete darkness. We report here that SOM, which encodes a nucleus-localized CCCH-type zinc finger protein whose molecular function is unknown, regulates hormone metabolic genes downstream of PIL5.
RESULTS
SOM, a CCCH-Type Zinc Finger Protein, Is a Negative Regulator of PHY-Mediated Promotion of Seed Germination
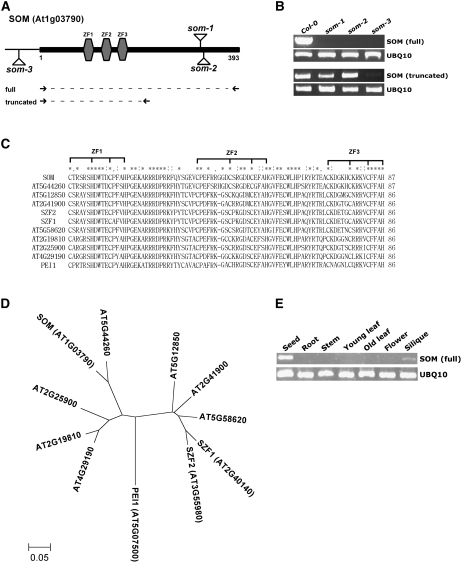
Only a few light signaling components that control seed germination have been identified. Thus, we screened Arabidopsis T-DNA insertion lines for mutants that germinate in total darkness and isolated a mutant (som-1) that germinates irrespective of light conditions. In this mutant, a T-DNA was inserted in the middle of a gene (At1g03790) that had not yet been characterized (Figure 1A). We obtained two additional som mutant alleles (som-2 and som-3) from the Arabidopsis Stock Center (SALK_090314 and SALK_008075, respectively) and included them in subsequent studies. The full-length SOM transcript was nearly undetectable by RT-PCR in three som mutants (som-1, som-2, and som-3) (Figure 1B). Unlike the full-length SOM transcript, an N-terminal fragment was expressed in two som alleles (som-1 and som-2) but was nearly undetectable in the third som allele (som-3).
Figure 1.
SOM Is a Seed-Specific Gene Encoding a Novel CCCH-Type Zinc Finger Protein.
(A) Diagram of the SOM protein, showing T-DNA insertion sites (som-1, som-2, and som-3) and three zinc finger motifs (gray hexagons). Numbers indicate amino acid numbers of SOM. Full indicates the position of a primer set amplifying the full-length SOM; truncated indicates the position of a primer set amplifying an N-terminal fragment of SOM. There are no introns in the SOM protein-coding sequence. Supplemental Figure 1 and Supplemental Table 1 online include detailed sequence information.
(B) Absence of expression of full-length SOM in the som-1, som-2, and som-3 mutants, as determined by RT-PCR. The N-terminal fragment is expressed in the som-1 and som-2 alleles, but not in som-3. Expression of UBQ10 (At4g05320) was used as an expression control.
(C) Multiple alignment of SOM and related Arabidopsis proteins for three zinc finger motifs (ZF1, ZF2, and ZF3). Asterisks indicate identical amino acid residues, and one or two dots indicate similar amino acid residues. Brackets indicate conserved Cys residues, characteristic of zinc finger motifs.
(D) Phylogenic tree of SOM and related Arabidopsis proteins.
(E) RT-PCR analysis showing that SOM is expressed mainly in seeds. Expression of UBQ10 was used as an expression control.
SOM contains a typical CCCH-type zinc finger motif (CX8CX5CX3H, ZF2 in Figure 1) that is between atypical zinc finger motifs (a CX5HX4CX3H motif at the N-terminal side and a CX5CX4CX3H motif at the C-terminal side; ZF1 and ZF3 in Figure 1). In the Arabidopsis genome, 11 proteins have three similar zinc finger motifs (Figures 1C and 1D), indicating that SOM belongs to a subset of the CCCH-type zinc finger family. Among these genes, At5g07500 (known as PEI1) has been characterized previously as a regulator of embryogenesis and At3g55980 and At2g40140 (known as SZF1 and SZF2, respectively) have been characterized as regulators of the salt response (Li and Thomas, 1998; Sun et al., 2007). The molecular function of the CCCH-type zinc finger motif is not yet clearly defined. Some CCCH-type zinc finger motifs regulate RNA stability by binding to AU-rich elements, some regulate RNA processing by binding to pre-mRNA, and some regulate transcription (Hudson et al., 2004; Barreau et al., 2005; Brown, 2005; Hall, 2005). PEI1 has been described as a DNA binding protein. Expression analysis showed that SOM is expressed mainly in seeds but not in adult organs and tissues (Figure 1E). Microarray data compiled in the Bio-Array Resource (http://www.bar.utronto.ca) also indicate that SOM is a seed-specific gene (see Supplemental Figure 2 online). Taken together, the analyses indicate that SOM is a seed-specific protein that belongs to a subset of CCCH-type zinc finger proteins.
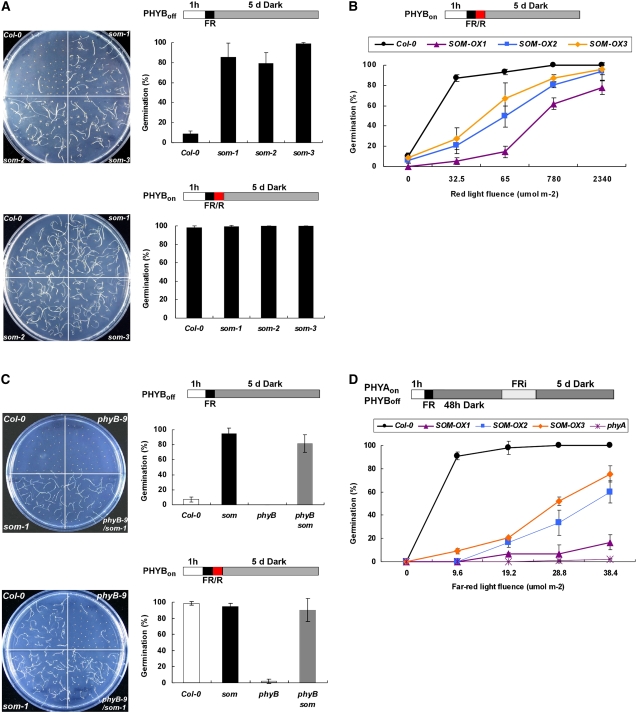
To test whether SOM is a negative regulator in PHYB-mediated promotion of germination, we analyzed the germination phenotypes of som mutants and SOM overexpression lines using a PHYB-dependent germination protocol. In this protocol, seed PHYB was inactivated by far-red light or activated by red light and seeds were then allowed to germinate in the dark for 5 d before scoring of germination. Wild-type seeds germinated when PHYB was activated by red light but not when PHYB was inactivated by far-red light (Figure 2A). As a control, phyB mutant seeds (phyB-9) did not germinate even after irradiation with red light. However, som mutant seeds germinated at rates of almost 100% irrespective of light conditions. Two other independent som alleles (som-2 and som-3) also germinated irrespective of light conditions. This indicates that mutations in the SOM gene alter germination behavior. To determine the effect of overexpression of SOM, we expressed intact SOM (lines SOM-OX1 and -OX2) and myc-tagged SOM (line SOM-OX3) under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The expression level of the SOM transcript was highest in the SOM-OX1 line and lowest in the SOM-OX3 line (see Supplemental Figure 3 online). All overexpression lines required more red light fluence for seed germination than did the wild type (Figure 2B). The germination phenotypes of both som mutants and SOM overexpression lines indicate that SOM is a negative regulator of seed germination.
Figure 2.
SOM Is a Negative Regulator in PHYA- and PHYB-Mediated Promotion of Seed Germination.
(A) Germination of som mutants (PHYB-dependent germination protocol). Light irradiation conditions are indicated by the diagrams above the graphs. FR, 5 min of far-red light (3.2 μmol·m−2·s−1); FR/R, 5 min of far-red light (3.2 μmol·m−2·s−1) and then 5 min of red light (13 μmol·m−2·s−1). The graphs show germination percentages of three replicates of 50 seeds each. Error bars indicate sd (n = 3). PHYBoff indicates a light scheme that inactivates PHYB by FR, and PHYBon indicates a light scheme that activates PHYB by FR/R.
(B) Germination of SOM-OX transgenic lines (PHYB-dependent germination protocol). Germination percentage is shown as a function of varying red light fluences that were given immediately after 5 min of far-red light (3.2 μmol·m−2·s−1). The SOM-OX1 line shows the highest SOM expression levels (endogenous plus transgene), and the SOM-OX3 line shows the lowest levels (see Supplemental Figure 3 online).
(C) Germination of Columbia (Col-0), som-1 mutant, phyB-9 mutant, and phyB-9 som-1 double mutant (PHYB-dependent germination protocol). Notations are as in (A).
(D) Germination of SOM-OX transgenic lines (PHYA-dependent germination protocol). Seeds were imbibed for 48 h to allow the accumulation of PHYA, varying fluences of FR light (FRi) were provided to imbibed seeds, and germination was scored after 5 d in darkness. PHYAon indicates a light scheme that activates PHYA by FRi. Error bars indicate sd (n = 3).
To further investigate the relationship between SOM and PHYB, we generated a phyB som double mutant (phyB-9 som-1) and studied germination using the same protocol. The phyB mutant failed to germinate irrespective of light conditions. The phyB som double mutant, however, germinated well (Figure 2C), indicating that the som mutation is epistatic to the phyB mutation under these conditions. These results support the hypothesis that SOM is a negative regulator in the PHYB-mediated promotion of the germination process and that PHYB promotes germination at least partly by inhibiting SOM.
We also tested whether SOM is a negative regulator in the PHYA-mediated promotion of germination. For the PHYA-dependent germination protocol, we inactivated PHYB with a far-red light pulse and allowed seeds to accumulate PHYA for 2 d at 22°C in the dark. Then we irradiated these imbibed seeds with prolonged far-red light to activate PHYA and incubated seeds in the dark for an additional 5 d. With this protocol, wild-type seeds germinate when PHYA is activated by prolonged far-red light but do not germinate without the prolonged far-red light (Figure 2D). We could not determine the germination phenotypes of the som mutant or the phyA som double mutant in this PHYA-dependent germination protocol because these mutants start to germinate during the imbibition period. Therefore, we used SOM overexpression lines to examine the role of SOM under these conditions. All three overexpression lines were hyposensitive to prolonged far-red light under this protocol (Figure 2D). Wild-type seeds germinated almost 100% even by 9.6 μmol·m−2·s−1 far-red light, whereas the phyA mutant did not germinate even by 38.4 μmol·m−2·s−1 far-red light. The three SOM overexpression lines required more far-red light than did the wild type for germination. These results indicate that SOM acts as a negative regulator in the PHYA-mediated promotion of germination.
The germination phenotypes of the som mutant are similar to those of the previously characterized pil5 mutant, and the phenotypes of SOM overexpression lines are similar to those of PIL5 overexpression lines (Oh et al., 2004). The negative role of SOM in seed germination suggests that PHYA and PHYB promote germination partly by inhibiting the action of SOM.
Phytochromes Inhibit SOM by Repressing Its Expression in Seeds
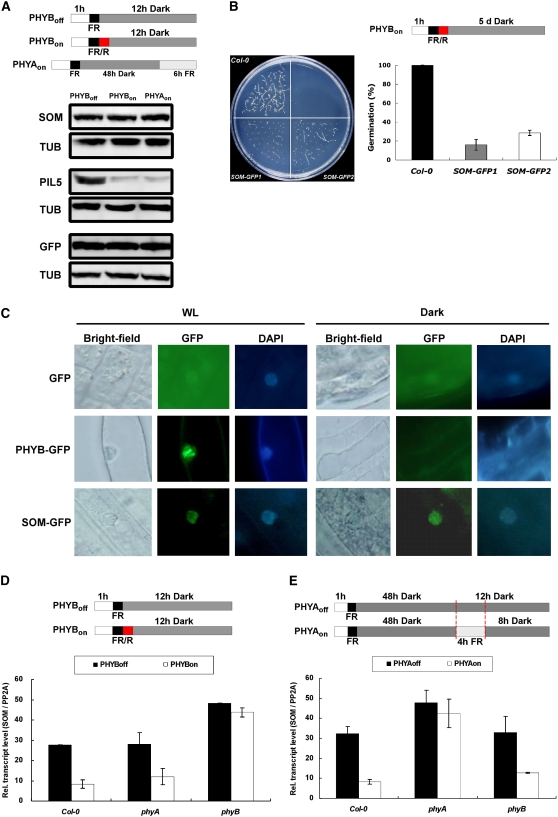
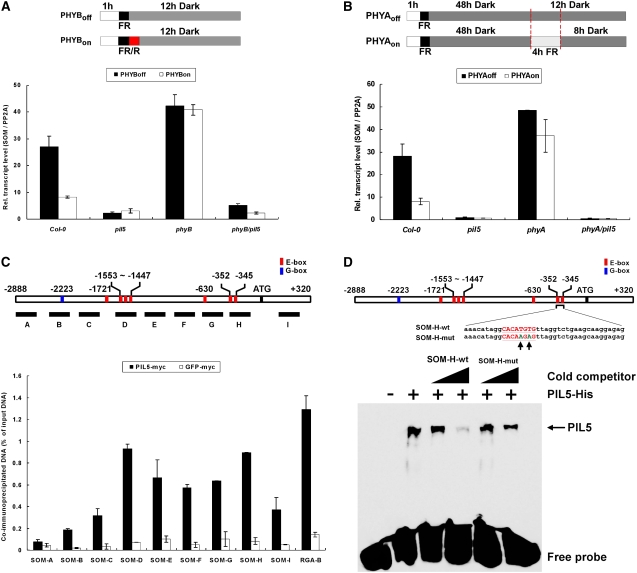
We investigated how phytochromes inhibit SOM and thereby promote germination. We first examined whether phytochromes regulate the stability of the SOM protein. For this study, we used transgenic lines that overexpress SOM-myc under the control of the CaMV 35S promoter, so that SOM is expressed under all light conditions. We first found that the myc-tagged SOM was functional (Figures 2B and 2D). We then analyzed the stabilities of SOM-myc under different light conditions in which PHYA and PHYB were either off or on (Figure 3A). We also examined myc-tagged green fluorescent protein (GFP) and myc-tagged PIL5 as controls. As reported previously, PHYA and PHYB activate the degradation of PIL5 but not GFP in imbibed seeds (Oh et al., 2006). By contrast, phytochromes did not alter the protein stability of SOM (Figure 3A). These results indicate that activation of phytochromes does not cause the degradation of SOM protein.
Figure 3.
Phytochromes Inhibit SOM Function at the Transcriptional Level.
Notations for light irradiation schemes are the same as those in Figure 2.
(A) No change in SOM protein stability under different light conditions. Proteins were extracted at the end of dark incubation from transgenic lines expressing myc-tagged SOM, myc-tagged PIL5, and myc-tagged GFP. Tubulin (TUB) was detected by anti-tubulin antibody as a loading control. Light conditions are indicated in the diagrams at top. Seeds were harvested for the protein extraction at the end of dark incubation (PHYBoff and PHYBon) or far-red irradiation (PHYAon).
(B) Germination of transgenic lines (SOM-GFP1 and SOM-GFP2) expressing the SOM-GFP fusion protein. Notations are the same as in Figure 2A.
(C) Subcellular localization of SOM-GFP, PHYB-GFP, and GFP determined using transgenic lines expressing these proteins. 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain the nucleus. WL, white light.
(D) Relative transcript levels of SOM in Col-0, phyA-211 mutant, and phyB-9 mutant seeds (PHYB-dependent germination protocol). Light conditions are indicated by the diagrams at top. Seeds were harvested for the RNA extraction at the end of dark incubation. The SOM transcript levels were normalized by the PP2A transcript level. Error bars indicate sd of three real-time PCR experiments of one biological sample.
(E) Relative transcript levels of SOM in Col-0, phyA-211 mutant, and phyB-9 mutant seeds (PHYA-dependent germination protocol). Light conditions are indicated by the diagrams at top. Seeds were harvested for RNA extraction at the end of dark incubation. SOM transcript levels were normalized by the PP2A transcript level. Error bars indicate sd of three real-time PCR experiments.
Next, we examined whether phytochromes regulate the subcellular localization of SOM using transgenic lines that overexpress GFP-tagged SOM under the control of the CaMV 35S promoter. The GFP-tagged SOM is functional (Figure 3B), and we detected a fluorescent signal in the nucleus irrespective of light conditions in transgenic plants (Figure 3C). By contrast, control transgenic plants that overexpress GFP alone emit fluorescence mainly in the cytosol. In the PHYB-GFP transgenic line, dark-grown plants had a diffuse GFP signal throughout the cytosol, but light-grown plants had a few discrete fluorescent spots in the nucleus (Kircher et al., 1999; Yamaguchi et al., 1999). These results indicate that SOM is a nucleus-localized protein and that its localization is independent of light conditions. This suggests that phytochromes do not alter the subcellular localization of SOM.
Next, we examined whether phytochromes inhibit SOM by repressing its gene expression in seeds. In the PHYB-dependent germination protocol, the red light pulse greatly reduced the levels of the SOM transcript in wild-type and phyA mutant seeds (Figure 3D; see Supplemental Table 2 online). By contrast, the level of the SOM transcript remained high in phyB mutant seeds irrespective of light conditions. This indicates that activation of PHYB represses SOM transcript expression in seeds. In the PHYA-dependent germination protocol, prolonged far-red light also greatly decreased the level of the SOM transcript in wild-type and phyB mutant seeds, but not in the phyA mutant seeds (Figure 3E). Taken together, these results indicate that phytochromes inhibit SOM at least partly by repressing the expression of the SOM transcript in seeds.
SOM Inhibits Seed Germination by Regulating the Expression of GA and ABA Metabolic Genes
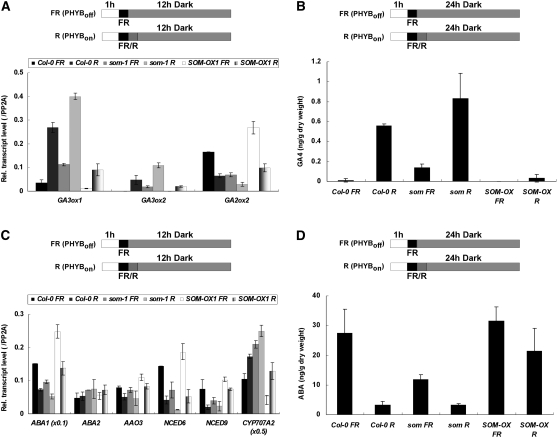
Phytochrome-mediated light signaling promotes germination partly by increasing the level of bioactive GA, decreasing the level of ABA, and increasing the responsiveness to GA (Oh et al., 2007). Since SOM regulates germination in response to light, we investigated whether SOM inhibits germination by altering the expression of hormone-related genes. Since light regulates mainly two GA anabolic genes (GA3ox1 and GA3ox2) and a catabolic gene (GA2ox2) (Ogawa et al., 2003; Oh et al., 2006), we determined the expressions of those three metabolic genes.
First, we measured levels of GA metabolic genes in the wild type, som mutants, and SOM overexpressers to test whether SOM inhibits germination partly by regulating GA metabolic genes (Figure 4A). As reported previously, red light increases the expression of GA3ox1 and GA3ox2 in wild-type seeds (Ogawa et al., 2003). In the som mutant, expression of these GA anabolic genes was already high, even in far-red-light-irradiated seeds, and red light further increased their expression. By contrast, expression of GA2ox2 was lower in the som mutant, and red light further decreased its expression. In contrast with the som mutant, the SOM overexpression line expressed lower levels of GA3ox1 and GA3ox2 and higher levels of GA2ox2 in seeds treated with far-red or red light (Figure 4A). These results indicate that SOM, like PIL5, inhibits the synthesis of bioactive GA partly by repressing two GA anabolic genes and activating a GA catabolic gene. However, the expression patterns of GA metabolic genes in the som mutant differ from those of the pil5 mutant. GA metabolic genes are expressed virtually independently of light conditions in the pil5 mutant (Oh et al., 2006) but are induced by red light in the som mutant. These results indicate that additional factors downstream of PIL5 influence the expression of GA metabolic genes. The expression patterns of GA metabolic genes that we observed suggest that the levels of bioactive GA would be higher in som mutant seeds and lower in the SOM overexpression line. Thus, we measured the levels of GA4 in different mutant seeds using the PHYB-dependent seed germination protocol (Figure 4B). The level of GA4 was low in wild-type seeds treated with far-red light but high in seeds treated with red light, as reported previously (Oh et al., 2006). Compared with the wild type, som mutant seeds had a higher level of GA4 when treated with far-red light, and red light further increased the level of GA4. In the SOM overexpression line, the level of GA4 was lower than in the wild type, and red light increased the level only slightly. These results are consistent with the expression patterns of GA metabolic genes.
Figure 4.
SOM Regulates Both GA and ABA Metabolic Genes.
Notations for light irradiation schemes are the same as those in Figure 2.
(A) Relative transcript levels of GA metabolic genes (GA3ox1, GA3ox2, and GA2ox2) in Col-0, som-1 mutant, and SOM-OX1 seeds (PHYB-dependent germination protocol). Light conditions are indicated by the diagrams at top. Error bars indicate sd of three PCR experiments.
(B) Bioactive GA levels in Col-0, som-1 mutant, and SOM-OX1 seeds under different light conditions. Seeds were harvested for quantification at the end of dark incubation. Error bars indicate sd of three biological samples.
(C) Relative transcript levels of ABA metabolic genes (ABA1, ABA2, AAO3, NCED6, NCED9, and CYP707A2) in Col-0, som-1 mutant, and SOM-OX1 seeds (PHYB-dependent germination protocol). Error bars indicate sd of three PCR experiments.
(D) ABA levels in Col-0, som-1, and SOM-OX1 seeds. Error bars indicate sd of three biological samples.
Next, we examined whether SOM also inhibits germination by regulating ABA metabolic genes (Figure 4C). As reported previously, red light decreased the expression of ABA anabolic genes (ABA1, NCED6, and NCED9) and increased the expression of an ABA catabolic gene (CYP707A2) in wild-type seeds (Seo et al., 2006; Oh et al., 2007). Compared with the wild type, the som mutant had lower expression of ABA anabolic genes and higher expression of the ABA catabolic gene. In this mutant, red light further decreased the expression of ABA anabolic genes and increased the expression of the ABA catabolic gene. By contrast, the SOM overexpression line had higher expression of ABA anabolic genes and lower expression of the catabolic gene compared with the wild type. These results indicate that SOM, like PIL5, activates ABA biosynthesis by increasing the expression of ABA anabolic genes and repressing an ABA catabolic gene. Unlike the pil5 mutant, however, ABA metabolic genes are still regulated by light in the som mutant.
The expression patterns of ABA metabolic genes suggest that ABA levels would be lower in the som mutant but higher in the SOM overexpression line compared with the wild type. Thus, we measured the levels of ABA in wild-type and mutant seeds (Figure 4D). The level of ABA was high in far-red-light-treated wild-type seeds but low in red-light-treated seeds, as reported previously (Oh et al., 2007). Far-red-light-irradiated som mutant seeds had lower levels of ABA than wild-type seeds, and red light treatment further decreased the level of ABA in this mutant. In the SOM overexpression line, seeds treated with far-red or red light had higher levels of ABA than did wild-type seeds. As with our GA studies (see above), these results are consistent with our studies of the expression patterns of ABA metabolic genes. Taken together, the results indicate that SOM regulates the levels of ABA and GA in seeds by regulating the expression of ABA and GA anabolic and catabolic genes.
SOM Does Not Regulate the Expression of DELLA Transcripts
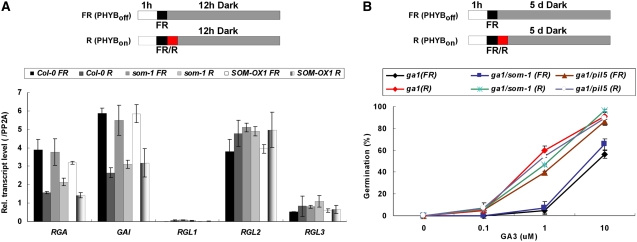
PIL5 inhibits seed germination by regulating GA and ABA metabolic genes and by activating the transcription of the DELLA genes GAI and RGA. We investigated whether SOM also regulates the expression of these DELLA transcripts using the PHYB-dependent germination protocol (Figure 5A). Red light decreased the levels of GAI and RGA transcripts in wild-type seeds but had no effect on the levels of the other three DELLA transcripts. By contrast, the pil5 mutant expressed constitutively low levels of GAI and RGA transcripts independent of light conditions. Both som mutation and SOM overexpression did not alter the expression levels of DELLA transcripts (Figure 5A). These results indicate that SOM does not regulate DELLA transcripts in seeds.
Figure 5.
SOM Does Not Regulate the Expression of GAI and RGA.
Notations for light irradiation schemes are the same as those in Figure 2.
(A) Relative transcript levels of GA signaling genes (RGA, GAI, RGL1, RGL2, and RGL3) in Col-0, som-1 mutant, and SOM-OX1 seeds (PHYB-dependent germination protocol). Error bars indicate sd of three PCR experiments.
(B) GA responsiveness of the ga1 mutant, the ga1 som-1 double mutant, and the ga1 pil5 double mutant (PHYB-dependent germination protocol). GA responsiveness is determined by the germination percentage in response to varying GA3 concentrations. Error bars indicate sd (n = 3).
Next, we investigated the effect of SOM on GA responsiveness. We determined GA responsiveness by measuring the germination response of the ga1 single mutant and the ga1 som double mutant in the presence of various concentrations of GA3 (Figure 5B). GA3 is a fungal GA that is not inactivated by GA2ox2. Thus, responsiveness to GA3 in the absence of GA biosynthesis (ga1 mutant background) can be used as an indicator of GA responsiveness (Oh et al., 2007). As reported previously, when we treated seeds with red light, the GA responsiveness of the ga1 mutant increased, whereas the GA responsiveness of the ga1 pil5 double mutant was constitutively high irrespective of light conditions (Oh et al., 2007). By contrast, the GA responsiveness of the ga1 som double mutant was similar to that of the ga1 single mutant (low under far-red light and high under red light; Figure 5B). These results indicate that SOM, unlike PIL5, does not regulate the GA responsiveness of seeds.
PIL5 Regulates the Expression of SOM by Binding Its Promoter
We previously showed that PIL5 is a master regulatory transcription factor that inhibits germination partly by directly regulating the expression of two DELLA genes and partly by indirectly regulating the expression of GA and ABA metabolic genes (Oh et al., 2007). In the som mutant, the germination characteristics and expression patterns of the GA and ABA metabolic genes suggest that SOM may be a PIL5 downstream component that regulates GA and ABA metabolic genes during germination.
Thus, we examined the role of PHY- and PIL5-mediated light signaling in the regulation of SOM transcript accumulation in seeds. In the PHYB-dependent seed germination protocol, red light decreased the expression of SOM, but SOM expression remained high in the phyB mutant irrespective of light conditions (Figure 6A). This indicates that in seeds, PHYB is required for repression of the expression of SOM transcript. By contrast, the expression of SOM remained low in the pil5 mutant irrespective of light conditions, indicating that higher expression of SOM in far-red-light-irradiated seeds requires the presence of PIL5. To further demonstrate the role of PIL5 in SOM expression, we measured the expression of SOM in the phyB pil5 double mutant. As shown in Figure 6A, the expression level of SOM in the double mutant was low irrespective of light conditions. This suggests that PHYB represses the expression of SOM by inhibiting PIL5. Similar to our results using the PHYB-dependent seed germination protocol, PIL5 was required for the expression of SOM transcript in the PHYA-dependent seed germination protocol (Figure 6B). Taken together, these results indicate that SOM is a downstream component of the PIL5 signaling pathway in PHYA- and PHYB-mediated light signaling during germination.
Figure 6.
PIL5 Regulates the in Vivo Expression of SOM by Binding the SOM Promoter.
Notations for light irradiation schemes are the same as those in Figure 2.
(A) SOM transcript levels in Col-0, pil5 mutant, phyB-9 mutant, and phyB-9 pil5 double mutant (PHYB-dependent germination protocol).
(B) SOM transcript levels in Col-0, pil5 mutant, phyA211 mutant, and phyA211 pil5 double mutant (PHYA-dependent germination protocol).
(C) PIL5 ChIP analysis of the SOM promoter. The diagram at top depicts the promoter sequence of the SOM gene. The red box represents an E-box element (CANNTG) and the blue box represents a G-box element (CACGTG). Underlying thick lines marked by uppercase letters indicate various promoter fragments used for PCR. The primer sequences used for ChIP-PCR are shown in Supplemental Table 1 online. The graph indicates the percentage of DNA fragments that coimmunoprecipitated with PIL5-myc (closed bars) or GFP-myc (open bars) relative to the input DNAs. The RGA-B fragment, identified as a PIL5 binding site in an earlier report (Oh et al., 2007), was used as a positive control. Shown are a typical result and sd from three PCR experiments. Similar results were obtained from three independent experiments.
(D) In vitro binding analysis of PIL5 and E-box elements in the SOM-H fragment of the SOM promoter. The diagram at top depicts the probe sequence corresponding to the SOM-H fragment of the SOM promoter. Mutated bases in the SOM-H-mut probe are represented by green letters and indicated by arrows. SOM-H-wt, probe with intact E-box elements; SOM-H-mut, probe with mutated E-box elements. Triangles indicate increasing amounts of unlabeled probes for competition. Error bars indicate sd from three PCR experiments.
PIL5 may regulate the expression of SOM either directly (as for GAI and RGA) or indirectly (as for GA and ABA metabolic genes). Thus, we performed a ChIP assay using the PIL5-OX line (which expresses functional myc-tagged PIL5) and a GFP-myc overexpression line to investigate whether PIL5 regulates SOM by directly binding to its promoter. For the ChIP assay, we designed eight primer sets for various parts of the 2.9-kb SOM promoter region and a primer set for a SOM coding region. We determined the binding of PIL5 to promoter fragments by enrichment of immunoprecipitated DNA fragments over input DNA fragments. If PIL5 binds the SOM promoter, then the PIL5-bound fragment should have the highest enrichment and other fragments (which are farther from the binding site) should have decreased enrichment. In addition, the enrichment by PIL5-myc for the PIL5 binding fragment should be much higher than that by GFP-myc.
Our results indicate that PIL5 binds to the promoter of SOM in vivo (Figure 6C). When we immunoprecipitated cross-linked GFP-myc with anti-myc antibody, all fragments showed very low enrichments. We consider this as nonspecific background enrichment. In contrast with GFP-myc, the ChIP of PIL5-myc showed high enrichment of an RGA promoter fragment (RGA-B), which was previously shown to be a binding site of PIL5 in vivo (Oh et al., 2007). In the same ChIP sample, some of the SOM promoter fragments also showed high enrichment. Among promoter fragments, a SOM-A fragment centered at −2220 showed the lowest enrichment and is comparable with the enrichment level of GFP-myc. The enrichment by PIL5-myc peaked at two positions, a SOM-D fragment centered at −1538 and a SOM-H fragment centered at −342. The enrichment of these two fragments was comparable to that of RGA-B. Compared with these two fragments, the enrichment of other fragments gradually decreased with distance from the SOM-D and SOM-H fragments. Taken together, our ChIP analyses indicate that PIL5 binds to two different parts of the SOM promoter.
Previous analyses demonstrated that PIL5 binds to G-box elements in the promoters of GAI and RGA (Oh et al., 2007). Sequence analysis of the SOM promoter shows that it also contains a G-box element. Curiously, however, the position of the G-box element (−2223) does not correspond to in vivo PIL5 binding promoter fragments (SOM-D and SOM-H) (Figure 6C). SOM-D and SOM-H contain E-box elements, which have been shown to serve as binding sites for many bHLH transcription factors (Ellenberger et al., 1994).We used an electrophoretic mobility shift assay (EMSA) to investigate whether PIL5 binds to E-box elements (Figure 6D). We made a double-stranded and labeled probe (38mer) containing the sequence of the part of the SOM-H fragment that contained E-box elements and studied its binding to the PIL5 protein using an EMSA. The binding was further competed out by the nonlabeled probe but was not competed out by the same probe with mutated E-box elements. These findings indicate that PIL5 can also bind to some E-box elements that are present in the SOM-H promoter fragments and suggest that PIL5 regulates the expression of the SOM transcript by binding to E-box elements in its promoter.
PIL5 Regulates ABA and GA Metabolic Genes Partly through SOM
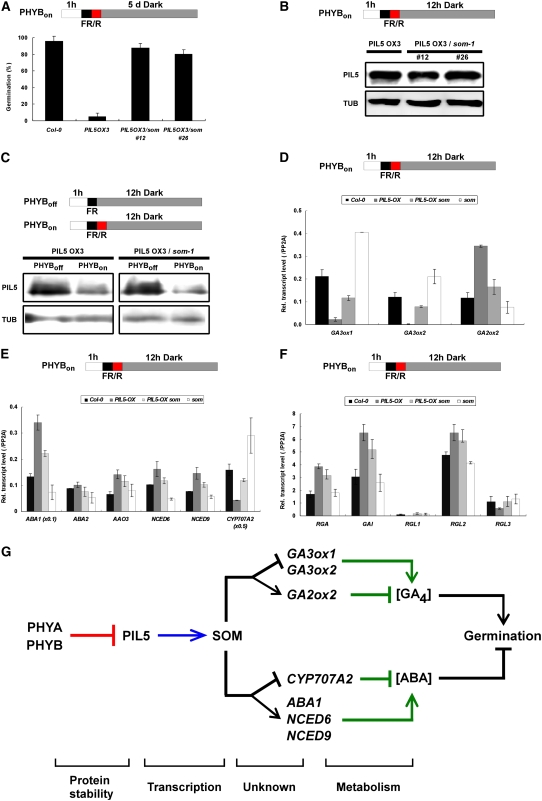
The activation of SOM expression by PIL5 via binding to the SOM promoter suggests that PIL5 may inhibit seed germination partly through SOM. To investigate this possibility, we established two PIL5-OX som mutant lines by crossing PIL5-OX and the som mutant. When we examined the germination response (Figure 7A), we found that PIL5-OX seeds did not germinate well in response to a short red light pulse, whereas almost all wild-type seeds germinated following this stimulus. Using the same protocol, the PIL5-OX som seeds germinated well. The higher germination frequency of PIL5-OX som lines was not due to the suppression of the PIL5 transgene, as shown by similar amounts of PIL5 protein in these lines (Figure 7B). The som mutation also did not affect the degradation of PIL5 by light in seeds (Figure 7C). PIL5 protein was degraded similarly both in the PIL5-OX and the PIL5-OX som lines when seeds were irradiated with red light or prolonged far-red light. Taken together, these results indicate that PIL5 requires SOM for the inhibition of germination.
Figure 7.
PIL5 Regulates ABA and GA Metabolic Genes Partly through SOM.
Notations for light irradiation schemes are the same as those in Figure 2.
(A) Germination of Col-0, PIL5-OX, and PIL5-OX som mutant seeds (PHYB-dependent germination protocol). Error bars indicate sd (n = 3).
(B) Similar levels of PIL5 protein in the seeds of PIL5-OX and two PIL5-OX som lines (lines 12 and 26).
(C) No change in PIL5 protein stability in the PIL5-OX som double mutant (line 12) seeds (PHYB-dependent germination protocol).
(D) Relative transcript levels of GA metabolic genes (GA3ox1, GA3ox2, and GA2ox2) in Col-0, PIL5-OX, PIL5-OX som, and som mutant seeds (PHYB-dependent germination protocol). Error bars indicate sd from three PCR experiments.
(E) Relative transcript levels of ABA metabolic genes (ABA1, ABA2, AAO3, NCED6, NCED9, and CYP707A2) in Col-0, PIL5-OX, PIL5-OX som, and som mutant seeds (PHYB-dependent germination protocol). Error bars indicate sd from three PCR experiments.
(F) Relative transcript levels of GA signaling genes (RGA, GAI, RGL1, RGL2, and RGL3) in Col-0, PIL5-OX, PIL5-OX som, and som mutant seeds (PHYB-dependent germination protocol). Error bars indicate sd from three PCR experiments.
(G) Proposed role of SOM in seed germination. Both PHYA and PHYB inhibit PIL5 by degrading the PIL5 protein, whereas PIL5 activates the expression of SOM by directly binding its promoter. SOM decreases GA4 levels by repressing GA anabolic genes and activating a GA catabolic gene. For ABA, SOM increases the level of ABA by activating ABA anabolic genes and repressing an ABA catabolic gene. Decreased GA4 cannot promote seed germination, whereas the increased ABA inhibits seed germination. Other unknown factors that also regulate ABA and GA metabolic genes downstream of PIL5 and DELLA proteins, including two PIL5 target DELLA proteins (GAI and RGA) that inhibit seed germination downstream of PIL5, are not indicated in the diagram. The mode of action for each step is indicated at bottom.
Since PIL5 regulates the expression of SOM and SOM regulates the expression of hormone metabolic genes (Figure 4), PIL5 might regulate the expression of hormone metabolic genes via SOM. We analyzed the expression patterns of hormone metabolic genes in the PIL5-OX som double mutant line (Figures 7D and 7E). As reported previously, PIL5 repressed the expression of two GA anabolic genes (GA3ox1 and GA3ox2) and activated the expression of a GA catabolic gene (GA2ox2). In addition, PIL5 activated the expression of ABA anabolic genes (ABA1, NCED6, and NCED9) and repressed an ABA catabolic gene (CYP707A2). The expression levels of ABA and GA metabolic genes in the PIL5-OX som double mutant were intermediate between those of PIL5-OX and the som mutant (Figures 7D and 7E). In contrast with hormone metabolic genes, the expression of GAI and RGA in the PIL5-OX som double mutant was similar to that of PIL5-OX (Figure 7F). Taken together, these results indicate that PIL5 regulates the expression of ABA and GA metabolic genes partly via SOM. However, the intermediate expression levels of metabolic genes in the PIL5-OX som double mutant suggest that there are additional PIL5 downstream components that also regulate ABA and GA metabolic genes.
DISCUSSION
We report here that SOM, a novel seed-specific CCCH-type zinc finger protein, negatively regulates seed germination by activating ABA biosynthesis and inhibiting GA biosynthesis. To determine the role of SOM in PHY-mediated promotion of germination, we investigated the functional relationship between SOM and PIL5, which has been characterized previously. ChIP analysis shows that PIL5 activates the expression of SOM by binding directly to its promoter, suggesting that SOM has a role downstream of PIL5 in the phytochrome-mediated transduction process. Our analysis of the PIL5-OX som double mutant also shows that PIL5 regulates hormone metabolic genes partly via SOM. Taken together, these results suggest that light regulates ABA and GA metabolic genes via PIL5 and then via SOM in seeds (Figure 7G and its legend for the detailed description). The molecular function of the CCCH-type zinc finger motif that is present in SOM is currently unknown. Further studies are needed to determine whether SOM is a transcription factor that regulates hormone metabolic genes, an RNA binding protein, or a protein with some other function.
PHY- and PIL5-Mediated Light Signaling Regulates ABA and GA Metabolic Genes Partly via SOM
During seed germination, phytochromes mediate the inhibition of ABA biosynthesis and the activation of GA biosynthesis via transcriptional regulation of ABA and GA metabolic genes (Ogawa et al., 2003; Oh et al., 2006, 2007; Seo et al., 2006). Our previous germination studies have shown that PIL5, an important negative regulator of germination, participates in the conversion of a phytochrome-mediated light signal into ABA and GA signals (Penfield et al., 2005; Oh et al., 2006, 2007). Following light absorption, phytochromes move into the nucleus (Kircher et al., 1999, 2002; Yamaguchi et al., 1999), interact with PIL5 (Huq et al., 2004; Oh et al., 2004), and activate the degradation of PIL5 (Shen et al., 2005; Oh et al., 2006). Since PIL5 activates ABA biosynthesis, represses GA biosynthesis, and increases the expression of two DELLA genes (GAI and RGA), the degradation of PIL5 by phytochromes decreases the level of ABA, increases the level of GA, and decreases GA negative signaling components, resulting in germination (Oh et al., 2006, 2007). ChIP analyses showed that PIL5 regulates the expression of GAI and RGA by binding directly to their promoters, but it regulates hormone metabolic genes indirectly (Oh et al., 2007). This suggested the presence of other signaling components that link PIL5 to the expression of hormone metabolic genes.
The results of this study show that SOM is one of the light signaling components that regulate the expression of ABA and GA metabolic genes. Similar to the pil5 mutant, the som mutant germinates irrespective of light conditions. The expression analyses show that SOM activates ABA biosynthesis by activating three ABA anabolic genes (ABA1, NCED6, and NCED9) and represses an ABA catabolic gene (CYP707A2), resulting in the increased ABA level. Our studies also show that SOM inhibits GA biosynthesis by repressing two GA anabolic genes (GA3ox1 and GA3ox2) and activating a GA catabolic gene (GA2ox2), resulting in the decreased GA level. In contrast with PIL5, however, SOM does not regulate the expression of GAI and RGA, so there is no change in the GA responsiveness of som mutants. The SOM regulation of hormone metabolic genes, but not DELLA genes, indicates that SOM regulates a subset of genes that are regulated by PIL5.
In imbibed seeds, light absorption by phytochromes alters the level of SOM transcripts via PIL5. Red light represses the expression of SOM in a PHYB-dependent manner, as supported by results showing that red light–dependent repression of SOM occurs in the wild type, whereas red light–independent constitutively high expression of SOM occurs in the phyB mutant. The repression of SOM by PHYB requires PIL5, as supported by results showing light-independent constitutively low expression of SOM in the pil5 mutant and the phyB pil5 double mutant. Prolonged far-red light also represses the expression of SOM in a PHYA-dependent manner. The light-dependent repression of SOM by prolonged far-red light is greatly decreased in the phyA mutant. The prolonged far-red light dependence of SOM expression is completely absent in the pil5 mutant and in the phyA pil5 double mutant. Taken together, these results suggest that PHYA and PHYB repress the expression of SOM via PIL5 in imbibed seeds.
The persistent light-dependent expression of ABA and GA metabolic genes in the som mutant suggests that PIL5 regulates these metabolic genes not just via SOM but also via other signaling components. The som mutant has altered expression levels of ABA and GA metabolic genes, but light also regulates these levels in this mutant. These expression patterns are paralleled by the concentrations of ABA and GA that we measured in imbibed seeds. ABA levels in the som mutant are already lower than in the wild type, even before red light treatment, but they are decreased even more by red light in the som mutant. Similarly, GA levels are already higher in the som mutant before red light treatment than in the wild type, but they are further increased by red light. This response differs from that of the pil5 mutant, which exhibits light-independent expression levels of hormone metabolic genes and light-independent hormone levels. Therefore, if PIL5 regulates these metabolic genes solely through SOM, the expression levels and hormone levels should also be light-independent in the som mutant. Taken together, our results suggest that, in addition to SOM, other components regulate the expression of ABA and GA metabolic genes downstream of PIL5.
PIL5 Regulates the Expression of SOM by Binding Its Promoter
The transmission of PHY-mediated light signals to SOM requires transcriptional and posttranslational regulation. Phytochromes transmit perceived light signals to PIL5 by interacting with PIL5 and activating its degradation, a posttranslational process (Shen et al., 2005; Oh et al., 2006). Other researches have shown that phytochromes also mediate the degradation of other negative components, such as PIF3, PIF4, and PIL6 (Bauer et al., 2004; Park et al., 2004; Shen et al., 2007; Lorrain et al., 2008). This suggests that a major mode of PHY-mediated light signaling involves activation of the degradation of specific proteins (Huq, 2006; Bae and Choi, 2008). Signal transmission from PIL5 to SOM occurs at the transcriptional level. ChIP analyses show that PIL5 binds to the promoter of SOM in vivo, indicating that PIL5 directly targets SOM as well as GAI and RGA. Curiously, the PIL5 binding sites in the SOM promoter do not correspond to a G-box element (CACGTG), an element to which PIL5 is known to bind (Huq et al., 2004; Oh et al., 2007). Instead, the binding sites are E-box elements (CANNTG), known binding elements of many bHLH transcription factors, indicating that PIL5 binds not only to the G-box element but also to some E-box elements. Irrespective of binding sequence elements, the binding of PIL5 to its target promoters activates the expression of target genes, including SOM, GAI, and RGA, and these regulate ABA and GA metabolic genes and GA responsiveness. In seeds, light activation of phytochromes decreases the level of the PIL5 protein and, consequently, the levels of PIL5 downstream target genes. PIL5-mediated regulation of some downstream target genes, such as SOM, alters the levels of ABA and GA inside the imbibed seeds. PIL5-mediated regulation of other downstream target genes, such as GAI and RGA, regulates hormone responsiveness.
The Molecular Function of SOM Is Unknown
We have not yet determined the exact molecular functions of SOM. Our analyses show that SOM is located in the nucleus and has three zinc finger motifs, including one tristetraprolin (TTP)-type zinc finger motif (CX8CX5CX3H) and two atypical zinc finger motifs (CX5HX4CX3H and CX5CX4CX3H). Other plant proteins have similar zinc finger motifs. These include PEI1 from Arabidopsis, which regulates embryogenesis (Li and Thomas, 1998), SZF1 and SZF2 from Arabidopsis, which regulate salt responses (Sun et al., 2007), and DOS from rice (Oryza sativa), which regulates leaf senescence (Kong et al., 2006). Another Arabidopsis zinc finger protein, FRIGIDA-ESSENTIAL1 (FES1), has a single zinc finger motif (CX7CX5CX3H) and has been shown to delay flowering in the presence of FRIGIDA by activating the expression of FLOWERING LOCUS C (Schmitz et al., 2005). The molecular functions of PEI1, DOS, and FES1 are also unknown.
Zinc finger proteins with tandem TTP-type zinc finger motifs regulate various developmental processes mainly by binding to RNA molecules and modulating their stabilities or processing (Blackshear, 2002). In the mouse, TTP regulates inflammation by binding to AU-rich elements that are in the 3′ untranslated region of tumor necrosis factor-α mRNA and destabilizing this mRNA (Taylor et al., 1996; Carballo et al., 1998; Lai et al., 2000). In Arabidopsis, HUA1 regulates floral organ development by binding to AGAMOUS pre-mRNA and facilitating its processing (Li et al., 2001). Taken together, the molecular functions of tandem TTP-type zinc finger proteins suggest that SOM may also act as an RNA binding protein. However, since TTP-type zinc finger proteins have other molecular functions, this requires experimental verification.
The Expression of SOM during Seed Maturation
Beyond its role in PHY- and PIL5-mediated light signaling, it will be interesting to determine the role of SOM in other seed germination conditions. The SOM promoter contains both a PIL5 binding G-box element and a putative RY element (CATGCAT/A). Since the RY motif is known to be a binding site for ABI3 (Finkelstein et al., 2002), the expression of SOM could be regulated by ABI3. According to the microarray data compiled in the Bio-Array Resource, expression levels of ABI3 increase dramatically during seed maturation and decrease during imbibition under white light (see Supplemental Figure 2 online). The increased expression of ABI3 during seed maturation likely plays a role in seed dormancy (Finkelstein et al., 2008). Curiously, the expression level of SOM also increases dramatically during seed maturation. Unlike SOM, however, the expression level of PIL5 remains low during seed maturation. Taken together, these data are consistent with a role for ABI3 and/or FUS3 in regulating SOM expression during seed maturation.
The Bio-Array Resource database also indicates, however, that the expression level of SOM at 24 h after imbibition under white light is not significantly different between the wild type and the abi3 mutant (see Supplemental Figure 2 online). Thus, the similar expression patterns of SOM and ABI3 transcripts during seed maturation could also be caused by shared regulatory factors. Alternatively, ABI3 may regulate SOM redundantly with other factors such as FUSCA3, such that a single mutation fails to dramatically affect SOM expression. Further studies are necessary to establish the functional relationships between SOM, ABI3, and FUS3 during seed maturation.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana plants were grown in a room with a 16-h-light/8-h-dark cycle at 22 to 24°C. Following seed harvesting, seeds were dried at 22°C in white paper bags for at least 1 month prior to germination assays. The first som mutant line (som-1) was identified by screening T-DNA insertion pools made in the laboratory. The second and the third som mutants (som-2 and som-3) were obtained from the Arabidopsis Stock Center (Salk_090314 and SALK_008075) (Alonso, 2003). T-DNA insertions sites were determined by amplifying the flanking regions and sequencing them (see Supplemental Figure 1 online). For generation of the SOM-OX1 and SOM-OX2 transgenic lines, full-length cDNA of SOM (At1g03790) was amplified with a primer set (5′-AACTCCTAGGATGGATGTCGTTTGTACGGAACATCAAA-3′ and 5′-ATCAGGATCCTTTCAAGTCAAGAGATCATTGACCCATC-3′), cloned into a binary vector (pBI121), and transformed into Arabidopsis, and homozygous lines were selected. Transgenic lines expressing myc-tagged SOM (SOM-OX3) and GFP-tagged SOM (SOM-GFP) under the control of the CaMV 35S promoter were generated with a primer set (5′-AACTCCTAGGATGGATGTCGTTTGTACGGAACATCAAA-3′ and 5′-ATCAGGATCCTTAGTCAAGAGATCATTGACCCATCCTAG-3′). Subcellular localization of SOM-GFP was determined by fluorescence microscopy (BX51; Olympus). All plants used in this study (the wild type, phyA-211, phyB-9, ga1 [Salk_109115], som-1, som-2, som-3, and SOM-OXs) were in the Col-0 background.
Germination Assay
Germination assays were performed as described previously (Oh et al., 2007). For the PHYB-dependent germination protocol, triplicate sets of ∼50 seeds for each mutant were surface-sterilized and plated on aqueous agar medium (0.6% phytoagar, pH 5.7) under white light. Plates were left on a clean bench before light irradiation. One hour after the beginning of seed sterilization, seeds were irradiated with 5 min of far-red light (3.2 μmol·m−2·s−1) or red light (13 μmol·m−2·s−1), unless specified otherwise, and incubated in the dark for 5 d, and then germination (protrusion of the radicle) was assayed. Growth chambers with 740-nm LEDs (far-red light) or 670-nm LEDs (red light) were used for the light treatment (Vision). For the PHYA-dependent germination assay, sterilized and plated seeds were irradiated by far-red light for 5 min and then incubated in the dark for 48 h to allow the accumulation of PHYA. After the dark incubation, seeds were irradiated with various fluence levels of far-red light and incubated in the dark for 5 d for germination scoring. Germination was scored after a 4-d dark incubation period. To determine the effect of GA3, triplicate sets of ∼50 seeds were placed on agar plates containing variable concentrations of GA3.
Gene Expression Analysis, Protein Analysis, and Sequence Analysis
For real-time PCR, total RNA was extracted from seeds that were incubated in the dark for 12 h after far-red or red light irradiation. We used the Spectrum Plant Total RNA kit (Sigma-Aldrich) according to the manufacturer's protocol. The total RNA was used to synthesize cDNA. The relative transcript level of each gene was determined by real-time PCR using specific primers (see Supplemental Table 1 online) by comparison with the level of PP2A (see Supplemental Table 2 online), SYBR Green Supermix (Bio-Rad), and the iCycler IQ PCR machine (Bio-Rad), as described previously (Oh et al., 2007). Expression patterns were confirmed using at least three different biological samples, and representative data are presented in the figures.
To determine the effect of light on the stability of SOM protein, dark-incubated imbibed seeds were treated with either red or far-red light. Light treatment schemes were identical to those for the seed germination assay except that seeds were harvested at 12 h after red light irradiation or immediately after 6 h of far-red light irradiation. Total protein was extracted from ∼50 μL of seeds, and myc-tagged protein levels were determined by protein gel blotting using an anti-myc antibody, as described previously (Oh et al., 2006).
Amino acid sequences of SOM and its homologs were retrieved from the database and aligned by Clustal in the MEGA4 program (www.megasoftware.net) using a defaulting setting. A bootstrap phylogenetic tree was constructed using the neighbor-joining method embedded in the MEGA4 program.
Quantification of Endogenous ABA and GA Levels
GA measurements were performed on a liquid chromatography–tandem mass spectrometry system consisting of a quadrupole/time-of-flight tandem mass spectrometer (Q-Tof Premier; Waters) and an Acquity Ultra Performance liquid chromatograph (Waters) equipped with a reverse-phase column (Acquity UPLC BEH-C18; Waters), as described previously (Varbanova et al., 2007). 2H-labeled GAs (purchased from Lewis Mander, Australian National University) were used as internal standards. Endogenous ABA levels were determined by liquid chromatography–tandem mass spectrometry using [2H6]ABA (Icon Services) as an internal standard, as reported previously (Saika et al., 2007).
ChIP Assay and EMSA
For the ChIP assay, PIL5-OX3 seeds were incubated in the dark for 6 h after far-red light irradiation. The ChIP assay was performed as described previously (Tai et al., 2005; Oh et al., 2007). After reverse cross-linking, the amount of each precipitated DNA fragment was determined by real-time PCR using specific primers (see Supplemental Table 1 online).
For the EMSA, recombinant His-tagged PIL5 protein was purified using a nickel-nitrilotriacetic acid agarose column. The ability of PIL5 to bind to biotin-labeled oligonucleotide probes (Forward, 5′-AAACATAGGCACATGTGTTAGGTCTGAAGCAAGGAGAG-3′; Reverse, 5′-CTCTCCTTGCTTCAGACCTAACACATGTGCCTATGTTT-3′) was determined with the Lightshift Chemiluminescent EMSA kit (Pierce). Briefly, PIL5 protein and biotin-labeled probes were incubated in the binding buffer (10 mM Tris and 10 ng/μL polydeoxy [inosinate-cytidylate], pH 7.5) for 20 min at room temperature, and the binding mixture was loaded onto a 5% native polyacrylamide gel. Detection of biotin-labeled probe was performed according to the manufacturer's instructions.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: GA1 (At4g02780), GA3ox1 (At1g15550), GA3ox2 (At1g80340); GA2ox2 (At1g30040), GAI (At1g14920), RGA (At2g01570), RGL1 (At1g66350), RGL2 (At3g03450), RGL3 (At5g17490), PIL5 (At2g20180), ABA1 (At5g67030), ABA2 (At1g52340), NCED6 (At3g24220), NCED9 (At1g78390), AAO3 (At2g23740), CYP707A2 (At2g29090), SOM (At1g03790), UBQ10 (At4g05320), and PP2A (At1g13320).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. T-DNA Insertion Sites of som-1, som-2, and som-3.
Supplemental Figure 2. Expression Profile of SOM during Seed Maturation.
Supplemental Figure 3. Expression Levels of SOM in Three SOM-OXs.
Supplemental Table 1. Primer Sequences.
Supplemental Table 2. Relative Expression Levels of Various Genes Used for Drawing Figures.
Supplemental Data Set 1. Text File Corresponding to the Alignment in Figures 1C and 1D.
Supplementary Material
Acknowledgments
We thank Chang Seob Kwon, Walton Johnson, and other laboratory members for critical reading of the manuscript. We also thank the Arabidopsis Stock Center for sending us valuable seeds. This work was supported in part by grants from the Korea Science Engineering Foundation (Grants R0A-2007-000-20024-0, PF06302-03, and M10601000088 to G.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Giltsu Choi (gchoi@kaist.ac.kr).
Online version contains Web-only data.
References
- Alonso, J.M. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Bae, G., and Choi, G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59 281–311. [DOI] [PubMed] [Google Scholar]
- Barreau, C., Paillard, L., and Osborne, H.B. (2005). AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 33 7138–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, D., Viczian, A., Kircher, S., Nobis, T., Nitschke, R., Kunkel, T., Panigrahi, K.C., Adam, E., Fejes, E., Schafer, E., and Nagy, F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear, P.J. (2002). Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30 945–952. [DOI] [PubMed] [Google Scholar]
- Borthwick, H.A., Hendricks, S.B., Parker, M.W., Toole, E.H., and Toole, V.K. (1952). A reversible photoreaction controlling seed germination. Proc. Natl. Acad. Sci. USA 38 662–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto, J.F., Sanchez, R.A., Whitelam, G.C., and Casal, J.J. (1996). Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol. 110 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, R.S. (2005). Zinc finger proteins: Getting a grip on RNA. Curr. Opin. Struct. Biol. 15 94–98. [DOI] [PubMed] [Google Scholar]
- Cao, D., Hussain, A., Cheng, H., and Peng, J. (2005). Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223 105–113. [DOI] [PubMed] [Google Scholar]
- Carballo, E., Lai, W.S., and Blackshear, P.J. (1998). Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281 1001–1005. [DOI] [PubMed] [Google Scholar]
- Dill, A., Thomas, S.G., Hu, J., Steber, C.M., and Sun, T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger, T., Fass, D., Arnaud, M., and Harrison, S.C. (1994). Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8 970–980. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.): S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R., Reeves, W., Ariizumi, T., and Steber, C.M. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59 387–415. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S., and Goodman, H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z.L., Powers, S.J., Gong, F., Phillips, A.L., Hedden, P., Sun, T.P., and Thomas, S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T.M. (2005). Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 15 367–373. [DOI] [PubMed] [Google Scholar]
- Hudson, B.P., Martinez-Yamout, M.A., Dyson, H.J., and Wright, P.E. (2004). Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 11 257–264. [DOI] [PubMed] [Google Scholar]
- Huq, E. (2006). Degradation of negative regulators: A common theme in hormone and light signaling networks? Trends Plant Sci. 11 4–7. [DOI] [PubMed] [Google Scholar]
- Huq, E., Al-Sady, B., Hudson, M., Kim, C., Apel, K., and Quail, P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305 1937–1941. [DOI] [PubMed] [Google Scholar]
- Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Matsuoka, M., and Steber, C.M. (2003). A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trends Plant Sci. 8 492–497. [DOI] [PubMed] [Google Scholar]
- Iuchi, S., Suzuki, H., Kim, Y.C., Iuchi, A., Kuromori, T., Ueguchi-Tanaka, M., Asami, T., Yamaguchi, I., Matsuoka, M., Kobayashi, M., and Nakajima, M. (2007). Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J. 50 958–966. [DOI] [PubMed] [Google Scholar]
- Jiang, C., and Fu, X. (2007). GA action: Turning on de-DELLA repressing signaling. Curr. Opin. Plant Biol. 10 461–465. [DOI] [PubMed] [Google Scholar]
- Karssen, C.M., Brinkhorst-van der Swan, D.L.C., Breekland, A.E., and Koornneef, A. (1983). Induction of dormancy during seed development by endogenous abscisic acid: Studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157 158–165. [DOI] [PubMed] [Google Scholar]
- Kircher, S., Gil, P., Kozma-Bognar, L., Fejes, E., Speth, V., Husselstein-Muller, T., Bauer, D., Adam, E., Schafer, E., and Nagy, F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., and Nagy, F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, J.H., Yang, S.H., and Han, K.H. (2006). Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 47 343–355. [DOI] [PubMed] [Google Scholar]
- Kong, Z., Li, M., Yang, W., Xu, W., and Xue, Y. (2006). A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 141 1376–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, A., Jorna, M.L., Brinkhorst-van der Swan, D.L.C., and Karssen, C.M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 61 385–393. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., and Van der Veen, J.H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana. Theor. Appl. Genet. 58 257–263. [DOI] [PubMed] [Google Scholar]
- Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S., Asami, T., Hirai, N., Koshiba, T., Kamiya, Y., and Nambara, E. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 23 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, W.S., Carballo, E., Thorn, J.M., Kennington, E.A., and Blackshear, P.J. (2000). Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 275 17827–17837. [DOI] [PubMed] [Google Scholar]
- Lee, S., Cheng, H., King, K.E., Wang, W., He, Y., Hussain, A., Lo, J., Harberd, N.P., and Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, V., North, H., Frey, A., Sotta, B., Seo, M., Okamoto, M., Nambara, E., and Marion-Poll, A. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45 309–319. [DOI] [PubMed] [Google Scholar]
- Li, J., Jia, D., and Chen, X. (2001). HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 13 2269–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., and Thomas, T.L. (1998). PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10 383–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain, S., Allen, T., Duek, P.D., Whitelam, G.C., and Fankhauser, C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53 312–323. [DOI] [PubMed] [Google Scholar]
- Mathews, S. (2006). Phytochrome-mediated development in land plants: Red light sensing evolves to meet the challenges of changing light environments. Mol. Ecol. 15 3483–3503. [DOI] [PubMed] [Google Scholar]
- McGinnis, K.M., Thomas, S.G., Soule, J.D., Strader, L.C., Zale, J.M., Sun, T.P., and Steber, C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi, K., Okamoto, M., Koshiba, T., Kamiya, Y., and Nambara, E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 41 697–709. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., Hanada, A., Yamauchi, Y., Kuwahara, A., Kamiya, Y., and Yamaguchi, S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E., Kim, J., Park, E., Kim, J.I., Kang, C., and Choi, G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E., Yamaguchi, S., Hu, J., Yusuke, J., Jung, B., Paik, I., Lee, H.S., Sun, T.P., Kamiya, Y., and Choi, G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E., Yamaguchi, S., Kamiya, Y., Bae, G., Chung, W.I., and Choi, G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47 124–139. [DOI] [PubMed] [Google Scholar]
- Okamoto, M., Kuwahara, A., Seo, M., Kushiro, T., Asami, T., Hirai, N., Kamiya, Y., Koshiba, T., and Nambara, E. (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, E., Kim, J., Lee, Y., Shin, J., Oh, E., Chung, W.I., Liu, J.R., and Choi, G. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45 968–975. [DOI] [PubMed] [Google Scholar]
- Penfield, S., Josse, E.M., Kannangara, R., Gilday, A.D., Halliday, K.J., and Graham, I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15 1998–2006. [DOI] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400 256–261. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saika, H., et al. (2007). Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol. 48 287–298. [DOI] [PubMed] [Google Scholar]
- Schmitz, R.J., Hong, L., Michaels, S., and Amasino, R.M. (2005). FRIGIDA-ESSENTIAL 1 interacts genetically with FRIGIDA and FRIGIDA-LIKE 1 to promote the winter-annual habit of Arabidopsis thaliana. Development 132 5471–5478. [DOI] [PubMed] [Google Scholar]
- Seo, M., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48 354–366. [DOI] [PubMed] [Google Scholar]
- Shen, H., Moon, J., and Huq, E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44 1023–1035. [DOI] [PubMed] [Google Scholar]
- Shen, Y., Khanna, R., Carle, C.M., and Quail, P.H. (2007). Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Chory, J., and Furuya, M. (1994). The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol. 104 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., and Furuya, M. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderman, E.M., Brocard, I.M., Lynch, T.J., and Finkelstein, R.R. (2000). Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 124 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J., Jiang, H., Xu, Y., Li, H., Wu, X., Xie, Q., and Li, C. (2007). The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 48 1148–1158. [DOI] [PubMed] [Google Scholar]
- Sun, T., Goodman, H.M., and Ausubel, F.M. (1992). Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.P., and Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55 197–223. [DOI] [PubMed] [Google Scholar]
- Sun, T.P., and Kamiya, Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, H.H., Tai, G.C., and Beardmore, T. (2005). Dynamic histone acetylation of late embryonic genes during seed germination. Plant Mol. Biol. 59 909–925. [DOI] [PubMed] [Google Scholar]
- Talon, M., Koornneef, M., and Zeevaart, J.A. (1990). Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc. Natl. Acad. Sci. USA 87 7983–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, G.A., Carballo, E., Lee, D.M., Lai, W.S., Thompson, M.J., Patel, D.D., Schenkman, D.I., Gilkeson, G.S., Broxmeyer, H.E., Haynes, B.F., and Blackshear, P.J. (1996). A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4 445–454. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz, G., Huq, E., and Quail, P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, L., Thomas, S.G., Hu, J., Dill, A., Alonso, J.M., Ecker, J.R., and Sun, T.P. (2004). Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.Y., Hsing, Y.I., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Nakajima, M., Motoyuki, A., and Matsuoka, M. (2007). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58 183–198. [DOI] [PubMed] [Google Scholar]
- Varbanova, M., et al. (2007). Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J., Phillips, A.L., Gaskin, P., and Hedden, P. (1998). Function and substrate specificity of the gibberellin 3β-hydroxylase encoded by the Arabidopsis GA4 gene. Plant Physiol. 117 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.L., Li, L., Wu, K., Peeters, A.J., Gage, D.A., and Zeevaart, J.A. (1995). The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: Molecular cloning and functional expression. Proc. Natl. Acad. Sci. USA 92 6640–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S.A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59 225–251. [DOI] [PubMed]
- Yamaguchi, S., Smith, M.W., Brown, R.G., Kamiya, Y., and Sun, T. (1998). Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, Y., Takeda-Kamiya, N., Hanada, A., Ogawa, M., Kuwahara, A., Seo, M., Kamiya, Y., and Yamaguchi, S. (2007). Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 48 555–561. [DOI] [PubMed] [Google Scholar]
- Zentella, R., Zhang, Z.L., Park, M., Thomas, S.G., Endo, A., Murase, K., Fleet, C.M., Jikumaru, Y., Nambara, E., Kamiya, Y., and Sun, T.P. (2007). Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.