Abstract
Shoot apical meristems (SAMs) are self-sustaining groups of cells responsible for the ordered initiation of all aerial plant tissues, including stems and lateral organs. The precise coordination of these processes argues for crosstalk between the different SAM domains. The products of YABBY (YAB) genes are limited to the organ primordium domains, which are situated at the periphery of all SAMs and which are separated by a margin of three to seven cells from the central meristem zone marked by WUSCHEL and CLAVATA3 expression. Mutations in the two related YAB1 genes, FILAMENTOUS FLOWER and YABBY3 (YAB3), cause an array of defects, including aberrant phyllotaxis. We show that peripheral YAB1 activity nonautonomously and sequentially affects the phyllotaxis and growth of subsequent primordia and coordinates the expression of SAM central zone markers. These effects support a role for YAB1 genes in short-range signaling. However, no evidence was found that YAB1 gene products are themselves mobile. A screen for suppression of a floral YAB1 overexpression phenotype revealed that the YAB1-born signals are mediated in part by the activity of LATERAL SUPPRESSOR. This GRAS protein is expressed at the boundary of organ primordia and the SAM central zone, distinct from the YAB1 expression domain. Together, these results suggest that YAB1 activity stimulates signals from the organs to the meristem via a secondary message or signal cascade, a process essential for organized growth of the SAM.
INTRODUCTION
Lateral organs of plants are generated from cells at the periphery of various shoot apical meristems (SAMs). SAM zonation is complex and dynamic (Carraro et al., 2006) but can be broadly demarcated to include a central zone of pluripotent cells that divide and enter into the flanking peripheral zone where cells are recruited into the growth and differentiation programs of the lateral organs. The cells destined to form organs are defined as primordia when they constitute a distinct bulge. These cells can also be recognized prior to primordium formation, either by extrapolation from position or by the presence of biochemical and anatomical markers (Steeves and Sussex, 1989). Molecular markers similarly reveal that the remainder of the SAM is highly organized with dynamic boundaries between structural and biochemical domains (Heisler et al., 2005).
The ordered positioning of organs relative to each other, and to the main shoot axis, is termed phyllotaxis and implies that there is feedback from the initiating organs to the meristem (reviewed in Reinhardt, 2005; Kuhlemeier, 2007). This pattern is highly conserved within a species, yet it can change dramatically during different developmental stages. For example, spiral initiation of leaves and bracts in the vegetative meristem (VM) and inflorescence meristem (IM) of Arabidopsis thaliana is followed by whorled initiation of floral organs in the flower meristem (FM). Various classical experiments and mathematical modeling led to fields of inhibition models for phyllotaxis (Snow and Snow, 1931; Mitchison, 1977; Veen and Lindenmayer, 1977). These models argue that the sum of the effects of inhibitory compounds secreted from emerging primordia prevent formation of a new primordium nearby (Steeves and Sussex, 1989). In this study, we will term such signals “sequential,” as the primordium initiates a signaling cascade that spreads and influences subsequently formed primordia. Recent studies suggest that auxin maxima specify the position of new anlagen, while surrounding regions are depleted of auxin and do not initiate primordia. In these models, anlagen and primordia factors might influence phyllotaxis through modulation of their auxin sink potential (Reinhardt et al., 2000, 2003; Jönsson et al., 2006; de Reuille et al., 2006; Smith et al., 2006).
A limited amount is understood about the molecular mechanisms governing SAM patterning and the organized, phyllotactic production of lateral organs. The phyllotactic pattern argues for sequential signaling to the meristem by organ primordia, which then specify specific groups of cells for primordium allocation. Organ-to-meristem signaling processes may also play a broader role in regulating the molecular zonation of the meristem as a component of coordinated meristem growth. A number of mutations have been identified in which changes in meristem size coincide with altered phyllotaxis, consistent with a link between these two processes. Generally, an increase in the size of the apical meristem accommodates more primordia. For example, in the abphyl1 mutant of maize (Zea mays), the transition of distichous to decussate phyllotaxis is associated with an increase in meristem size (Jackson and Hake, 1999). Likewise, in the Arabidopsis clavata (clv), fasciata, wuschel (wus), pennywise, and jabba-1D mutants, where the circumference of the meristem is altered, phyllotaxis also is disrupted and becomes irregular (Leyser and Furner, 1992; Clark et al., 1993; Laux et al., 1996; Smith and Hake 2003; Williams et al., 2005). The affected genes in these mutants are expressed at the center of the meristem, although not necessarily exclusively, suggesting that central zone expressed genes that are involved in maintaining SAM size are also involved in the establishment or maintenance of phyllotaxis (Giulini et al., 2004; Reinhardt, 2005). This implies that central zone factors may be targets of the primordia-derived signals. It could also mean that physical parameters, such as the number of cells the signal must traverse, constrain the effective range or direction of inhibitory fields or auxin flux regulating phyllotaxis. Short-range communication between adjacent cells has been shown to play an important role in the formation of the radial patterning of the Arabidopsis root. In this case, translocation of the SHORT ROOT protein into an adjacent cell layer and the activation of its target SCARECROW was demonstrated (Nakajima et al., 2001). Whether a similar type of communication exists between the primordia, peripheral zone, and central zone for proper apical meristem patterning and growth homeostasis is unknown. Analysis of anlagen and primordial factors that affect phyllotaxis and SAM zonation is essential to approach this question.
A small group of HMG-like proteins found in seed plants and termed YABBY (YAB) after their founding member CRABS CLAW is made up of five basic members, some of which recently underwent duplication in different lineages (Yamada et al., 2004; Lee et al., 2005). Two members of this family have been shown to have overlapping functions in patterning of all lateral organs in Antirrhinum majus (Golz et al., 2004). For the sake of clarity, we will refer to the proteins most closely related to these as members of the YAB1 or YAB5 groups. The YAB1 group comprises both FILAMENTOUS FLOWER (FIL) and YAB3 in Arabidopsis but only GRAMINOFOLIA (GRAM) in Antirrhinum (Navarro et al., 2004; Golz et al., 2004). The YAB5 group includes YAB5 in Arabidopsis and PROLONGATA in Antirrhinum. Expression analyses indicated that transcripts of all Arabidopsis family members are restricted to lateral organ primordia and excluded from the central region of SAMs (Bowman and Smyth, 1999; Eshed et al., 1999; Sawa et al., 1999b; Siegfried et al., 1999; Villanueva et al., 1999; Watanabe and Okada, 2003). YABBY expression is also generally restricted to abaxial domains in lateral organs (Siegfried et al., 1999; Golz et al., 2004; Navarro et al., 2004), although some specific extensions have been reported (Bowman and Smyth, 1999; Juarez et al., 2004; Yamaguchi et al., 2004; Lee et al., 2005). However, in no case have YABBY transcripts been detected in the inner domains of the SAM.
While YAB1 RNA is restricted to the SAM periphery in flowering plants, the loss of YAB1 alone or in combination with the loss of YAB5 activity results in phyllotactic alterations (Chen et al., 1999; Sawa et al., 1999a; Siegfried et al., 1999; Figures 1A to 1H) and even SAM maintenance failure (Golz et al., 2004). In addition, leaves with reduced YAB1 activity develop ectopic SAMs, indicating that YAB1 activity prevents expression of SAM programs in the developing primordia (Kumaran et al., 2002). Thus, YAB1-mediated non-cell-autonomous signaling appears to contribute to both phyllotaxis maintenance and boundary establishment in the SAM periphery. In this study, we show that YAB1 genes can indeed stimulate short-range signals when expressed from the primordia domain of the meristem periphery and that neither YAB1 RNA nor protein is mobile; therefore, neither is likely to constitute the signal. YAB1-induced signals sequentially maintain wild-type phyllotaxis, and the response to their absence involves dramatic changes in the expression of the meristem center markers WUS and CLV3, illustrating that central SAM genes are targets of this signaling pathway. Finally, we show that these signals are mediated by the activity of LATERAL SUPRESSOR, a GRAS protein expressed at the boundary of organ primordia and the central meristem domain. Together, these results suggest that YAB1 proteins act to define the organ primordia domain and stimulate signals necessary for the dynamic partitioning of SAMs.
Figure 1.
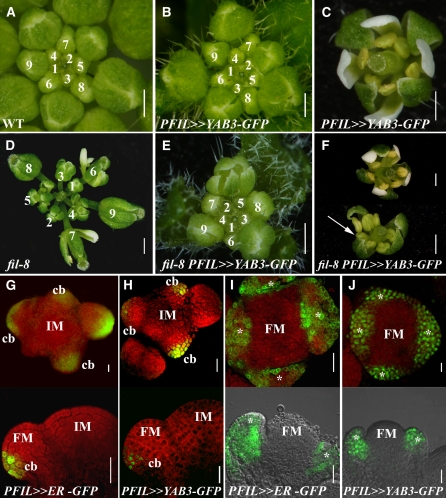
Loss of YAB1, Which Is Expressed in Organ Primordia, Results in Aberrant Phyllotaxis.
(A) and (B) Recognizable domains within floral Arabidopsis apices. (A) shows the side view, and (B) shows the top view. Yellow color marks the SHOOTMERISTEMLESS expression domain, and green marks cryptic bract primordia. The youngest primordium is marked as p1 and later ones as p2, p3, etc. Incipient primordia are labeled i1, i2, etc. The red color marks the organ primordia boundary, the purple color marks flower domains differentiating into sepals and pedicels, and blue color marks the flower B class domain.
(C) Wild-type inflorescence shoot. Flowers and siliques are produced in spiral phyllotaxis with progressive, even, internodal growth between them.
(D) and (E) Disrupted phyllotactic patterns in the fil-8 (D) and fil-8 yab3-2 (E) mutant shoots showing clustered (arrows) or dispersed flowers positioned at variable distances and angles relative to each other.
(F) to (H) Phyllotaxis of IMs. Scanning electron micrographs of wild-type (F), fil-8 mutant (G), and fil-8 yab3 double mutant (H) IMs producing flowers and bract-like structures. Numbers represent phyllotactic order of developing primordia: 0 marks oldest incipient primordia, and 1 marks first morphologically detectible primordia (p1). In fil-8 and fil-8 yab3 mutants ([G] and [H]), relative positions of adjacent primordia, marked by two-headed arrows, are different from relative positions of same primordia pairs in wild-type apex (F).
(I) and (J) FIL mRNA distribution. Transverse (I) or longitudinal (J) sections of apices from wild-type flowering plants. Expression is limited to the organ domain, either cryptic bracts (cb) at IM periphery or sepal primordia in stage 3 flowers.
(K) to (M) Phyllotaxis of FMs. Aerial-view, scanning electron micrographs of wild-type (K), fil-8 mutant (L), and fil-8 yab3 double mutant (M) flowers. (K) shows the stereotypic organization of a stage 3 to 4 wild-type flower, and disrupted phyllotactic patterns of organ initiation can be seen in the mutant flowers ([L] and [M]). Asterisks mark sepal primordia and f marks filamentous organs.
Bars = 1 mm in (C) to (E), 100 μm in (A), (B), and (F) to (J), and 50 μm (K) to (M).
RESULTS
Disruption of YAB1 Expression in Organ Primordia Alters Phyllotaxis
The vegetative and inflorescence SAMs of Arabidopsis produce lateral appendages in a stereotypic spiral phyllotaxis approximating the classical 137°. The angle between, shape, size, and identity of these appendages changes with age: from opposite small juvenile leaves to spiral large rosette ones produced by the VM, and from narrow cauline leaves subtending the elongated flowering shoot to miniature modified leaves (termed cryptic bracts) that subtend flowers produced by the IM (Figures 1A and 1B; Long and Barton, 2000). Subsequently, in the FM, the phyllotaxis of the organs, sepals excluded, becomes whorled. FIL and YAB3, together termed here YAB1, are expressed in the lateral primordia derived from all of these apical meristems. In this study, we focused on the effects of YAB1 activity in the inflorescence and flower apices. In the wild type, the regular, spiral phyllotaxis of cryptic bracts leads to regular angles between adjacent siliques on the mature stem (Figures 1A to 1C). By contrast, fil-8 or fil-8 yab3-2 mutant plants exhibited irregular phyllotaxis as evidenced by variable angles between mature flowers (Figures 1D and 1E) or initiating bract primordia (marked by arrows in Figures 1F to 1H; Chen et al., 1999; Sawa et al., 1999a). These phyllotactic defects were incongruent with FIL or YAB3 mRNA distribution, which was restricted to cryptic bract primordia, marking cells destined to leave the SAM (Figures 1I and 1J; Sawa et al., 1999b; Siegfried et al., 1999; Watanabe and Okada, 2003; Dinneny et al., 2004; Heisler et al., 2005).
In the Arabidopsis flower, four sepals, four petals, six stamens, and two carpels are organized in four concentric whorls (Figure 1K and schematically marked in Figures 1A and 1B). Irregular phyllotaxis in floral apices characterizes fil-8 and fil-8 yab3-mutants, which exhibited improperly positioned organ primordia and variable numbers of organs. In addition, they had poorly defined whorl boundaries, resulting in chimeric or radial organs (Figures 1D, 1E, 1L, and 1M; Chen et al., 1999; Sawa et al., 1999a). Thus, in both IM and FM peripheries, the primordial-restricted YAB1 RNA is involved in developmental decisions at the central meristem domain.
Peripheral YAB1 Activities Regulate Expression at the Central Meristem Domain
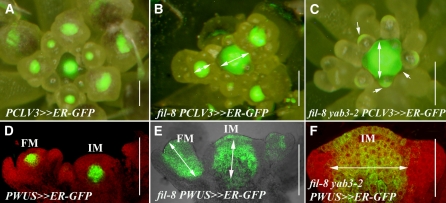
The observation that a reduction in YAB1 expression at the SAM periphery altered organ positioning suggests that YAB1 activities pattern the meristem. Therefore, the expression of central meristem growth regulator markers, PCLV3≫ER-GFP or PWUS≫ER-GFP, where the driver lines transactivated an endoplasmic reticulum (ER) targeted green fluorescent protein (GFP), were examined in wild-type and yab1 mutant apices. In wild-type IMs, fluorescence of the PCLV3≫ER-GFP marker demarcated a broad disc-shaped area comprised of the two to three uppermost cell layers and is excluded from the peripheral region of the SAM, similar to reported CLV3 RNA distribution (Fletcher et al., 1999). Within floral meristems, expression was initiated at stage 2 flowers and maintained at the center of the FM (Figure 2A). When YAB1 activities were compromised, as in the fil-8 or fil-8 yab3-2 mutants, PCLV3≫ER-GFP expression in the IM expanded into the initiating cryptic bract primordia. In FMs, delayed expression was evident in flowers that initiated organs, whereas filamentous structures that likely represented radial bracts maintained brief abaxial expression (Figures 2B and 2C).
Figure 2.
Loss of YAB1 Expression in Organ Primordia Stimulates Altered Distribution of Meristem-Specific Markers.
(A) to (C) Aerial view of inflorescences expressing the PCLV3≫ER-GFP marker. The green fluorescent signal is superimposed on the light image viewed by a stereoscope. Wild-type (A), fil-8 (B), and fil-8 yab3-2 (C) apices are shown. White two-headed arrows mark outward expansion of PCLV3≫ER-GFP expression in the mutants compared with the wild type, and arrows mark abaxial expression in fil-8 yab3-2 filaments.
(D) to (F) Longitudinal sections through inflorescences expressing the PWUS≫ER-GFP marker. In the wild type (D), the PWUS≫ER-GFP signal marks a limited domain above the rib meristem. In the fil-8 mutant apex (E), PWUS≫ER-GFP expression has considerably expanded in both IM and FM. In fil-8 yab3-2 mutant apices (F), it has expanded in the IM, while filamentous flowers fail to initiate PWUS≫ER-GFP expression. White two-headed arrows mark expansion of marker expression in the mutants compared with the wild type.
Bars = 100 μm in (A) to (E) and 50 μm in (F).
PWUS≫ER-GFP was expressed in the center of the wild-type IMs and FMs, below the L3 cell layer in the IM and at the L3 in developing flowers, matching the described WUS RNA distribution (Figure 2D; Leibfried et al., 2005). In fil-8 or fil-8 yab3-2 mutant apices, a dramatic expansion in the expression domain was observed in both IMs and FMs. This included expansion into the L1 and L2 layers as well as to internal layers below the rib meristem (Figures 2E and 2F). Thus, altered YAB1 expression outside the meristem central domain stimulates a dramatic change in the expression patterns of genes regulating meristem activity in the central region.
YAB1 Molecules Are Not Detectibly Mobile
YAB1 non-cell-autonomous signals may be mediated by short-range, cell-to-cell protein, or RNA trafficking (Gallagher and Benfey, 2005). To test if YAB1 RNA or protein was the signal, we followed in planta YAB1 distribution by driving either FIL or YAB3 cDNA in either native or GFP-tagged forms using the 5′ promoter of FIL. Wild-type plants expressing any of the PFIL≫YAB1 constructs had normal patterns of flower and floral organ positioning (Figures 3A to 3C), but when strong driver lines were used, had slightly narrower curling leaves, sepals, and petals (see Supplemental Figures 1A and 1B online).
Figure 3.
Autonomous Distribution of Functional GFP-Tagged YAB1 Proteins.
(A) to (F) Complementation of fil-8 by GFP-tagged YAB1. Inflorescences ([A], [B], [D], and [E]) and flowers ([C] and [F]) are shown from wild-type ([A] to [C]) and fil-8 mutant ([D] to [F]) plants with ([B], [C], [E], and [F]) or without ([A] and [D]) PFIL≫YAB3-GFP. The numbers mark the order of flower initiation and outline the phyllotactic pattern. Note the partial complementation in older flowers (arrow in [F]).
(G) to (J) GFP fluorescence activated by the FIL promoter. Sections through inflorescences ([G] and [H]) and flowers ([I] and [J]) showing the distribution of fluorescence emitted from the ER-localized GFP ([G] and [I]) or from GFP-tagged YAB1 ([H] and [J]) reporter lines driven by the same FIL promoter line. The top rows of images are transverse sections, and the bottom rows are longitudinal sections, each at different magnifications. All apices were counterstained by propidium iodide (PI), excluding the longitudinal sections in (I) and (J). The asterisk indicates the sepal primordia. cb, cryptic bract.
Bars = 1 mm in (A) to (F) and 20 μm in (G) to (J).
All versions of FIL or YAB3 proteins, whether native or GFP tagged, partially rescued the fil-8 mutant when expressed by the FIL promoter (Figures 3D to 3F; see Supplemental Figures 1C to 1E online; additional comparisons between various FIL and YAB3 responder lines expressed by other promoters are shown in Supplemental Figure 2 online). These results support the functional similarity between the YAB1 genes FIL and YAB3 and the functional equivalence of their GFP-tagged reporter lines to their nontagged versions. Further analyses of YAB1-GFP localization were performed with responder lines that complemented fil mutants, while the phenotypic effects of overexpression were monitored in independent single insertion lines consistently producing phenotypes ranging from weak to strong (see Supplemental Figure 2 online).
The distribution of fluorescence induced by activation of YAB1-GFP proteins in floral shoot apices by the PFIL:LhG4 driver line was compared with that emitted from cell autonomous GFP reporters with either ER or nuclear localization signaling (NLS) peptides. In all cases, identical patterns were observed (Figures 3G to 3J; see also Figures 4J and 4K). All fluorescence patterns approximated the RNA expression pattern of FIL in the cryptic bract primordia and floral organs, although in some cases initial fluorescence was not abaxially restricted (Figures 3G to 3J). Significantly, YAB1-GFP fluorescence was not detected in cells that did not express the cell autonomous GFP reporter.
Figure 4.
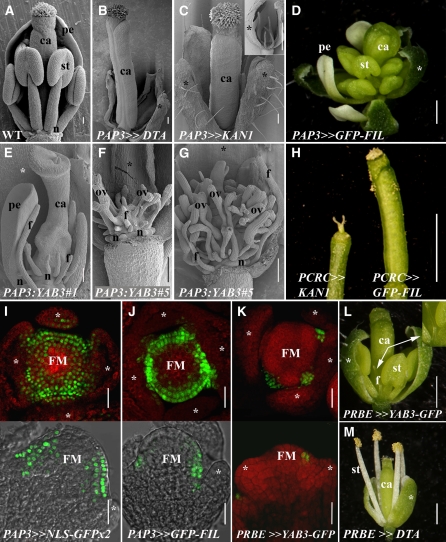
Autonomous YAB1 Expression Initiates Morphogenic Cues.
(A) to (C) Scanning electron micrographs of near-anthesis flowers. Wild-type flowers (A) have a stereotypic phyllotaxis (three sepals and two petals were removed), while PAP3≫DTA flowers ([B]; one sepal removed) have arrested petal and stamen development. PAP3≫KAN1 flowers (C) lack petals and have radialized stamens, as also illustrated in the inset (here, two sepals and the gynoecium have been removed).
(D) to (H) Flowers with graded effects of ectopic YAB1 expression. Flowers of a weak PAP3≫FIL-GFP line (D) have normal petals and an excess of stamens and carpels. An intermediate (E) and a strong ([F] and [G]) line of PAP3:YAB3 have abnormal, filamentous organs that occupy the 2nd and 3rd whorls in addition to naked placental mounds bearing malformed ovules at the center of the flowers. Sepals but no other organs were removed. The effects of PAP3:YAB3 on carpel development are likely sequential, as ectopic YAB1 expression throughout carpel primordia, as in PCRC≫GFP-FIL, caused minor defects only ([H], right) by sharp contrast with the miniature, abnormal carpels of CRC≫KAN1 plants (left).
(I) to (K) GFP fluorescence in floral apices. Distribution of the NLS-GFPx2 (I) or GFP-FIL (J) fluorescent reporters transactivated by the PAP3:LhG4 driver line in stage 4 to 5 flowers. Top images are transverse sections, and bottom images are longitudinal. (K) shows the specific distribution in petal primordia of YAB3-GFP expressed by the PRBE:LhG4 driver.
(L) and (M) Petal-specific responses. PRBE≫YAB1-GFP flowers (L) have small radial petals (arrows and inset), while other organs are normal. Similarly, PRBE≫DTA flowers (M) lack petals.
Asterisks indicate sepals. FM, floral meristem; pe - petals; st, stamens; ca, carpels; f, filaments; n, nectaries; ov, ovules. Bars = 100 μm in (A) to (C), (E), and (G), 1 mm in (D), (H), (L), and (M), and 20 μm in (I) to (K).
These observations demonstrate that YAB1 molecules are not detectably mobile. Therefore the non-cell-autonomous YAB1 effects on the meristem may be via a secondary messenger or a signaling cascade initiated from the domain expressing YAB1.
Ectopic YAB1 Expression in the 2nd and 3rd Floral Whorls Stimulates Nonautonomous, Sequential Effects on the 4th Whorl
The simplest model to explain the non-cell-autonomous YAB1 effects is through short-distance trafficking of the YAB1 gene products. Our inability to detect movement of YAB1 gene products cannot exclude limited translocation of morphogenic molecules into central meristem cells, a region responsive to ubiquitous YAB1 expression (Sawa et al., 1999b; Siegfried et al., 1999). We therefore established an experimental system to characterize YAB1-born signals in greater details. Effects of YAB1 ectopic expression were first monitored using the B class floral driver APETALA3 (AP3). Expression from a 500-bp AP3 promoter initiates immediately after sepal primordia emergence, in a ring spanning the 2nd and 3rd floral whorls, and is maintained later in the developing petals and stamens (Tilly et al., 1998). The use of this promoter allowed the bypassing of lethality constraints encountered with ubiquitous promoters and produced ectopic YAB1 activity in a discrete domain. In control plants, promoter PAP3≫DIPHTHERIA TOXIN A (DTA; Collier, 1975; Pappenheimer, 1977) and promoter PAP3≫KANADI1 (KAN1), petal initiation was abolished and the formation of normal gynoecia followed the production of no stamens in PAP3≫DTA or four filamentous stamens in PAP3≫KAN1 (Figures 4A to 4C; Day et al., 1995; Pekker et al., 2005). By contrast, the gynoecia as well as the number, position, and shape of petals and stamens were modified in PAP3≫YAB1 flowers (Figures 4D to 4G; see Supplemental Table 1 online).
Notably, petals were the most resistant primordia to the YAB1 misexpression meditated by the AP3 promoter. By contrast, increased numbers of stamens and carpels were evident in a weak PAP3≫GFP-FIL line that maintained normal petal morphology (Figure 4D; see Supplemental Table 1 online). Progeny from the self-cross of this line that were homozygous for the transgenes or strong lines of YAB1 transcriptionally fused with the AP3 promoter (PAP3:YAB3) exhibited more severe phenotypes. Flowers with miniature radial stamens, disrupted ovary walls, and a proliferation of a central placental mound carrying abnormal naked ovules were produced (Figures 4E to 4G). These observations are in sharp contrast with effects stimulated by other abaxial promoting factors, such as KAN and miR165, where dramatic effects on organs were restricted to the manipulated domain (Figure 4C; Alvarez et al., 2006). This difference was further emphasized by comparison of the carpels of PCRC≫KAN1 line with those of PCRC≫YAB1 (Figure 4H). An early dramatic arrest of carpel development was stimulated by KAN1, whereas the effects of YAB1 were minor. Thus, the dramatic effects of PAP3≫YAB1 activity on carpels suggests a nonautonomous mechanism initiated by YAB1 activity that sequentially impacts the inner meristem and initiating organs.
Nonautonomous YAB1 Effects in Flowers Are Stimulated by Autonomous Interorgan Expression
The dramatic non-cell-autonomous effects observed in PAP3≫YAB1 flowers could reflect some unexpected feedback effects of the transgene on the promoter expression domain. The expression of PAP3≫NLS-GFP and PAP3≫GFP-YAB1 was therefore compared. In PAP3≫NLS-GFP flowers, the earliest fluorescence was detected as a ring in stage 3 flowers just internal to sepal primordia. The ring of fluorescent signal was maintained until after stamen initiation and was not observed to expand into the center of the FM or into the carpel primordia (Figure 4I). A similar pattern was observed in PAP3≫GFP-YAB1 plants, although in many cases, the ring of fluorescent cells was narrower, likely reflecting the altered flower development of these lines (Figure 4J).
The differences between endogenous YAB1 expression and the expression originating from the AP3 promoter occurred in the meristem periphery at the 2nd and 3rd floral whorl boundaries and in the adaxial domains of the developing petal and stamen primordia. To further define this domain with respect to nonautonomous YAB1 signals, we used the RABBIT EARS (RBE) promoter (Takeda et al., 2004), which is active throughout petals but not between organs (Figure 4K). In strong PRBE≫YAB1 lines, the phenotype was restricted to the petals, which developed as radial filaments (Figure 4L). From our analyses, the RBE and AP3 promoters were activated at the same time in stage 3 flowers (cf. Figures 4J and 4K) yet stimulated different effects upon YAB1 transactivation. Expression from the RBE promoter was restricted to cells that would solely give rise to petals as confirmed by the RBE-mediated DTA expression, which resulted in the arrest of petal development only (Figure 4M). Thus, YAB1 expression from an interorgan domain mediated by promoter AP3 likely accounted for the nonautonomous carpel phenotypes seen in PAP3≫YAB1 flowers. These observations demonstrated that organ-specific YAB1 expression could dramatically change autonomous development without altering floral phyllotaxis.
Sequential Effects of a Domain-Specific YAB1 Reduction by MicroRNA
Expanded YAB1 expression demonstrates the potential of these genes to stimulate short-range signals. Investigating the source and range of such signals in loss-of-function mutant lines of YAB1 is difficult because of developmental epistasis. To investigate the effects of specific and regulated elimination of YAB1 activities, an artificial microRNA (miRNA) targeting both FIL and YAB3 mRNAs (amiR-YAB1) was designed (see Alvarez et al., 2006 and Schwab et al., 2006 for design principles and Supplemental Figure 3 online for actual design) and introduced into plants. Plants expressing amiR-YAB1 under the control of the AINTEGUMENTA promoter (PANT≫amiR-YAB1 plants) were indistinguishable from fil-8 yab3-2 plants (Figures 5A to 5D; Siegfried et al., 1999), confirming the specificity and potency of amiR-YAB1. Expressing amiR-YAB1 under the control of the PAP1:LhG4 driver line allowed the removal of YAB1 activities from initiating flowers while maintaining normal activity in their hosting cryptic bract (the cryptic bract does not express promoter AP1 but expresses promoter ANT; see Supplemental Figure 4A online; Long and Barton, 2000; Grandjean et al., 2004). Thus, in fil-8 yab3-2 and PANT≫amiR-YAB1 plants, FMs arise in the axils of cryptic bracts lacking YAB1 activities, whereas in PAP1≫amiR-YAB1, FMs arise form in the axils of cryptic bracts with normal YAB1 activity. The phyllotactic patterns of flowers in PAP1≫amiR-YAB1 inflorescences remained the same as in the wild type (cf. Figures 1A and 1B with 5E); all flowers were initiated normally, and no bracts or filamentous flowers were generated (Figure 5E). By contrast, fil-8 yab3-2 and PANT≫amiR-YAB1 plants exhibited disrupted flower initiation phyllotactic patterns (Figures 5A to 5D). In addition, the formation of filamentous flowers (Siegfried et al., 1999; Figure 5C) and cryptic bract outgrowth were common to both types of gene knockdown plants (Figures 5B and 5D). Notably, when flowers of fil-8 yab3-2 or PANT≫amiR-YAB1 were initiated, they were almost identical to those of PAP1≫amiR-YAB1 plants (cf. Figures 5F to 5G with 5D). The only difference was a slightly higher number of floral organs in PAP1≫amiR-YAB1 flowers, including a greater number of sepals (see Supplemental Figures 3C to 3G and Supplemental Table 1 online).
Figure 5.
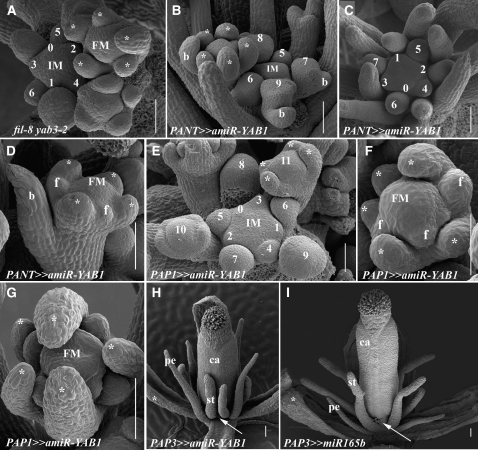
Effects of a Domain-Specific Reduction in Floral YAB1 Activity Generated by an Artificial miRNA.
(A) to (C) Scanning electron micrographs of phyllotactic patterns in apices lacking YAB1 activity in bracts. Disrupted phyllotactic patterns and abnormal flower initiation are seen either in fil-8 yab3-2 (A) or PANT≫amiR-YAB1 inflorescences ([B] and [C]).
(D) PANT≫amiR-YAB1 flower with reduced numbers and altered positions of organ primordia.
(E) PAP1≫amiR-YAB1 inflorescences have normal flower initiation phyllotaxis.
(F) and (G) Phyllotactic patterns in PAP1≫amiR-YAB1 flowers. Disrupted numbers and positions of floral organs are evident (cf. Figure 1K).
(H) Mature PAP3≫amiR-YAB1 flower. The radial petals are formed at the correct position, whereas the adjacent medial stamens are closer to each other than in wild-type or PAP3≫miR165 flowers (cf. stamen proximity marked by the arrows in [H] and [I]).
(I) Mature PAP3≫miR165 flower. The radial petals and stamens are formed in the correct positions.
Asterisks indicate the sepal. Numbers represent phyllotactic order. b, bract; f, filament; pe, petal; st, stamen. Bars = 50 μm in (A) to (F) and 100 μm in (G) to (I).
We next examined the effect of reducing YAB1 activity exclusively in the B class domain and compared the defects of PAP3≫amiR-YAB1 flowers with those of PAP3≫miR165 (Figures 5H and 5I; Alvarez et al., 2006). In both cases, four filamentous petals and six filamentous stamens were formed, as determined by organ position. However, in PAP3≫miR165 flowers, petal filaments were smaller then stamen filaments (Figure 5I), while this relationship were reversed in PAP3≫amiR-YAB1 (Figure 5H). In addition, the spacing of stamen filaments was abnormal in PAP3≫amiR-YAB1, where medial ones were closer to each other (see arrows in Figures 5H and 5I). Notably, expressing YAB1 in the B domain only, as in fil8 yab3-2 PAP3:YAB3 plants, failed to stimulate normal initiation of petals while still promoting a non-cell-autonomous gynoecium defect (see Supplemental Figure 3H online). Thus, patterning defects stimulated by the absence of YAB1 are most evident in primordia formed immediately following YAB1 elimination.
An Organ Boundary Gene Mediates YAB1-Derived Signals
The consequences of the loss or directed overexpression of YAB1 activity suggest that YAB1 proteins perform non-cell-autonomous functions but do not act as mobile molecules themselves. To genetically characterize the YAB1-born signaling network, a screen for second-site suppressors of the increased floral organ number was performed in the background of hemizygous PAP1:YAB3 plants (Figure 6A). In PAP1:YAB3 plants, sepals were generally normal, while the effects on the subsequent flower whorls were dramatic. (see Supplemental Figures 2 and 4 and Supplemental Table 1 online). In weak lines, a large increase in the number of petals, stamens, and carpels was observed. More severe lines or homozygous progenies of the weaker lines initiated numerous petals that subsequently failed to expand and mature normally, whereas stamen and carpel initiation was completely abolished (see Supplemental Figures 4E to 4H online).
Figure 6.
The LAS Gene Mediates YAB1-Derived Signals.
(A) to (F) Effect of the las-11 mutant on flowers with altered levels of YAB1 activity.
(A) PAP1:YAB3 flower with one sepal removed revealing additional petals, additional stamens, and disrupted gynoecium development.
(B) las-11 PAP1:YAB3 flower with a nearly normal number of petals and stamens (one sepal removed).
(C) PAP3:YAB3 flower with additional stamens and disrupted gynoecium development (one sepal removed).
(D) las-11 PAP3:YAB3 flower with a near wild-type appearance (one sepal removed).
(E) fil-8 inflorescence.
(F) las-11 fil-8 inflorescence, comprising primarily filamentous flowers.
(G) to (R) Distribution of PLAS≫LAS-GFP fluorescence. Transverse ([G] and [I]) and longitudinal ([H] and [J]) sections through LAS≫LAS-GFP inflorescences ([G] and [H]) and flowers ([I] and [J]). Transverse section of a fil-8 PLAS≫LAS-GFP inflorescence (K) and longitudinal section of a flower (L) with expression between the floral organs and the FM (b, bract). Transverse section of a fil-8 yab3-2 PLAS≫LAS-GFP inflorescence (M) and longitudinal section of a flower (N) with almost no expression between the sepal and the FM. Transverse section of a PAP1:YAB3 PLAS≫LAS-GFP inflorescence (O) and longitudinal section of a flower (P) with significant expansion of the LAS≫LAS-GFP expression domain marked by arrows. Longitudinal section through stage 4 wild-type (Q) and PAP3:YAB3 (R) flowers, where expansion of PLAS≫LAS-GFP expression is marked with arrows.
(S) A model of IM expression territories. A representation of the expression domains of genes examined in this study and the direction of influence (arrows) established or speculated based on the results. PLAS≫LAS-GFP signal is present, while the others have been painted in as representative domains. Primordial YAB1 activity (pink) nonautonomously communicates with the floral (FM) and inflorescence (IM) meristems and regulates the expression of CLV3 (purple) and WUS (yellow). LAS, acting at the organ-meristem boundary (bou), mediates this signaling process. cb, cryptic bract.
Asterisks indicate the sepal. Numbers represent the order of flower initiation. Bars = 1 mm in (A) to (F) and 50 μm in (G) to (R).
Three independent suppressors of the PAP1:YAB3 phenotype were identified as new alleles of LATERAL SUPPRESSOR (LAS; Figure 6B). LAS RNA is found in all boundaries between lateral organ primordia and the SAM from which they originated (Greb et al., 2003). Notably, this LAS expression domain does not overlap with that of YAB1, and las defects were limited to rosette axillary meristem production (see Supplemental Figures 5A and 5B online). To examine the role of LAS in non-cell-autonomous YAB1 signals, all new las mutant alleles were crossed to PAP3:YAB3. In the las background, suppression of the nonautonomous PAP3:YAB3 gynoecia defects was evident (Figures 6C and 6D). Significantly, the las-11 mutant strongly enhanced fil-8 inflorescence and flower defects (Figures 6E and 6F). Numerous filaments arose instead of flowers in the fil-8 las-11 double mutant, and the flowers typically lacked 2nd and 3rd whorl organs, reminiscent of fil yab3-2 flowers.
We monitored LAS expression under conditions of altered YAB1 activity by analysis of a functional PLAS≫LAS-GFP (that complemented the las-11mutant; see Supplemental Figure 5C online). In the wild type, the expression of this marker matched the reported distribution of LAS mRNA and did not overlap with the YAB1 expression domains (Figures 6G to 6J and 6Q; Greb et al., 2003). The same basic pattern of PLAS≫LAS-GFP signal was maintained in fil-8, but prolonged expression was detected between the IM and FMs of fil-8 inflorescences (cf. Figures 6G and 6K), followed by delayed, though spatially normal, expression within fil-8 flowers (cf. Figures 6J and 6L). By contrast, a dramatic reduction in PLAS≫LAS-GFP expression was observed in fil-8 yab3-2 flowers, with a milder reduction observed at the fil-8 yab3-2 IM-FM boundary (Figures 6M and 6N). Thus, expression of the LAS marker is strongly altered in yab1 mutant apices even though it does not overlap with detectable YAB1 gene products.
When YAB1 was present ectopically, as in PAP1:YAB3 flowers, the expression of the meristem markers PCLV3≫ER-GFP and PWUS≫ER-GFP was dramatically altered (see Supplemental Figures 4I to 4M online). Likewise, the PLAS≫LAS-GFP expression domain also exhibited an early and dramatic change. PLAS≫LAS-GFP expression in stage 2 to stage 6 flowers expanded into the entire FM, while normal expression at the FM-IM boundary was maintained (Figures 6O and 6P). PLAS≫LAS-GFP expression was also broader in PAP3:YAB3 flowers, where the normal one to two cell layer boundary between the FM and sepals marked by PLAS≫LAS-GFP expanded to four to seven cells (Figures 6Q and 6R). Thus, altered YAB1 expression in organ primordia or in the meristem periphery stimulated dramatic alterations in the expression of a meristem boundary gene.
DISCUSSION
The Multiple Functions of the YAB Genes
The expression of the YAB genes in the abaxial side of lateral organs is important for establishment of abaxial cell fates (Eshed et al., 1999; Siegfried et al., 1999), promotion of lateral organ growth and expansion (Bowman and Smyth, 1999; Villanueva et al., 1999; Eshed et al., 2004; Golz et al., 2004), and suppression of SAM gene activity in the organ domain (Kumaran et al., 2002). Analysis of fil mutant plants suggested involvement of the YAB1 genes in phyllotaxis regulation and signaling (Chen et al., 1999; Sawa et al., 1999a, 1999b; Siegfried et al., 1999). In this study, this function was studied in greater detail. Our analysis suggests that cell-autonomous YAB1 proteins in the lateral organs stimulate a non-cell-autonomous, centripetal signal (or, possibly, signals) to help maintain robust meristem organization and regular phyllotaxis. In addition, the demonstration that the organ-meristem boundary gene LAS mediates the role of YAB1 in organ initiation suggests that LAS and potentially other factors in this domain help transduce these peripheral, YAB1 organ-based signals to the meristem.
The abaxial expression pattern, typical of all Arabidopsis YAB genes, and their mutant phenotypes led to their classification as abaxial-promoting factors (Eshed et al., 1999; Sawa et al., 1999b; Siegfried et al., 1999; Bowman et al., 2002). Here, we have shown that Arabidopsis plants respond to altered YAB1 activities differently than to the alteration of other abaxial factors, such as KAN or miR165/6 (Figure 4). While the effects of ectopic of KAN or miR165 were cell autonomous, ectopic YAB1 expression was accompanied by sequential effects on the meristem. We therefore propose that YAB1 proteins have multiple functions that depend on different cellular contexts during the different phases of organ development at which they operate.
During the early stages of primordium initiation, the PI phase (Poethig and Sussex, 1985), YAB1 activities participate in marking the primordium domain, and are the source of a signal that patterns the adjacent meristem peripheral and central zones. Since no evidence was found for Arabidopsis YAB1 RNA or protein trafficking (Figures 3G to 3J), it is most probable that YAB1 generates a secondary messenger or initiates a signaling cascade to mediate this non-cell-autonomous effect. Once organ primordia have initiated and organ polarity has been established, YAB1 activities are recruited to promote laminar growth (Eshed et al., 2004; Golz et al., 2004), a function reflected in postinitiation YAB1 expression, which is highly correlated with extensive cell divisions. These functions have some overlap with the growth stimulation mediated by AINTEGUMENTA, as evident in their combined loss of function (Nole-Wilson and Krizek, 2006). Lastly, recruitment of YAB1 for the specification of abaxial cell types was evident by the altered specification of adaxial epidermis in P35S:FIL cotyledons (Sawa et al., 1999b; Siegfried et al., 1999). While the polarity role is shared by other YAB genes, such as CRC (Eshed et al., 1999), it may be the least conserved function of YAB gene activity in diverse angiosperm species (Juarez et al., 2004). Indeed, in Arabidopsis, all studied YAB genes exhibit non-cell-autonomous activities, including INO in the developing ovule (Villanueva et al., 1999) and CRC in the FM (Alvarez and Smyth, 1999; Bowman and Smyth, 1999; Prunet et al., 2008), suggesting this as a central, and potentially ancestral, function of the YAB clade.
YAB1 Activities Define the Primordia Domain of the SAM Periphery and Promote Robust Partitioning of the SAM
SAMs maintain a dynamic zonation but in simple terms have a peripheral domain, where organ primordia are initiated, and a central domain responsible for SAM self-maintenance (Steeves and Sussex, 1989). It has been shown that ABPHYL (Jackson and Hake, 1999), CLV1, CLV3, WUS, and other genes acting in the central domain of the SAM influence the relative positioning of organ primordia (Leyser and Furner, 1992; Clark et al., 1993; Laux et al., 1996). In this study, we have demonstrated the significance of peripheral, early primordial YAB1 activity for proper positioning of organ primordia and for normal expression of central SAM domain markers (Figure 6S), establishing a regulatory link between domains separated by three to seven cells. The sequential and nonautonomous effects of the loss of YAB1 activities were evident in the analysis of amiR-YAB1 (Figure 5) and were corroborated by the sequential effects of ectopic YAB1 expression on the central meristem, including on the position and number of subsequently produced organs as well as on central SAM gene expression (Figures 4D to 4F; see Supplemental Figures 2A to 2F online). These observations illustrate the central importance of nonautonomous YAB1-derived signals not only in proper SAM maintenance but also in axillary FM establishment. A non-cell-autonomous role for YAB1 genes in meristem maintenance was similarly observed in Antirrhinum, where the loss of the primordia-autonomous YAB1-class protein GRAM resulted in altered SAM cell number and density (Golz et al., 2004; Navarro et al., 2004).
The YAB1 genes are responsible for a signal that is emitted from primordia and exerts its effects on expression of central meristem genes, including CLV3 and WUS, which balance meristem maintenance and differentiation programs (Figure 2; Doerner, 2003). In root tips, the WUS-like gene WOX5 is required to maintain meristem activity. It performs this function along with the PLETHORA-group genes, which also provide an instructional gradient for different cellular responses (Galinha et al., 2007; Sarkar et al., 2007). It is possible that the nonautonomous YAB1 signals provide equivalent spatially coordinating instructions for the SAM. Since the YAB1 gene products themselves are not the signal, how might this non-cell-autonomous effect be transduced? Via a mutant screen for suppressors of ectopic YAB1 activity, we determined that such signals are perceived and/or transduced by the organ-meristem boundary factor LAS. Conversely, when YAB1 activity is lost in the flower, the domain marked by LAS activity is poorly specified (Figures 6G to 6N), and chimeric organs, a reflection of mixed homeotic domains, are generated (Chen et al., 1999; Sawa et al., 1999a). From these observations, we suggest that concentric expression domains of factors that include LAS insulate the SAM periphery from the center (Figure 6S). These domains filter or amplify morphogenic signals involved in the crosstalk between meristem domains and are essential for proper meristem organization and maintenance. Other components of this program may include the other GRAS protein HAIRY MERISTEM (Stuurman et al., 2002), which, like LAS, is expressed at the periphery of the meristem, albeit between primordia anlagen. Indirect support for these genes playing such a role comes from the stable partitioning of cell layers in the root meristem, which involves the concentrically expressed related GRAS proteins SCR and SHR (Sabatini et al., 2003).
By the same rationale, other meristem-primordia boundary factors, such as the CUP SHAPED COTYLEDON, RAX, and LOB genes, may play a role in processing YAB1 signals (Aida et al., 1999; Takada et al., 2001; Vroemen et al., 2003; Aida and Tasaka, 2006; Keller et al., 2006; Muller et al., 2006). In this respect, it is notable that the mutations in the UNUSUAL FLORAL ORGANS (UFO) gene, which, like LAS, has a meristem-boundary expression domain that does not overlap with that of the YAB1 genes, significantly enhance the inflorescence phenotype of fil to the point that only filamentous structures are produced by the fil ufo inflorescence (Chen et al., 1999; Sawa et al., 1999a). The similarities of this ufo fil phenotype with that of fil-8 las (Figure 6F) implies that UFO, like LAS, may modulate crosstalk between the outer and inner regions of the SAM and, in this role, mediate YAB1 signaling.
The Role of YAB1 in Phyllotactic Patterning
The requirement of primordial YAB1 activity for normal phyllotaxis along with the strong sequential effects on younger primordia and the meristem indicate that YAB1 proteins act nonautonomously to affect the meristem. The YAB1 targets might contribute to the proposed mobile inhibitory signal or act locally to deplete peripheral auxin, a principle component of mathematical models accounting for the regular phyllotactic pattern of organ initiation (Mitchison, 1977; Veen and Lindenmayer, 1977; de Reuille et al., 2006; Jönsson et al., 2006; Smith et al., 2006; Kuhlemeier, 2007). However, the phenotypes caused by both the loss and misexpression of YAB1 functions defy simple interpretation in this respect. Either a reduction in YAB1 or its ectopic expression can result in fewer organ primordia in some contexts and more in others. For instance, PAP1≫amiR-YAB1 flowers have more sepals but fewer petals and stamens, while, depending on expression levels, PAP1:YAB3 can produce a greater number of organs interior to the sepals or exhibit a partial meristem arrest (Figures 5 and 6; see Supplemental Figure 4 online). It is conceivable that the multifactorial role of YAB1 activity obscures a simple interpretation of its primary function. Likewise, the complexity of phyllotaxis maintenance means that altered levels of YAB1 signal could change multiple parameters of SAM partitioning and influence phyllotaxis indirectly. Indeed, the significant deregulation of the normally tightly coordinated and spatially separated CLV3 and WUS markers that occurred when levels of YAB1 expression were altered (Figure 2; see Supplemental Figure 4 online) raises the possibility that regulation of meristem patterning is the primary process affected by the nonautonomous YAB1 signal and that phyllotaxis regulation is secondary. In any case, the involvement of the meristem-organ boundary factor LAS in perceiving the YAB1-mediated signal (Figure 6S) links the developing primordia and the more central meristem. That las mutations do not alter the large meristems of clv3 flowers (see Supplemental Figures 5D to 5F online) suggests that there may be multiple such signaling networks. Further understanding of the role of LAS in this process and the identification of additional factors contributing to the YAB1 signaling pathway promises fertile ground to enrich our understanding of the complex interactions underlying the ordered initiation of lateral organs from the SAM.
METHODS
Plant Material, Growth, Transformation, and Mutagenesis
All Arabidopsis thaliana plants used were in the Landsberg erecta background, are described in Supplemental Table 2 online, and were grown under 18-h cool-white fluorescent light at 18 to 22°C. fil-8 and fil-8 yab3-2 seeds (Kumaran et al., 2002) were provided by Venkatesen Sundaresen (University of California, Davis). Transactivation lines were generated by transcriptional fusion of promoters in front of the chimeric LhG4 in the BJ36 plasmid to generate driver lines, and cDNAs were subcloned behind an operator array in the BJ36 plasmid to generate responder lines (Moore et al., 1998). AP1 and AP3 promoters were transcriptionally fused to YAB3 cDNA to obtain PAP1:YAB3 and PAP3:YAB3 constructs in the BJ36 plasmid. Next, all constructs were subcloned into the NotI site of the pMLBART binary plasmid and transformed into plants by floral dipping using the Agrobacterium tumefaciens GV3101 strain. Transformants were selected on soil on the basis of resistance to 0.1% of the herbicide BASTA. PCLV3:LhG4 was generated from a plasmid gift from Eliot Meyerowitz containing the promoter 5′ and 3′ regulatory sequences between which was cloned the LhG4 gene using XhoI and BamHI sites. Additional lines were generated with PCR-based cloning using primers described in Supplemental Table 3 online. Generally, promoter:LhG4 lines were crossed to different OP:cDNA responder lines to generate transactivated F1s (marked as ≫ in the text). Mutant and transgene combinations were generated through conventional breeding.
PAP1:YAB3 seeds were mutagenized with 15 mM ethyl methanesulphonate for 12 h, and phenotypic suppressors were selected in the M2 generation. The suppressor mutations were backcrossed to Landsberg erecta, and only those in which the transgene phenotype was restored were analyzed further. Novel LAS (At1g55580) alleles were identified due to phenotypic similarity to the previously described las mutants (Greb et al., 2003). The new alleles were as follows: las-11, G443→ A (stop), las-12, G1297→A 1297 (stop), las-13, G1066 → A (Glu to Lys), where bases are numbered relative to the cDNA ATG. The las-11 allele was used for further analyses.
Preparation of Tissue for Detection of Fluorescent Signals
Intact plants or inflorescences were collected and immediately placed in vials with ice-cold 2.5% paraformaldehyde (PFA; Sigma-Aldrich) at pH 7.0 (0.4 pH fluctuations were considered acceptable) and vacuum infiltrated for ∼30 min until all tissue ceased floating. Vials were stored overnight at 4°C and washed the next morning with 10% sucrose and 1% PFA, pH 7.0, for 20 min, with 20% sucrose and 1% PFA, pH 7.0, for 20 min, and finally with 30% sucrose and 1% PFA, pH 7.0, for 30 min. In parallel, 7% LM-GQT agarose (Conda) water-based gel was prepared and heated for 30 to 40 min in boiling water bath. Ten to fifteen minutes before fixation was finished, the 7% LM agarose solution was cooled down to 30°C and poured into small Petri dishes, where fixed samples were dipped in the liquid gel. Dishes were then placed at 4°C for 10 to 15 min to solidify. A block with the tissue was then cut out of the gel and glued at the desired angle onto the sliding microtome stage using Tissue-tek O.C.T. compound. The tissue was sliced with a Leica Sliding Microtome SM-2000 after the stage was cooled to −15 to −25°C, aided by dry ice powder to ensure fast and homogenous freezing. The frozen blocks were sliced to 50 to 60 μm thick (transverse sections) or 30 to 40 μm thick (longitudinal sections), immediately placed into drops of buffer (0.5% PBS, 50% glycerol, and 0.01% NaN3) on glass microscope slides (25 × 75 × 1 mm Menzel-Glaser, Super Frost Plus) and allowed to defrost. Selected slices were chosen using a dissecting microscope, and the buffer solution was removed with a Pasteur pipette. Tissue was counter stained with propidium iodide (Sigma-Aldrich) 1 mg/mL solution, mounted with ProLong Gold antifade reagent (Invitrogen), covered with cover slips (24 mm in diameter, 0.08 to 0.13 mm thick), and sealed with nail polish.
Microscopy and Confocal Imaging
Confocal images were taken by an Olympus IX-70 microscope with the argon laser set at 488 nm for excitation, a 505- to 525-nm filter for GFP emission, and a 560- to 600-nm filter for PI emission. Images were captured and processed with the FW-500 image analysis system. Scanning electron microscopy was performed using an XL30 ESEM FEG microscope (FEI) after standard tissue preparations (Alvarez et al., 2006).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: FIL (At2g45190), YAB3 (At4g00180), LAS (At1g55580), KAN1 (At5g16560), and miR165B (At4g00885).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Equivalence of FIL and YAB3 Proteins.
Supplemental Figure 2. Phenotypic Range of Effects Induced by Ectopic Expression of Various Forms of YAB1.
Supplemental Figure 3. Sequential Effects of YAB1 Revealed by the Use of a Synthetic miRNA Targeting the YAB1 Genes.
Supplemental Figure 4. Unique Responses of Flower Meristems to Ectopic YAB1 Expression.
Supplemental Figure 5. Additional Analyses of lateral suppressor.
Supplemental Table 1. Mean Number of Floral Organs in Plants with Altered YAB1 Expression.
Supplemental Table 2. Arabidopsis Mutant and Transgenic Lines Used in This Study.
Supplemental Table 3. Primers Used for Construction of Responder and Driver Lines.
Supplementary Material
Acknowledgments
The dedicated work of Amy Hamilton, Michelle T. Juarez, Olga Sonkin, Efrat Arbiv, and Maria Goldshmidt is greatly appreciated. We thank Eugenia Klein and the electron microscopy facility for help with scanning electron microscopy, Raya Eilam for help with tissue preparation techniques, and Vladimir Kiss for assistance with confocal laser scanning microscopy. We also thank the late Alex Levitan, Gideon Grafi, Friedrich Kragler, Patricia Zambryski, Venkatesan Sundaresan, Thomas Laux, Eliot Meyerowitz, Roger Tsien, and Chuck Gasser for providing plasmids and plant material. We thank members of the Eshed lab for comments and discussions. This work was made possible with funding from Research Grant 386-02 from the Israel Science Foundation and from MINERVA (Y.E.) and support from a U.S. National Science Foundation grant (IOB 0342253 to J.L.B.). Y.E. is an incumbent of the Judith and Martin Freedman Career Development Chair.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yuval Eshed (yuval.eshed@weizmann.ac.il).
Online version contains Web-only data.
References
- Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126 1563–1570. [DOI] [PubMed] [Google Scholar]
- Aida, M., and Tasaka, M. (2006). Morphogenesis and patterning at the organ boundaries in the higher plant shoot apex. Plant Mol. Biol. 60 915–928. [DOI] [PubMed] [Google Scholar]
- Alvarez, J., and Smyth, D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126 2377–2386. [DOI] [PubMed] [Google Scholar]
- Alvarez, J.P., Pekker, I., Goldshmidt, A., Blum, E., Amsellem, Z., and Eshed, Y. (2006). Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18 1134–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., and Smyth, D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126 2387–2396. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Eshed, Y., and Baum, S.F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18 134–141. [DOI] [PubMed] [Google Scholar]
- Carraro, N., Peaucelle, A., Laufs, P., and Traas, J. (2006). Cell differentiation and organ initiation at the shoot apical meristem. Plant Mol. Biol. 60 811–826. [DOI] [PubMed] [Google Scholar]
- Chen, Q., Atkinson, A., Otsuga, D., Christensen, T., Reynolds, L., and Drews, G.N. (1999). The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation. Development 126 2715–2726. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119 397–418. [DOI] [PubMed] [Google Scholar]
- Collier, R.J. (1975). Diphtheria toxin: Mode of action and structure. Bacteriol. Rev. 39 54–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, C.D., Galgoci, B.F.C., and Irish, V.F. (1995). Genetic ablation of petal and stamen primordia to elucidate cell interactions during floral development. Development 121 2887–2895. [DOI] [PubMed] [Google Scholar]
- de Reuille, P.B., Bohn-Courseau, I., Ljung, K., Morin, H., Carraro, N., Godin, C., and Traas, J. (2006). Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny, J.R., Yadegari, R., Fischer, R.L., Yanofsky, M.F., and Weigel, D. (2004). The role of JAGGED in shaping lateral organs. Development 131 1101–1110. [DOI] [PubMed] [Google Scholar]
- Doerner, P. (2003). Plant meristems: A merry-go-round of signals. Curr. Biol. 13 R368–R374. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Izhaki, A., Baum, S.F., Floyd, S.K., and Bowman, J.L. (2004). Leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131 2997–3006. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 1911–1914. [DOI] [PubMed] [Google Scholar]
- Galinha, C., Hofhuis, H., Luijten, M., Willemsen, V., Blilou, I., Heidstra, R., and Scheres, B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449 1053–1057. [DOI] [PubMed] [Google Scholar]
- Gallagher, K.L., and Benfey, P.N. (2005). Not just another hole in the wall: understanding intercellular protein trafficking. Genes Dev. 19 189–195. [DOI] [PubMed] [Google Scholar]
- Giulini, A., Wang, J., and Jackson, D. (2004). Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430 1031–1034. [DOI] [PubMed] [Google Scholar]
- Golz, J.F., Roccaro, M., Kuzoff, R., and Hudson, A. (2004). GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development 131 3661–3670. [DOI] [PubMed] [Google Scholar]
- Grandjean, O., Vernoux, T., Laufs, P., Belcram, K., Mizukami, Y., and Traas, J. (2004). In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell 16 74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb, T., Clarenz, O., Schafer, E., Muller, D., Herrero, R., Schmitz, G., and Theres, K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler, M.G., Ohno, C., Das, P., Sieber, P., Reddy, G.V., Long, J.A., and Meyerowitz, E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15 1899–1911. [DOI] [PubMed] [Google Scholar]
- Jackson, D., and Hake, S. (1999). Control of phyllotaxy in maize by the ABPHYL1 gene. Development 126 315–323. [DOI] [PubMed] [Google Scholar]
- Jönsson, H., Heisler, M., Shapiro, B.E., Meyerowitz, E.M., and Mjolsness, E. (2006). An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. USA 103 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez, M.T., Kui, J., Thomas, J., Heller, B.A., and Timmermans, M.C. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428 84–88. [DOI] [PubMed] [Google Scholar]
- Keller, T., Abbott, J., Moritz, T., and Doerner, P. (2006). Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlemeier, C. (2007). Phyllotaxis. Trends Plant Sci. 12 143–150. [DOI] [PubMed] [Google Scholar]
- Kumaran, M.K., Bowman, J.L., and Sundaresan, V. (2002). YABBY genes mediate down-regulation of KNOTTED-like genes during leaf development in Arabidopsis thaliana. Plant Cell 14 2761–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F.X., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 87–96. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Baum, S.F., Oh, S.H., Jiang, C.Z., Chen, J.C., and Bowman, J.L. (2005). Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development 132 5021–5032. [DOI] [PubMed] [Google Scholar]
- Leibfried, A., To, J.P.C., Busch, W., Stehling, S., Kehle, A., Demar, M., Kieber, J.J., and Lohmann, J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 22 1172–1175. [DOI] [PubMed] [Google Scholar]
- Leyser, O., and Furner, I.J. (1992). Characterization of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116 397–403. [Google Scholar]
- Long, J.A., and Barton, M.K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218 341–353. [DOI] [PubMed] [Google Scholar]
- Mitchison, G.J. (1977). Phyllotaxis and the Fibonacci series. Science 196 270–275. [DOI] [PubMed] [Google Scholar]
- Moore, I., Galweiler, L., Grosskopf, D., Schell, J., and Palme, K. (1998). A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl. Acad. Sci. USA 95 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, D., Schmitz, G., and Theres, K. (2006). Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, K., Sena, G., Nawy, T., and Benfey, P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413 307–311. [DOI] [PubMed] [Google Scholar]
- Navarro, C., Efremova, N., Golz, J.F., Rubiera, R., Kuckenberg, M., Castillo, R., Tietz, O., Saedler, H., and Schwarz-Sommer, Z. (2004). Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development 131 3649–3659. [DOI] [PubMed] [Google Scholar]
- Nole-Wilson, S., and Krizek, B.A. (2006). AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol. 141 977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappenheimer, A.M. (1977). Diphtheria toxin. Annu. Rev. Biochem. 46 69–94. [DOI] [PubMed] [Google Scholar]
- Pekker, I., Alvarez, J.P., and Eshed, Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17 2899–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig, R.S., and Sussex, I.M. (1985). The cellular parameters of leaf development in tobacco: A clonal analysis. Planta 165 170–184. [DOI] [PubMed] [Google Scholar]
- Prunet, N., Morel, P., Thierry, A.-M., Eshed, Y., Bowman, J.L., Negrutiu, I., and Trehin, C. (2008). REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 20: 901–919. [DOI] [PMC free article] [PubMed]
- Reinhardt, D., Mandel, T., and Kuhlemeier, C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Reinhardt, D. (2005). Regulation of phyllotaxis. Int. J. Dev. Biol. 49 539–546. [DOI] [PubMed] [Google Scholar]
- Sabatini, S., Heidstra, R., Wildwater, M., and Scheres, B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, A.K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., Scheres, B., Heidstra, R., and Laux, T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446 811–814. [DOI] [PubMed] [Google Scholar]
- Sawa, S., Ito, T., Shimura, Y., and Okada, K. (1999. a). FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell 11 69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Liu, Y.G., Shibata, D., Kanaya, E., Morita, E.H., and Okada, K. (1999. b). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128. [DOI] [PubMed] [Google Scholar]
- Smith, H.M., and Hake, S. (2003). The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R.S., Guyonmach, S., Mandel, T., Reinhardt, D., Kuhlemeier, C., and Prusinkiewicz, P. (2006). A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. USA 103 1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow, M., and Snow, R. (1931). Experiments on phyllotaxis. I. The effect of isolating a primordium. Philos. Trans. R. Soc. Lond. B Biol. Sci. 221 1–43. [Google Scholar]
- Steeves, T.A., and Sussex, I.M. (1989). Patterns in Plant Development. 2nd ed. (Cambridge, UK: Cambridge University Press).
- Stuurman, J., Jaggi, F., and Kuhlemeier, C. (2002). Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 16 2213–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128 1127–1135. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Matsumoto, N., and Okada, K. (2004). RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131 425–434. [DOI] [PubMed] [Google Scholar]
- Tilly, J., Allen, D.W., and Jack, T. (1998). The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125 1647–1657. [DOI] [PubMed] [Google Scholar]
- Veen, A.H., and Lindenmayer, A. (1977). Diffusion mechanism for phyllotaxis: Theoretical, physico-chemical and computer study. Plant Physiol. 60 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva, J.M., Broadhvest, J., Hauser, B.A., Meister, R.J., Schneitz, K., and Gasser, C.S. (1999). INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 13 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroemen, C.W., Mordhorst, A.P., Albrecht, C., Kwaaitaal, M.A., and De Vries, S.C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 7 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K., and Okada, K. (2003). Two discrete cis elements control the Abaxial side-specific expression of the FILAMENTOUS FLOWER gene in Arabidopsis. Plant Cell 15 2592–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, L., Grigg, S.P., Xie, M., Christensen, S., and Fletcher, J.C. (2005). Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132 3657–3668. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, T., Nagasawa, N., Kawasaki, S., Matsuoka, M., Nagato, Y., and Hirano, H.Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, T., Ito, M., and Kato, M. (2004). YABBY2-homologue expression in lateral organs of Amborella trichopoda (Amborellaceae). Int. J. Plant Sci. 165 917–924. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.