Abstract
N6-Methyladenosine is a ubiquitous modification identified in the mRNA of numerous eukaryotes, where it is present within both coding and noncoding regions. However, this base modification does not alter the coding capacity, and its biological significance remains unclear. We show that Arabidopsis thaliana mRNA contains N6-methyladenosine at levels similar to those previously reported for animal cells. We further show that inactivation of the Arabidopsis ortholog of the yeast and human mRNA adenosine methylase (MTA) results in failure of the developing embryo to progress past the globular stage. We also demonstrate that the arrested seeds are deficient in mRNAs containing N6-methyladenosine. Expression of MTA is strongly associated with dividing tissues, particularly reproductive organs, shoot meristems, and emerging lateral roots. Finally, we show that MTA interacts in vitro and in vivo with At FIP37, a homolog of the Drosophila protein FEMALE LETHAL2D and of human WILMS' TUMOUR1-ASSOCIATING PROTEIN. The results reported here provide direct evidence for an essential function for N6-methyladenosine in a multicellular eukaryote, and the interaction with At FIP37 suggests possible RNA processing events that might be regulated or altered by this base modification.
INTRODUCTION
In eukaryotic DNA, methylation most commonly occurs as 5-methylcytosine. This is often in blocks of heterochromatin or in CpG islands surrounding genes (http://www.methdb.de) and is recognized as playing a fundamental role in regulating gene expression. In the case of mRNA, a number of modifications are possible, such as C-to-U editing in some chloroplast and mitochondrial transcripts (Shikanai, 2006) or A-to-I deamination, found in some animal RNAs (Zhang and Carmichael, 2001). Methylation of the N6 position of the adenosine base has been observed in many RNA species, including tRNA, rRNA, and small nuclear RNA (snRNA) (Bjork et al., 1987; Maden, 1990; Shimba et al., 1995; Gu et al., 1996; Agris et al., 2007; Piekna-Przybylska et al., 2008), but the functional importance of its occurrence in mRNA has remained unclear since its discovery >30 years ago (Desrosiers et al., 1974; Perry and Kelley, 1974).
N6-Methyladenosine (m6A) is found in the mRNA of some viruses (Beemon and Keith, 1977; Aloni et al., 1979) and all multicellular eukaryotes examined, including mammals (Adams and Cory, 1975; Perry et al., 1975; Wei et al., 1976), insects (Levis and Penman, 1978), and the monocot plants maize (Zea mays; Nichols, 1979), wheat (Triticum aestivum; Kennedy and Lane, 1979), and oat (Avena sativa; Haugland and Cline, 1980). Ribonuclease fragmentation and labeling studies on mRNA from animal cells and maize tissues show m6A to be present only at the central A within the defined sequence context GAC and AAC, with a 75% preference for GAC (Wei et al., 1976; Schibler et al., 1977; Nichols and Welder, 1981; Harper et al., 1990).
Unlike C-to-U or A-to-I conversions, m6A does not result in a change following reverse transcription, so it is not revealed in cDNA libraries. Precise mapping of m6A has only been reported for two mRNAs, Rous sarcoma virus and bovine prolactin (Horowitz et al., 1984; Kane and Beemon, 1985). A 1865-nucleotide region of Rous sarcoma virus genomic RNA contains seven m6A sites, all of which are found within the GAC context. Most of these are within the sequence GGACU, with some also present as UGACU and AGACU. In bovine prolactin mRNA, methylation occurs at a single AGACU site within the 3′ untranslated region. These mapped methylated sites are consistent with an extended consensus RRACH (where R = purine and H = A, C, or U) proposed earlier by Schibler et al. (1977). However, the frequency of this extended consensus within a population of mRNAs is far higher than the observed frequency of m6A (typically 0.1 to 0.2% of the nucleotides). Thus, it appears that additional sequence or structural cues are likely to play a role in the choice of methylation sites.
Within a cell, different types of mRNA can contain different amounts of m6A. For example, the mouse dihydrofolate reductase transcript contains 3 m6A residues and simian virus 40 viral mRNA has >10, although in both cases the methylation sites were not mapped (Canaani et al., 1979; Rana and Tuck, 1990). In contrast, m6A is absent from histone and globin transcripts (Tuck, 1992). In human HeLa cell extracts, the formation of m6A is catalyzed by a multiprotein complex (Bokar et al., 1997). The methyltransferase activity requires 200- and 875-kD components, which are separable under nondenaturing conditions. The 200-kD component contains the methyltransferase function on a 70-kD subunit. Following further purification and microsequencing, the protein corresponding to this subunit was identified and named MT-A70 (Bokar et al., 1997). The other components of the enzymatic complex remain unknown.
A phylogenetic analysis of proteins homologous with MT-A70 has identified four lineages (A to D) of proteins with similar motifs and a clear common ancestry (Bujnicki et al., 2002). Lineages A, B, and C are unique to eukaryotes. Lineage A contains MT-A70 itself, while lineages B and C are represented by MT-A70 homologous sequences corresponding to two predicted proteins originally derived from human cDNA clones from brain and testis tissue. Among sequenced eukaryote genomes, humans, mice, pufferfish, Drosophila, and Arabidopsis thaliana each contain representatives of the A, B, and C lineages. Thus, it would appear that duplication of the ancestral sequence that gave rise to these three lineages occurred very early in eukaryote evolution.
However, while the B and C lineages appear to share a common ancestry, their role in mRNA methylation has not been demonstrated. Lineage D is the most distantly related and consists of a small cluster of bacterial DNA m6A methyltransferases associated with restriction/modification systems. Saccharomyces cerevisiae contains just A and B orthologous genes (IME4 and KAR4, respectively; Bujnicki et al., 2002). The presence of a MT-A70 ortholog (IME4) in S. cerevisiae is surprising, as budding yeast were not believed to contain m6A within their mRNAs. IME4 had previously been characterized as encoding a product that regulated the entry of diploid cells into meiosis by elevating the levels of IME1 and IME2 mRNAs via an unknown posttranscriptional mechanism. Subsequently, low levels of m6A were shown to appear in sporulating yeast poly(A) RNA, and this methylation is IME4-dependent (Clancy et al., 2002).
Arabidopsis possesses a single homolog of MT-A70 (encoded by At4g10760), which we refer to as mRNA adenosine methylase (MTA). The gene at this locus was previously designated Embryo-Defective1706 (EMB1706) following a global screen for embryo-defective mutants (Tzafrir et al., 2003). In this article, we show that m6A is present in Arabidopsis poly(A) RNA. MTA is required for this methylation, and MTA disruption results in embryo lethality. We also show that MTA interacts in vivo with At FIP37, a plant homolog of the Drosophila gene female-lethal(2)d required for sex-specific splicing in flies (Ortega et al., 2003; Vespa et al., 2004). The results reported here provide direct evidence for an essential function for m6A in a multicellular eukaryote, and the identification of At FIP37 as an interacting partner of MTA suggests a possible role for m6A in alternative splicing.
RESULTS
m6A Is Present within Arabidopsis poly(A) RNA
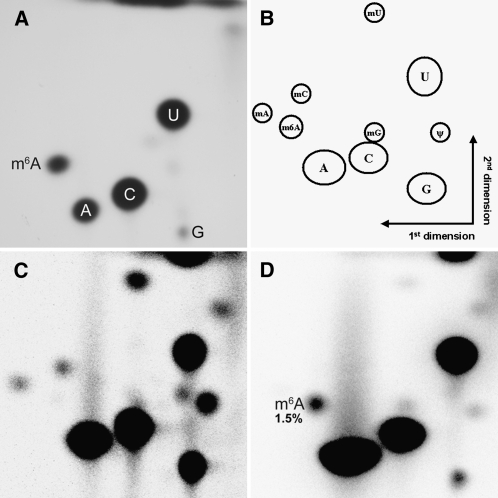
In order to establish whether m6A is present within Arabidopsis mRNA, a two-dimensional thin layer chromatography (TLC) method was adapted from Keith (1995) to detect and quantify m6A levels. In this method, mRNA is first digested with T1 ribonuclease, which cuts after every guanosine residue, leaving short polynucleotides that can be readily labeled with 32P at their 5′ ends. Thus, only those nucleotides that immediately follow a guanosine will be labeled. The labeled polynucleotides are further digested to give nucleotide 5′ monophosphates, which are separated by TLC as described in Methods.
Using mixtures of in vitro transcribed RNA either containing or not containing m6A, the relative positions of the nucleotides following TLC separation were established and the efficient labeling of m6A was demonstrated (Figures 1A and 1B). As expected, the spot corresponding to guanosine 5′-monophosphate (pG) is relatively weak; this is because the only Gs available for labeling following T1 digestion are mononucleotides, which are not efficient substrates for the kinase reaction. Thus, pG will be underrepresented relative to the other nucleotides. By combining in vitro transcribed methylated and nonmethylated RNA in known ratios, we demonstrated that this detection method, combined with phosphor imaging, gives quantitative values for pm6A relative to pA over the biological range observed (see Supplemental Figure 1 online).
Figure 1.
Two-dimensional TLC Detection of m6A in Arabidopsis Poly(A) RNA.
(A) Two-dimensional TLC analysis of in vitro transcribed RNA containing m6A and normal adenosine.
(B) Schematic diagram of the relative positions of nucleotide spots.
(C) Two-dimensional TLC analysis of total RNA extracted from 2-week-old Arabidopsis seedlings.
(D) Two-dimensional TLC analysis of poly(A) RNA from 2-week-old Arabidopsis seedlings. The m6A:A ratio is 1.5%.
Total RNA was extracted from 2-week-old Arabidopsis seedlings, and the poly(A) fraction was purified by oligo(dT) chromatography. Both the total and the poly(A) RNA fractions were subjected to TLC analysis. As expected, due to the abundance of rRNA and tRNA species containing modified nucleotides, the total RNA sample gave spots in addition to pG, pA, pC, and pU following TLC separation (Figure 1C). The relative positions of these additional spots are consistent with the established positions of pseudouridine and the 2′ methylated forms of the four nucleotides (Keith, 1995). A spot corresponding to pm6A was not readily detectable in the total RNA sample. In contrast, the poly(A) sample gave a clear spot in the expected position for pm6A, while the additional spots seen in the total RNA sample were absent or very much reduced (Figure 1D). Measuring the intensity of the pm6A spot relative to the pA spot gave an m6A-to-A ratio of 1.5%, which is consistent with reported values in animal systems.
Disruption of MTA Leads to an Arrest at the Globular Stage of Embryo Development
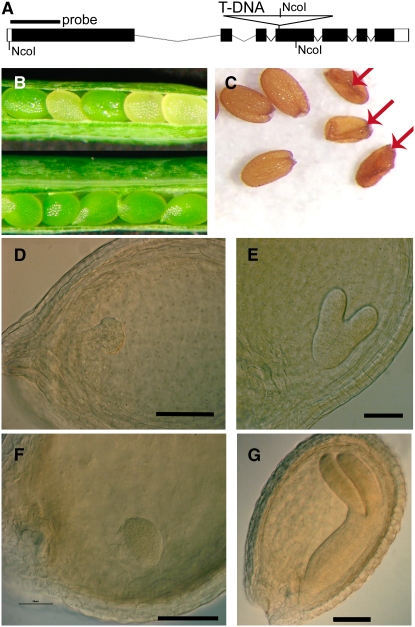
To investigate the functional importance of MTA during Arabidopsis development, we characterized a T-DNA insertion mutant in At4g10760. SALK_074069 contains a T-DNA within an annotated exon (exon 4) of MTA (Figure 2A). Plants containing this insertion could only be isolated as hemizygotes and produced green and white seeds in immature siliques. The white seeds failed to develop further, indicating an embryo-lethal phenotype for the homozygous insertion (Figures 2B and 2C). This ratio does not differ significantly (P = 0.26) from the 3:1 ratio that would be predicted for a recessive embryo-lethal mutation. Of the normally developing seeds, approximately two-thirds produced plants containing the SALK_074069 insertion. A second T-DNA insertion line (SALK_114710) in exon 6 also gave the same embryo-lethal phenotype (see Supplemental Figure 2 online). Further examination of the green and white seeds from siliques of different developmental stages in the SALK_074069 plants revealed that the embryos were not progressing from the globular stage to the heart stage (Figure 2D). However, a limited number of additional cell divisions occurred as the embryo aged (Figure 2F).
Figure 2.
Seeds of the SALK_074069 Insertion in MTA Arrest at the Globular Stage.
(A) Arrangement of At4g10760 showing the location of the SALK_074069 T-DNA insertion in exon 4. Exons are shown in black, and introns are shown in white.
(B) Siliques from a plant hemizygous for SALK_074069 (top) and from a wild-type control plant (bottom). Seeds homozygous for the SALK_074069 insertion appear white and fail to develop normally.
(C) White seeds become shrivelled and nonviable at maturity (arrows).
(D) Arrested embryo of the hemizygous SALK_074069.
(E) A phenotypically normal embryo from the same silique as in (D) that has reached the heart stage.
(F) Although the arrested embryo has not developed past the globular stage, some cell division has continued.
(G) A phenotypically normal embryo from the same silique as in (F).
Bars = 100 μm.
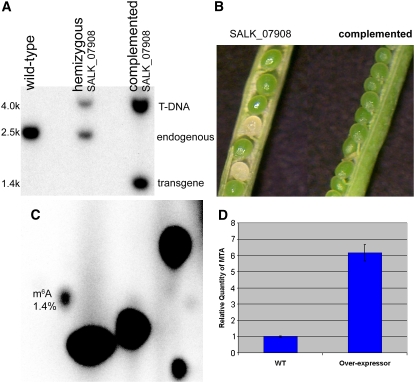
To confirm that the disruption to MTA was the sole cause of the embryo-lethal phenotype, plants hemizygous for the SALK_074069 insertion were transformed with a full-length MTA cDNA under the control of the constitutive cauliflower mosaic virus 35S promoter. From these lines, progeny were selected that were homozygous for the SALK_074069 insertion, as confirmed by DNA gel blot analysis (Figure 3A). The complemented homozygous lines gave only green seeds in their siliques, confirming that the insertion in MTA is the cause of the embryo-lethal phenotype (Figure 3B). Poly(A) RNA was purified from 2-week-old seedlings of the complemented line and subjected to TLC analysis as described above. The presence of m6A was readily detected (Figure 3C), and the levels were not significantly different from those in the wild type, even though the MTA cDNA transgene was expressed at a sixfold higher level than the wild type, as confirmed by quantitative real-time RT-PCR (qRT-PCR) (Figure 3D; see Supplemental Figure 3 online).
Figure 3.
Complementation of SALK_074069 with 35S:MTA.
(A) DNA gel blot analysis of wild-type, hemizygous SALK_074069, and homozygous SALK_074069 lines complemented with the MTA cDNA construct.
(B) Silique phenotype of plants hemizygous for SALK_074069 (left) and homozygous for both SALK_074069 and the complementing cDNA transgene (right).
(C) TLC analysis of the poly(A) RNA purified from the complemented line.
(D) qRT-PCR analysis shows a sevenfold higher expression of the MTA transgene in the complemented line. Error bars show sd from three replicates.
MTA Expression and m6A Levels in Different Arabidopsis Tissues
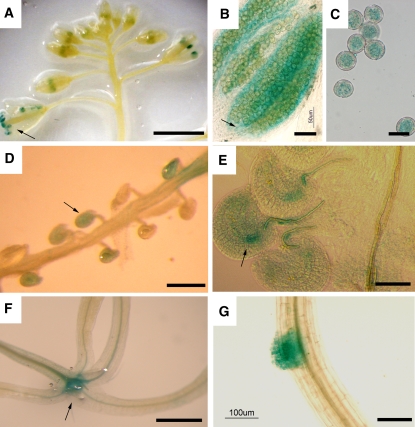
Published Affymetrix data suggest that MTA is expressed at relatively low levels in most tissues, with the highest levels of expression being found in seeds, pollen microspores, and meristems (Craigon et al., 2004). We constructed a promoter–β-glucuronidase (GUS) reporter using the 1.5-kb promoter region of MTA. This promoter is sufficient to drive the expression of MTA to complement the SALK_074069 knockout line (see Supplemental Figure 4 online). Twenty-eight plants containing this reporter were examined, and all of them showed essentially identical expression patterns, which were consistent with published Affymetrix data sets. Strong GUS staining occurred in just a few limited locations: in the apical meristems, ovules, anthers, and developing seeds (Figure 4). Some expression was also seen at the lateral root primordia. The expression in anthers was initially seen only in tapetal cells, but as they matured, strong expression was also found in the pollen microspore.
Figure 4.
Strong MTA Promoter Activity Is Limited to Discrete Developmental Tissues.
GUS expression in various organs of transformed plants is shown.
(A) Expression in anthers is initially seen only in tapetal cells (arrow). Bar = 1 cm.
(B) Close-up of the staining in (A). Bar = 50 μm.
(C) Pollen microspore in a mature anther. Bar = 10 μm.
(D) Developing seeds (arrow). Bar = 1 mm.
(E) Weak expression is seen in the ovule prior to fertilization (arrow). Bar = 25 μm.
(F) In young seedlings, GUS expression is concentrated in the apical meristem (arrow). Bar = 5 mm.
(G) In roots, GUS expression is observed primarily during lateral root initiation. Bar = 100 μm.
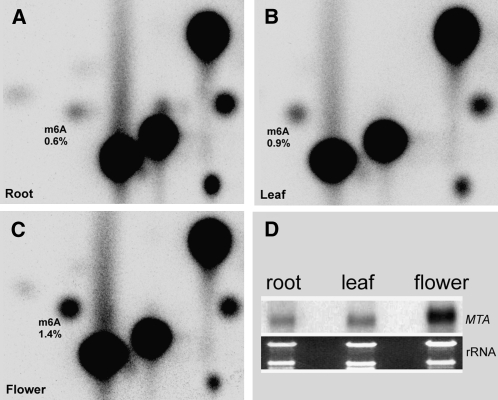
We purified mRNAs from Arabidopsis roots, leaves, and flower buds and subjected them to TLC analysis. A high ratio of m6A to A was found in the floral mRNA sample (1.4%; Figure 5C), whereas the leaf and root mRNAs contained less m6A (0.9 and 0.6%, respectively; Figures 5A and 5B). Values for m6A:A were consistent between biological replicates, differing by <0.1% from mean values. RNA gel blot analysis confirmed MTA expression in all tissues, with the highest levels in flower buds (Figure 5D).
Figure 5.
m6A Levels in Root, Leaf, and Floral Tissues.
(A) Root.
(B) Fully expanded leaf.
(C) Flower buds.
(D) RNA gel blot assay of MTA expression.
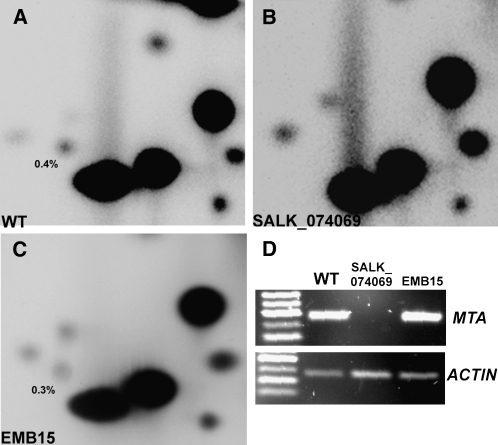
Clancy et al. (2002) demonstrated that a null mutation of the MTA ortholog, IME4, in S. cerevisiae results in a loss of m6A from sporulating yeast mRNA. To test the role of MTA in mRNA methylation in Arabidopsis, we assayed m6A levels in the white embryo-defective seeds. Microgram quantities of total RNAs were extracted from ∼1000 white seeds dissected from the SALK_074069 siliques, and the mRNA was enriched using oligo(dT) magnetic beads and subjected to TLC analysis, as described in Methods. As expected, mRNA prepared from the white seeds of the MTA null mutant does not contain quantifiable levels of m6A (Figure 6B), whereas this modification is readily detectable in both green seeds and in the white seeds of the embryo-defective control mutant EMB15 (N6307), which also arrests at the globular stage (Figures 6A and 6C). RT-PCR from the purified mRNA samples confirmed the absence of MTA transcript in the SALK_074069 white seeds (Figure 6D). Similar results were also obtained for white seeds of the SALK_114710 allele of MTA (see Supplemental Figure 2 online). While the white seeds of an additional embryo-lethal control line, SALK_072168, contained wild-type levels of m6A (see Supplemental Figure 5 online), SALK_072168 was mutated for At5g60540 (EMB2407), a gene required for vitamin B6 synthesis (Tambasco-Studart et al., 2005).
Figure 6.
Disruption of MTA Results in the Loss of m6A from the mRNA of Embryo-Defective Seeds.
(A) m6A is readily detectable in the mRNA from wild-type seeds.
(B) m6A is not detectable in the mRNA from white embryo-defective seeds of SALK_074069, even after prolonged exposure.
(C) m6A is readily detectable in the mRNA from white seeds of the control embryo development mutant emb15.
(D) RT-PCR showing the absence of MTA transcript in the poly(A) RNA from SALK_074069 white seeds.
MTA Interacts with a Homolog of a Human and Drosophila Protein Required for Alternative Splicing
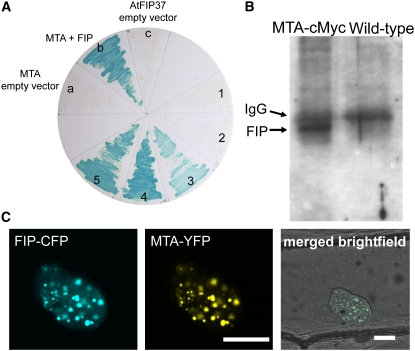
Human MT-A70 was originally isolated as part of a 200-kD complex that, together with an 875-kD component, is required for mRNA methylation activity in vitro (Bokar et al., 1997). We sought to identify potential interacting partners of the Arabidopsis MTA by screening an Arabidopsis yeast two-hybrid library prepared from anther tissues, in which MTA is highly expressed (a gift from Z. Wilson). One strongly interacting clone was obtained (Figure 7A), which was identified as encoding At FIP37 (At3g54170). This protein had been characterized previously as a plant homolog of the human WTAP (for WILMS' TUMOUR1-ASSOCIATING PROTEIN) and Drosophila FL(2)D (for FEMALE LETHAL2) and a target of the 12-kD Arabidopsis immunophilin, FKBP12 (Faure et al., 1998). In addition, disruption of At FIP37 by T-DNA insertion also results in an embryo-lethal phenotype with developmental arrest at the globular stage (Vespa et al., 2004).
Figure 7.
MTA Interacts with FIP37.
(A) MTA interacts with FIP37 (b) but does not interact with the empty prey vector, pDEST22 (a). FIP37 does not transactivate GUS expression in the presence of the empty bait vector, pDEST32 (c). ProQuest (Invitrogen) controls for assessing the relative interaction strengths are indicated 1 to 5.
(B) Proteins of wild-type and transgenic Arabidopsis plants expressing MTA-c-Myc were extracted under nondenaturing conditions and immunoprecipitated using anti-c-Myc antibody. After SDS-PAGE separation, the anti-FIP37 antibody detected a protein of 48 kD (bottom arrow) in the precipitated extracts of plants expressing the MTA-c-Myc fusion but not in those of the wild-type control.
(C) Transient expression of fluorescent protein fusions in onion epidermal peels. FIP37-CFP (left) and MTA-YFP (middle) colocalized to the nucleus. YFP and CFP images were superimposed on the bright-field image to show the position of the nucleus (right). Bars = 10 μm.
We tested whether the interaction between MTA and FIP37 occurs in vivo by carrying out coimmunoprecipitation experiments. The MTA coding sequence was fused at its C-terminus to four copies of the c-Myc epitope tag. This construct was used to transform hemizygous SALK_074069 plants, and complementing lines were isolated. Coimmunoprecipitation was carried out using an anti-c-Myc antibody (Invitrogen), and protein gel blot analysis of the precipitated proteins was performed using a polyclonal antibody previously raised against FIP37 (Vespa et al., 2004). A band corresponding to FIP37 was only detectable in extracts from the MTA-c-Myc–tagged Arabidopsis plants and was not present in the wild type (Figure 7B). The band detected with the anti-FIP37 antibody appears to have a molecular mass of 48 kD compared with the protein markers, a mass that is ∼11 kD larger than the predicted molecular mass of FIP37. Interestingly, a similar observation was reported for the Drosophila FL(2)D protein, which runs with an apparent molecular mass 38% greater than its predicted size (Ortega et al., 2003). The coimmunoprecipitation was repeated using plants expressing a hemagglutinin (HA)-tagged version of MTA, and identical results were obtained (see Supplemental Figure 6 online).
It has been reported that both the human MT-A70 and Arabidopsis FIP37 plus its Drosophila homolog FL(2)D are found in speckle-like nuclear bodies (Bokar et al., 1997; Vespa et al., 2004; Ortega, 2005). We investigated the interaction and subcellular localization of MTA and FIP37 in Arabidopsis by generating C-terminal fusions to yellow and cyan fluorescent protein tags (MTA-YFP and FIP37-CFP, respectively). As expected, FIP37 was targeted to nuclear speckles when transiently expressed in onion (Allium cepa) epidermal cells (Figure 7C). The YFP-tagged MTA proteins clearly colocalize to the same discrete spots within the nucleus when coexpressed with FIP37-CFP (Figure 7C), further supporting their in vivo interaction.
DISCUSSION
Despite its apparent abundance in mRNA, the role of adenosine methylation has received little attention in recent years. Unlike C-to-U editing, which is widespread in plant chloroplast and mitochondrial transcripts (Shikanai, 2006), or A-to-I deamination, which is found in some animal RNAs (Zhang and Carmichael, 2001), reverse transcription of an m6A-containing message does not result in a sequence that is altered from the original template. This makes the detection of individual methylation events very difficult. The importance of cytosine methylation of DNA is well recognized, and studies using methylation-sensitive restriction enzymes or bisulfite sequencing are common practice. However, such techniques cannot be used for identifying or mapping m6A in individual RNA species.
TLC Detection of m6A in Arabidopsis Tissues
The TLC results demonstrate that Arabidopsis mRNAs contain m6A. Because the method labels 5′ ends that are made available following T1 ribonuclease digestion, the m6A detected using this assay must be in the sequence context GA, which is consistent with the preferred m6A consensus sequence of GAC reported in other species. Thus, by measuring the m6A:A ratio, we are actually comparing the abundance of Gpm6A relative to GpA. The amount of m6A (if any) present in other sequences (such as AAC) cannot be determined with this method; thus, estimations of the true abundance of m6A within mRNA are likely to be underestimates. Our observed value of 1.5% for the m6A:A ratio in young seedlings would be close to a 0.1% m6A frequency in the mRNA sample as a whole if we assume that all 16 dinucleotide pairs occurred at equal frequency in the mRNA population and if m6A were only to occur in Gpm6A dinucleotides.
When the Arabidopsis total RNA was assayed, m6A was not detectable (Figure 1C). This does not mean that Arabidopsis rRNA, tRNAs, or snRNAs do not contain m6A, but if they do, it is presumably not in a GA sequence context. Indeed, this is true for the majority of mapped m6A sites in noncoding RNAs from other species. For example, m6A has been found in Escherichia coli tRNAVal (Cm6A), human U2 (Am6A), and U6 (Cm6A) snRNA; human 25S and 18S rRNA contains m6A within the sequence context of Um6A and Am6A, respectively (Gu et al., 1996; Agris et al., 2007; Piekna-Przybylska et al., 2008). In both human and yeast 18S rRNA, an adenosine with methylation at the N6 position is found following a guanosine (Piekna-Przybylska et al., 2008). However, in both cases, this residue is doubly methylated and so will not migrate at the same position as m6A in the TLC assay.
In mammals, two splice forms of MT-A70 have been reported; the shorter splice variant lacks a complete second exon and may be nonfunctional. Cultured cancer cells have an increased ratio of the longer MT-A70 splice variant relative to the shorter form (Leach and Tuck, 2001). This is consistent with the observation that extracts of immortalized human and mouse cell lines have an increased mRNA N6-adenosine methyltransferase activity (Tuck et al., 1996). However, in neither of these experiments were the endogenous m6A levels measured directly. The modified TLC method we used allows m6A to be detected and quantified in different cell types, tissues, or developmental stages. Our values for the dinucleotide Gpm6A range from 0.6 to 1.5%, and within the mRNA samples assayed, the highest levels of m6A were detected in young seedlings and flower buds. This is consistent with the MTA promoter–GUS fusion data and may indicate a function for m6A in dividing tissues (Figure 4).
MTA Interaction with At FIP37 and Possible Functions of m6A in mRNA
FIP37 was isolated as a strong interacting partner of MTA when screening the yeast two-hybrid library (Figure 7), and T-DNA knockout of either FIP37 or MTA resulted in arresting of embryo development at the globular stage (Figure 2A) (Vespa et al., 2004). At FIP37 is a homolog of the Drosophila FL(2)D, which is required for the accumulation of correctly spliced forms of sex lethal and transformer RNA (Granadino et al., 1990, 1996), two critical genes involved in Drosophila sexual determination and dosage compensation. WTAP, the human homolog of FL(2)D and At FIP37, interacts with WILMS' TUMOUR1 (WT1). WT1 exists in two major isoforms, +KTS and −KTS. The +KTS WT1 binds RNA and is incorporated into spliceosomes (Larsson et al., 1995; Davies et al., 1998). Strikingly, transgenic male mice engineered to be deficient in one WTAP isoform developed female reproductive tissues (Hammes et al., 2001). Both FL(2)D and WTAP are suggested to be associated with the spliceosome (Zhou et al., 2002; Penn et al., 2008), and consistently, our results also showed that the MTA can interact and colocalize with FIP37 in splicing speckle-like nuclear compartments (Figure 7).
In both plant and mammal mRNA, m6A occurs only within sequences matching the consensus GAC or AAC (Wei et al., 1976; Nichols and Welder, 1981). It has been demonstrated that there is a preferred five-nucleotide consensus sequence, RRACH (Schibler et al., 1977). However, from the frequency of this degenerate sequence and the estimated m6A content of mRNA, it is clear that only a minority of sequences matching the consensus are actually methylated. This suggests that there are other constraints in addition to the primary sequence that affect m6A formation. Our results showed that MTA is clearly essential in Arabidopsis, at least for embryogenesis. Similarly, the yeast homolog of MTA, IME4, is not needed for vegetative growth but is required for meiosis (Clancy et al., 2002). This, together with the conservation of the methylation consensus site across kingdoms, suggests an important biological role for this base modification.
A possible role in regulating splicing is particularly intriguing. During splicing, the 5′ end of the intron forms a 2′-5′ linkage to an adenosine upstream of the polypyrimidine tract. Cleavage at the 3′ end of the intron follows, and the intron lariat is removed and rapidly degraded. This branch point adenosine is usually found within the sequence CURAY (where R = purine and Y = pyrimidine) (Brown et al., 2002), a pattern consistent with the invariant GAC or AAC methylation target, but not as close a match with the extended RRACH consensus. If m6A acted as a positive signal for branch formation, it would be removed along with the intron and would not appear in the mature mRNA.
However, a role as a branch site suppressor is also possible. In this case, methylation could perform a housekeeping function, in which aberrant lariat formation at cryptic branch sites within exons is suppressed. N6-Adenosine methylation might also regulate some alternative splicing events by preventing the use of a default 3′ splice site, presumably resulting in the use of an alternative downstream 3′ consensus. In this latter role, the m6A would again be retained in the spliced lariat. Since both the fly and human homologs of At FIP37 are RNA binding proteins involved in regulating RNA splicing and stability, it is possible that the FIP37/FL(2)D/WTAP might recruit the adenosine methylase to a specific mRNA target site via direct protein interaction. While m6A has only modest effects on the strength of A:U pairing, it disrupts the ability of adenosines to form non–Watson-Crick G:A pairs (Micura et al., 2001; Dai et al., 2007). Thus, the presence of m6A has the potential to affect mRNA secondary structure as well as mRNA–protein and mRNA–snRNA interactions. Little is known of the distribution of m6A in nascent mRNA transcripts, but one case of its presence within an intron has been reported (Carroll et al., 1990).
The Drosophila homolog of At FIP37 appears to be required for specific alternative splicing events, but it is a formal possibility that FL(2)D acts through stabilizing or destabilizing one of the alternatively spliced products, ensuring that only one form predominates. Indeed, a role in regulating mRNA stability has been reported for its human homolog WTAP (Horiuchi et al., 2006). Alternative functions for m6A could be envisaged in modulating translatability, mRNA turnover, RNA transport, or susceptibility to posttranscriptional silencing (Revel and Groner, 1978). Whatever its mechanism of action, our results clearly indicate an essential role for m6A in cell function and development in a plant system.
METHODS
mRNA Purification, Labeling, and TLC Analysis
Qualitative analysis of m6A in Arabidopsis thaliana mRNA was performed by two-dimensional TLC. Briefly, 100 μg of total RNA was extracted from Arabidopsis tissue samples using the Plant RNeasy Mini kit (Qiagen), and the poly(A) fraction was purified using the PolyAttract mRNA isolation kit (Promega). The quality of the mRNA was checked by Agilent Bioanalyzer (Ambion), and samples analyzed containing 50 ng of mRNA were digested with 1 μL of Ribonuclease T1 (1000 units/μL; Fermentas) in a final volume of 20 μL (1× polynucleotide kinase buffer, 1 unit/μL RNase inhibitor). The 5′ end of the digested mRNA fragments were then labeled using 10 units of T4 polynucleotide kinase (Fermentas) in the presence of 1 μL of [γ-32P]ATP (6000 Ci/mmol; Perkin-Elmer). After ethanol precipitation, the labeled RNA was resuspended in 10 μL of 50 mM sodium acetate (pH 5.5) and digested to monophosphonucleotides by RNase P1 (Sigma-Aldrich). Two microliters of the samples was applied to cellulose TLC plates (20 × 20 cm; Merck) and developed in a solvent system composed of isobutyric acid:0.5 M NH4OH (5:3, v/v) in the first dimension and isopropanol:HCl:water (70:15:15, v/v/v) in the second dimension. The identification of labeled nucleotide spots was carried out using synthetic methylated and nonmethylated RNAs and by comparison with the published reference map of nucleotides for this solvent system (Keith, 1995). Quantification was carried out using a storage phosphor screen (K-Screen; Kodak) and Bio-Rad Molecular Imager FX in combination with Quantity One software (Bio-Rad).
For analyzing the m6A levels in aborted Arabidopsis seeds, 2 μg of total RNA was extracted from ∼1000 white embryo-defective seeds collected from siliques with a stereomicroscope, and mRNA was isolated using oligo(dT) Dynabeads (Invitrogen) or latex beads (Qiagen). The RNA was first hybridized to 5 μL of oligo(dT) beads, then washed five times with low-salt buffer A (150 mM LiCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 0.1% SDS) and three times with low-salt buffer B (100 mM NaCl, 10 mM Tris-HCl, pH 7.5, and 1 mM EDTA). Half of the mRNA-bound beads were used for T1 digestion and subsequent two-dimensional TLC analysis as described above, and the remaining beads were used for RT-PCR.
Plant Transformation
Following reverse transcription of total Arabidopsis seedling RNA, a 2079-bp cDNA fragment of MTA was amplified using forward primer 5′-CACCGAAGCCATGGAAACTGAATCTGATG-3′ and reverse primer 5′-AGCTGTGATTGAGTCAATAGCCATTGGTTC-3′ (without stop codon) or 5′-TTGGAATTGAACTAAGCTGTGATTGAGTC-3′ (with stop codon) and cloned into pENTR/D-TOPO (Invitrogen). Following the LR reaction, the MTA cDNA (minus stop codon) was transferred to the plant binary transformation vectors pGWB14 and pGWB17 to create MTA-HA and MTA-c-Myc (Nakagawa et al., 2007). The MTA cDNA (with stop codon) was also transferred to the binary vector pGWB8 to create 35S:MTA. After transfer to Agrobacterium tumefaciens C58, these vectors were used for floral dip transformation of Arabidopsis ecotype Columbia plants heterozygous for the SALK_074069 insertion. The presence of the SALK_074069 insertion in potentially complementing lines was initially tested by PCR and confirmed by DNA gel blot analysis.
Gene Expression Analysis
For RNA gel blot analysis, 3 μg of total RNA was loaded per lane and transferred to a HyBond-N membrane (GE Healthcare). The ethidium bromide–stained membrane was imaged under UV light as a record of equal loading and transfer. The MTA cDNA was labeled (Rediprime; GE Healthcare) and used for hybridization. The membrane was then washed according to the manufacturer's recommendations.
Reverse transcription was carried out using SuperScriptII (Invitrogen) and oilgo(dT)25. Real-time PCR was carried out using the MX3005P qPCR machine and the Brilliant SYBR Green qPCR Master Mix (Stratagene). MAXpro software was used for data analysis. Samples were run in duplicate, and relative expression levels were determined compared with actin expression. All measurements were taken in the log phase of amplification (see Supplemental Figure 3 online). MTA primers were 5′-GGAACCTTTGGAGTTGTTATG-3′ and 5′-CAAAGCTCCAAACATTCACG-3′, and normalizer gene β-ACTIN2 primers were 5′-GTACAACCGGTATTGTGCT-3′ and 5′-ATCAGTAAGGTCACGTCCA-3′.
DNA Analysis
Genomic DNA from Arabidopsis Columbia wild-type, hemizygous SALK_074069, and complemented lines was digested with NcoI (Fermentas). After gel electrophoresis, DNA was transferred to a HyBond-N membrane (GE Healthcare) and hybridized to an MTA exon 1 gene-specific probe as described above. NcoI cuts the MTA genomic sequence in exon 1 upstream of the probe fragment (Figure 2A) and also cuts within the SALK T-DNA and in exon 4. Thus, the genomic DNA fragment containing the SALK T-DNA migrates at a higher molecular weight than the endogenous MTA.
MTA Promoter–GUS Fusion
The 1.5-kb MTA promoter sequence was PCR amplified with primers 5′-CACCATGCTCGACCCATAACCTACGAC-3′ and 5′-GGCTTCGAAACAAAAGAATTCGAGAC-3′. Following cloning into pENTR/D-TOPO (Invitrogen), it was recombined into the destination vector pGWB3, which contains a GUS reporter gene. This construct was transferred to Agrobacterium C58 for transformation of Arabidopsis ecotype Columbia. The GUS assay was performed in a GUS staining solution containing 50 mM sodium phosphate buffer, pH 7, 1 mM EDTA, pH 8, 0.1% Triton X-100, 2 mM potassium ferricyanide and ferrocyanide, and 2 mM X-glucuronic acid for 24 to 48 h at 37°C. After termination of staining, tissues were cleared with ethanol and imaged.
Yeast Two-Hybrid Screening
For the construction of the bait vector, the MTA coding sequence was cloned into bait plasmid pDEST32 (Invitrogen). A ProQuest pEXP-AD502 (Invitrogen) prey library, prepared from developing Arabidopsis anthers, was kindly provided by Z. Wilson (Nottingham Arabidopsis Stock Center). A total of 4 × 106 colonies were screened for their ability to grow on plates lacking His (with 15 mM 3-amino-1,2,4-triazole). Putative positive colonies were further tested for uracil-independent growth, β-galactosidase expression, and lack of growth on 5-fluoroorotic acid containing medium to eliminate any false-positive interactions.
Coimmunoprecipitation
Arabidopsis proteins were extracted by grinding 4 g of aerial tissues at 4°C in 6 mL of extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 0.2% Nonidet P-40, and a protease inhibitor cocktail [Sigma-Aldrich]). Cell lysates were cleared of debris by centrifugation at 14,000g for 15 min at 4°C. The protein concentration in each lysate was determined using the Bradford protein assay kit (Bio-Rad). An equal volume containing 2 mg of total protein of each extract was precleared by adding 100 μL of protein G magnetic beads (New England Biolabs). After incubation at 4°C for 1 h, the protein G magnetic beads were collected, the supernatants were transferred to new tubes, and 6 μL of anti-c-Myc antibody (mouse monoclonal; Invitrogen) was added. After incubation at 4°C for 1 h, 100 μL of protein G magnetic beads (New England Biolabs) was added to bind and precipitate the antigen–antibody complex. The protein G magnetic beads were collected after 3 h of incubation at 4°C and washed four times with 10 volumes of extraction buffer. The antigen–antibody complex was eluted by adding 30 μL of SDS sample loading buffer without β-mercaptoethanol and incubated at 70°C for 5 min. Following this, DTT was added to a final concentration of 100 mM and the sample was incubated at 37°C for 1 h. The magnetic beads were removed, and the supernatant was separated on a 10% SDS-PAGE gel and then transferred to a nitrocellulose membrane by electroblotting. FIP37 was detected using an anti-FIP37 polyclonal rabbit antibody (Vespa et al., 2004) and the WesternBreeze kit (Invitrogen).
Transient Expression and Imaging
Transient gene expression was carried out using the Biolistic PDS-1000/He particle delivery system (Bio-Rad) as described previously (Zhong et al., 2008). The MTA and FIP37 cDNA (without stop codon) in pENTR vector were cloned into pDH51-GW-CFP (AM773751) and pDH51-GW-YFP (AM773752), respectively. Two micrograms of each plasmid was mixed, coated onto gold particles (sphere, 0.8 to 1.5 μm; AlfaAesar), and bombarded into onion (Allium cepa) epidermal peels placed on Murashige and Skoog medium. The onion peels were then incubated overnight at room temperature. Images were obtained using line-by-line sequential scan mode (Leica TCS SP2 AOBS laser confocal scanning microscope). CFP was excited using a 458-nm laser, and its emission was measured from 465 to 505 nm. The excitation wavelength for YFP was 514 nm, and its emission was measured from 525 to 600 nm.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: β-ACTIN2, At3g18780; FIP37, At3g54170; FKBP12, At5g64350; EMB2407, At5g60540; FL(2)D, AJ243599 (EMBL); KAR4, YCL055W; MTA, At4g10760; and WTAP, NM_152858. Germplasm information for the mutant and T-DNA insertion lines used in this study can be found in the Nottingham Arabidopsis Stock Center under accession numbers N6307 (EMB15), N570469 (SALK_070469), N614710 (SALK_114710), and N572168 (SALK_072168).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. TLC Detection of m6A in Synthetic RNA.
Supplemental Figure 2. Embryo Lethality and Absence of m6A in MTA SALK_114710.
Supplemental Figure 3. Amplification Plots for qRT-PCR and RNA Gel Blot Analysis.
Supplemental Figure 4. Complementation of SALK_074069 with the Endogenous Promoter-Driven Construct.
Supplemental Figure 5. mRNA from Control White Seeds of the Embryo-Defective Mutant SALK_072168 Contains Wild-Type Levels of m6A.
Supplemental Figure 6. Coimmunoprecipitation of MTA-HA and FIP37.
Supplementary Material
Acknowledgments
This work was supported by Biotechnology and Biological Science Research Council Grant BB/C523369/1 awarded to R.G.F. The gift from Z. Wilson of the yeast two-hybrid prey library is gratefully acknowledged. We thank A. Littlehales, G. Kahaka, and S. Mehra for technical assistance.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Rupert G. Fray (rupert.fray@nottingham.ac.uk).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adams, J.M., and Cory, S. (1975). Modified nucleosides and bizarre 5′-termini in mouse myeloma messenger-RNA. Nature 255 28–33. [DOI] [PubMed] [Google Scholar]
- Agris, P.F., Vendeixa, F.A.P., and Grahama, W.D. (2007). tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 366 1–13. [DOI] [PubMed] [Google Scholar]
- Aloni, Y., Dhar, R., and Khoury, G. (1979). Methylation of nuclear simian virus-40 RNAs. J. Virol. 32 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemon, K., and Keith, J. (1977). Localization of N6-methyladenosine in Rous-sarcoma virus genome. J. Mol. Biol. 113 165–179. [DOI] [PubMed] [Google Scholar]
- Bjork, G.R., Ericson, J.U., Gustafsson, C.E., Hagervall, T.G., Jonsson, Y.H., and Wikstrom, P.M. (1987). Transfer RNA modification. Annu. Rev. Biochem. 56 263–287. [DOI] [PubMed] [Google Scholar]
- Bokar, J.A., Shambaugh, M.E., Polayes, D., Matera, A.G., and Rottman, F.M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Brown, J.W.S., Simpson, C.G., Thow, G., Clark, G.P., Jennings, S.N., Medina-Escobar, N., Haupt, S., Chapman, S.C., and Oparka, K.J. (2002). Splicing signals and factors in plant intron removal. Biochem. Soc. Trans. 30 146–149. [DOI] [PubMed] [Google Scholar]
- Bujnicki, J.M., Feder, M., Radlinska, M., and Blumenthal, R.M. (2002). Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. J. Mol. Evol. 55 431–444. [DOI] [PubMed] [Google Scholar]
- Canaani, D., Kahana, C., Lavi, S., and Groner, Y. (1979). Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. 6 2879–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, S.M., Narayan, P., and Rottman, F.M. (1990). N6-Methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol. Cell. Biol. 10 4456–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy, M.J., Shambaugh, M.E., Timpte, C.S., and Bokar, J.A. (2002). Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: A potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 30 4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigon, D.J., James, N., Okyere, J., Higgins, J., Jotham, J., and May, S. (2004). NASCArrays: A repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 32 575–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Q., Fong, R., Saika, M., Stephenson, D., Yu, Y., Pan, T., and Piccirilli, J.A. (2007). Identification of recognition residues for ligation-based detection and quantification of pseudouridine and N6-methyladenosine. Nucleic Acids Res. 35 6322–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, R.C., Calvio, C., Bratt, E., Larsson, S.H., Lamond, A.I., and Hastie, N.D. (1998). WT1 interacts with the splicing factor U2AF65 in an isoform-dependent manner and can be incorporated into spliceosomes. Genes Dev. 12 3217–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 71 3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure, J.D., Gingerich, D., and Howell, S.H. (1998). An Arabidopsis immunophilin, AtFKBP12, binds to AtFIP37 (FKBP interacting protein) in an interaction that is disrupted by FK506. Plant J. 15 783–789. [DOI] [PubMed] [Google Scholar]
- Granadino, B., Campuzano, S., and Sanchez, L. (1990). The Drosophila melanogaster fl(2)d gene is needed for the female-specific splicing of Sex-lethal RNA. EMBO J. 9 2597–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granadino, B., Penalva, L.O., and Sanchez, L. (1996). The gene fl(2)d is needed for the sex-specific splicing of transformer pre-mRNA but not for double-sex pre-mRNA in Drosophila melanogaster. Mol. Gen. Genet. 253 26–31. [DOI] [PubMed] [Google Scholar]
- Gu, J., Patton, J.R., Shimba, S., and Reddy, R. (1996). Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: Modified nucleotides are clustered in functionally important regions. RNA 2 909–918. [PMC free article] [PubMed] [Google Scholar]
- Hammes, A., Guo, J.K., Lutsch, G., Leheste, J.R., Landrock, D., Ziegler, U., Gubler, M.C., and Schedl, A. (2001). Two splice variants of the Wilms' tumor 1 gene have distinct functions during sex determination and nephron formation. Cell 106 319–329. [DOI] [PubMed] [Google Scholar]
- Harper, J.E., Miceli, S.M., Roberts, R.J., and Manley, J.L. (1990). Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res. 18 5735–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugland, R.A., and Cline, M.G. (1980). Post-transcriptional modifications of oat coleoptile ribonucleic acids. 5′-Terminal capping and methylation of internal nucleosides in poly(A)-rich RNA. Eur. J. Biochem. 104 271–277. [DOI] [PubMed] [Google Scholar]
- Horiuchi, K., Umetani, M., Minami, T., Okayama, H., Takada, S., Yamamoto, M., Aburatani, H., Reid, P.C., Housman, D.E., Hamakubo, T., and Kodama, T. (2006). Wilms' tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc. Natl. Acad. Sci. USA 103 17278–17283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz, S., Horowitz, A., Nilsen, T.W., Munns, T.W., and Rottman, F.M. (1984). Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc. Natl. Acad. Sci. USA 81 5667–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane, S.E., and Beemon, K. (1985). Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: Implications for RNA processing. Mol. Cell. Biol. 5 2298–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, G. (1995). Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie 77 142–144. [DOI] [PubMed] [Google Scholar]
- Kennedy, T.D., and Lane, B.G. (1979). Wheat embryo ribonucleates. XIII. Methyl-substituted nucleoside constituents and 5′-terminal dinucleotide sequences in bulk poly(AR)-rich RNA from imbibing wheat embryos. Can. J. Biochem. 57 927–931. [DOI] [PubMed] [Google Scholar]
- Larsson, S.H., Charlieu, J.P., Miyagawa, K., Engelkamp, D., Rassoulzadegan, M., Ross, A., Cuzin, F., Vanheyningen, V., and Hastie, N.D. (1995). Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell 81 391–401. [DOI] [PubMed] [Google Scholar]
- Leach, R.A., and Tuck, M.T. (2001). Expression of the mRNA (N6-adenosine)-methyltransferase S-adenosyl-L-methionine binding subunit mRNA in cultured cells. Int. J. Biochem. Cell Biol. 33 984–999. [DOI] [PubMed] [Google Scholar]
- Levis, R., and Penman, S. (1978). 5′-Terminal structures of poly(A)+ cytoplasmic messenger RNA and of poly(A)+ and poly(A)− heterogeneous nuclear RNA of cells of the dipteran Drosophila melanogaster. J. Mol. Biol. 120 487–515. [DOI] [PubMed] [Google Scholar]
- Maden, B.E. (1990). The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 39 241–303. [DOI] [PubMed] [Google Scholar]
- Micura, R., Pils, W., Höbartner, C., Grubmayr, K., Ebert, M.O., and Jaun, B. (2001). Methylation of the nucleobases in RNA oligonucleotides mediates duplex-hairpin conversion. Nucleic Acids Res. 29 3997–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., Toyooka, K., Matsuoka, K., Jinbo, T., and Kimura, T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104 34–41. [DOI] [PubMed] [Google Scholar]
- Nichols, J.L. (1979). N6-Methyladenosine in maize poly(A)-containing RNA. Plant Sci. Lett. 15 357–361. [Google Scholar]
- Nichols, J.L., and Welder, L. (1981). Nucleotides adjacent to N6-methyladenosine in maize poly(A)-containing RNA. Plant Sci. Lett. 21 75–81. [Google Scholar]
- Ortega, A. (2005). Localization of the Drosophila protein FL(2)D in somatic cells and female gonads. Cell Tissue Res. 320 361–367. [DOI] [PubMed] [Google Scholar]
- Ortega, A., Niksic, M., Bachi, A., Wilm, M., Sanchez, L., Hastie, N., and Valcarcel, J. (2003). Biochemical function of female-lethal (2)D/Wilms' tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. J. Biol. Chem. 278 3040–3047. [DOI] [PubMed] [Google Scholar]
- Penn, J., Graham, P., Deshpande, G., Calhoun, G., Chaouki, A., Salz, H., and Schedl, P. (2008) Functioning of the Drosophila Wilms' Tumor 1 Associated Protein (WTAP) homolog, Fl(2)d, in Sex-lethal dependent alternative splicing. Genetics 178 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, R.P., and Kelley, D.E. (1974). Existence of methylated messenger-RNA in mouse L cells. Cell 1 37–42. [Google Scholar]
- Perry, R.P., Kelley, D.E., Friderici, K., and Rottman, F. (1975). Methylated constituents of L cell messenger-RNA—Evidence for an unusual cluster at 5′ terminus. Cell 4 387–394. [DOI] [PubMed] [Google Scholar]
- Piekna-Przybylska, D., Decatur, W.A., and Fournier, M.J. (2008). The 3D rRNA modification maps database: With interactive tools for ribosome analysis. Nucleic Acids Res. 36 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana, A.P., and Tuck, M.T. (1990). Analysis and in vitro localization of internal methylated adenine residues in dihydrofolate reductase mRNA. Nucleic Acids Res. 18 4803–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel, M., and Groner, Y. (1978). Post-transcriptional and translational controls of gene expression in eukaryotes. Annu. Rev. Biochem. 47 1079–1126. [DOI] [PubMed] [Google Scholar]
- Schibler, U., Kelley, D.E., and Perry, R.P. (1977). Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 115 695–714. [DOI] [PubMed] [Google Scholar]
- Shikanai, T. (2006). RNA editing in plant organelles: Machinery, physiological function and evolution. Cell. Mol. Life Sci. 63 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba, S., Bokar, J.A., Rottman, F., and Reddy, R. (1995). Accurate and efficient N-6-adenosine methylation in spliceosomal U6 small nuclear RNA by HeLa cell extract in vitro. Nucleic Acids Res. 23 2421–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambasco-Studart, M., Titiz, O., Raschle, T., Forster, G., Amrhein, N., and Fitzpatrick, T.B. (2005). Vitamin B6 biosynthesis in higher plants. Proc. Natl. Acad. Sci. USA 102 13687–13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck, M.T. (1992). The formation of internal 6-methyladenine residues in eucaryotic messenger RNA. Int. J. Biochem. 24 379–386. [DOI] [PubMed] [Google Scholar]
- Tuck, M.T., James, C.B., Kelder, B., and Kopchick, J.J. (1996). Elevation of internal 6-methyladenine mRNA methyltransferase activity after cellular transformation. Cancer Lett. 103 107–113. [DOI] [PubMed] [Google Scholar]
- Tzafrir, I., Dickerman, A., Brazhnick, O., Nguyen, Q., McElver, J., Frye, C., Patton, D., and Meinke, D. (2003). The Arabidopsis SeedGenes Project. Nucleic Acids Res. 31 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vespa, L., Vachon, G., Berger, F., Perazza, D., Faure, J.D., and Herzog, M. (2004). The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 134 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, C.M., Gershowitz, A., and Moss, B. (1976). 5′-Terminal and internal methylated nucleotide sequences in HeLa cell messenger-RNA. Biochemistry 15 397–401. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., and Carmichael, G.G. (2001). The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106 465–475. [DOI] [PubMed] [Google Scholar]
- Zhong, S., Lin, Z., and Grierson, D. (2008). Tomato ethylene receptor-CTR interactions: Visualization of NEVER-RIPE interactions with multiple CTRs at the endoplasmic reticulum. J. Exp. Bot. 59 965–972. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., and Reed, R. (2002). Comprehensive proteomic analysis of the human spliceosome. Nature 419 182–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.