Abstract
Mutations in a number of cardiac sarcomeric protein genes cause hypertrophic cardiomyopathy (HCM). Previous findings indicate that HCM-causing mutations associated with a truncated cardiac troponin T (TnT) and missense mutations in the β-myosin heavy chain share abnormalities in common, acting as dominant negative alleles that impair contractile performance. In contrast, Lin et al. [Lin, D., Bobkova, A., Homsher, E. & Tobacman, L. S. (1996) J. Clin. Invest. 97, 2842–2848] characterized a TnT point mutation (Ile79Asn) and concluded that it might lead to hypercontractility and, thus, potentially a different mechanism for HCM pathogenesis. In this study, three HCM-causing cardiac TnT mutations (Ile79Asn, Arg92Gln, and ΔGlu160) were studied in a myotube expression system. Functional studies of wild-type and mutant transfected myotubes revealed that all three mutants decreased the calcium sensitivity of force production and that the two missense mutations (Ile79Asn and Arg92Gln) increased the unloaded shortening velocity nearly 2-fold. The data demonstrate that TnT can alter the rate of myosin cross-bridge detachment, and thus the troponin complex plays a greater role in modulating muscle contractile performance than was recognized previously. Furthermore, these data suggest that these TnT mutations may cause disease via an increased energetic load on the heart. This would represent a second paradigm for HCM pathogenesis.
Hypertrophic cardiomyopathy (HCM) is an autosomal dominant disorder of cardiac muscle resulting from mutations in genes encoding sarcomeric proteins (1). Genetic analyses have revealed that mutations in the following seven different genes can cause this disease: the β-myosin heavy chain (2), the cardiac myosin essential light chain (3), the cardiac myosin regulatory light chain (3), α-tropomyosin (4), cardiac troponin T (TnT) (4), cardiac troponin I (5), and cardiac myosin-binding protein C genes (6, 7). We and others have suggested that the fact that the same gross phenotype is associated with a number of mutations affecting the proteins of the cardiac contractile apparatus indicates that a common mode of pathogenesis, involving a primary contractile dysfunction, exits (8, 9).
It might be anticipated that this diversity at the molecular level would manifest in significant differences in disease phenotype. Indeed, clinical analyses do reveal quantitatively significant differences in phenotype resulting from mutations in different disease genes. Missense mutations in the β-myosin heavy chain gene are associated with widely varying prognosis and risk of sudden death; those associated with a poor prognosis tend also to be associated with quite marked hypertrophy (10). In contrast, mutations in cardiac TnT produce much less clinical hypertrophy despite a typically poor prognosis (11). These clinical findings point to a dissociation between the pathways leading to hypertrophy and leading to the ventricular arrhythmias thought to underlie sudden death.
From a functional standpoint, the best characterized mutations associated with HCM are those in the β-myosin heavy chain. These mutations are missense mutations clustered in the head and head–rod junction (12). A number of studies involving a variety of approaches suggest that the primary consequence of the myosin heavy chain mutations is diminished power production by the cardiac cells that express the mutant proteins (13–18). Thus the myocardial hypertrophy seen in patients with these mutations is presumed to reflect a compensatory phenomenon, where this diminished power output likely creates a stimulus for hypertrophy. We recently examined a truncated cardiac TnT resulting from a mutation that causes abnormal splicing (8). Contractile measurements on myotubes that expressed and incorporated the truncated human cardiac TnT suggested that a mode of pathogenesis similar to that for myosin heavy chain mutations was involved, because the truncated cardiac TnT was associated with markedly reduced force production.
However, other studies call into question whether or not decreased power output is the only manifestation of the HCM-causing mutations. Mutations in the myosin light chains have been shown to cause increased speed of actin filament sliding in an in vitro motility assay (3). A missense mutation in cardiac TnT (Ile79Asn) also has been reported to cause increase in velocity in an in vitro motility assay (19), without any affect on the in vitro ATPase activity of myosin interacting with regulated actin filaments. Missense mutations in α-tropomyosin that cause HCM were also associated with increased velocity under activating conditions in the motility assay (20), whereas human skeletal muscle expressing a mutant α-tropomyosin associated with HCM displayed increased calcium sensitivity of force production (21). These data have lead to the hypothesis that a primary hypercontractile state underlies some forms of HCM (19, 21).
Other than the C-terminal truncation, all of the other HCM-linked cardiac TnT mutations that have been described result in single amino acid changes. These result from point mutations, and in one case, a single codon deletion (4, 11, 22–24). These cardiac TnT mutations would be predicted to have a lesser impact on protein structure than does the C-terminal truncation. To ascertain whether the contractile phenotype resulting from these mutations is fundamentally different from that associated with the myosin heavy chain mutations that have been examined, either wild-type or mutant (Ile79Asn, Arg92Gln or ΔGlu160) human cardiac TnT cDNA constructs were transfected into quail myotubes for structural and contractile characterization.
MATERIALS AND METHODS
Expression Constructs.
A full-length wild-type human cardiac TnT cDNA was prepared from reverse transcription-coupled PCR of mRNA from normal heart tissue as described (8). Regions containing missense mutations (Ile79Asn and Arg92Gln) and the in-frame single codon deletion (ΔGlu160) were amplified from leukocyte RNA from individuals with HCM carrying these mutations. Restriction enzyme fragments containing these mutations were then subcloned into the full-length cardiac TnT cDNA. The nucleotide sequence of each insert was confirmed. Each mutant TnT cDNA subclone was then transferred to the pCMV vector (Invitrogen), which uses the cytomegalovirus (CMV) promoter and includes a neomycin-resistance gene as a selectable marker. Resultant clones were designated by the relevant vector and mutation, e.g., pCMV.Arg92Gln.
Myocyte Transfection and Selection.
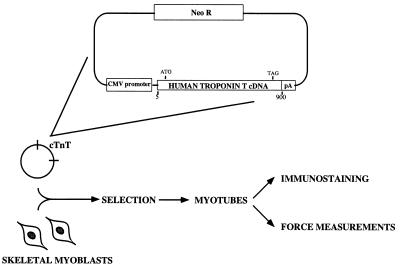
This study used a myotube assay system that was recently described in detail (25). Briefly, normal myoblasts were prepared from day 8 quail embryonic pectoral muscle and plated at a density of 5 × 105 cells per 35-mm Petri dish on a collagen- and Matrigel-coated aclar coverslip. These primary cultures of skeletal myoblasts were transfected with vector containing the wild-type, one of the two missense mutations (Ile79Asn; Arg92Gln), or the codon deletion (ΔGlu160) of human cardiac TnT cDNA. After transfection, the transfected myoblasts were selected with G418 [under bromodeoxyuridine (BrdUrd) conditions that blocked differentiation; see ref. 25], and maintained under these conditions for 7 days. After selection, BrdUrd was withdrawn, allowing myocyte differentiation into myotubes. Twelve days after withdrawal of BrdUrd, the dishes of myotubes were prepared for either immunostaining or mechanical study. These procedures are represented schematically in Fig. 1.
Figure 1.
Flow diagram of the experimental procedures. At the top of the figure is a schematic representation of the expression vector into which the human cardiac TnT cDNAs were cloned. The vector is then transfected into undifferentiated, primary skeletal myoblasts. Selection occurs under nondifferentiating conditions before allowing myotube formation from transfected myoblasts. Either microscopic or mechanical measurements are then made on the myotubes.
Western Blots.
Protein extracts from permeabilized myotubes were analyzed by Western blotting. One-dimensional denaturing PAGE was followed by electrophoretic transfer to Immobilon P membrane. The resulting transfers were probed with either an anti-TnT monoclonal antibody that recognizes all vertebrate TnT isoforms (Serotec product T1/61, Harlan Bioproducts for Science, Indianapolis, IN; see ref. 8) or one that reacts with human but not quail TnT (Sigma product JLT-12). The secondary antibody was conjugated with horseradish peroxidase to allow visualization by diaminobenzidene staining.
Immunostaining.
A monoclonal anti-TnT (clone JLT-12, Sigma) was used in immunofluorescent assays to detect the transfected human cardiac TnT in quail myotubes. Cultures were fixed in 2% formaldehyde then permeabilized and soluble proteins extracted with 0.5% Triton X-100 in PBS. The primary antibody was incubated in a humidified chamber at 37°C for 90 min, and the secondary antibody (an affinity-purified rhodamine-conjugated goat-anti-mouse IgG; Jackson ImmunoResearch) was incubated for 60 min. Preparations were examined, imaged, and photographed with a Leitz DMR microscope and a cooled charge-coupled devise camera (RTE/CCD-1317-K/1; Princeton Instruments, Trenton, NJ).
Functional Assessment.
Isolated myotubes were obtained from culture dishes of native control, wild-type, and mutant transfection groups. Medium was removed and replaced with a permeabilizing solution (170 mM potassium propionate/5 mM EGTA/2.5 mM MgCl2/2.5 mM ATP/10 mM imidazole/0.2 mM phenylmethylsulfonyl fluoride, pH 7.0) at 4°C overnight. The cultures were subsequently stored in 170 mM potassium propionate/5 mM EGTA/2.5 mM MgCl2/2.5 mM ATP/10 mM imidazole/1 mM NaN3/2.5 mM glutathione/50% glycerol, pH 7.0, at −20°C. A single permeabilized myotube was mounted with miniature aluminum “T-clips” between a servo-controlled motor and a force transducer (Series 400 Force Transducer System, Cambridge Technology, Watertown, MA) for measurement of isometric force. The entire set-up was mounted on the stage of a Leitz Fluovert equipped with Nomarski interference-contrast objectives for direct observation of sarcomere patterns and measurement of myotube width and depth to calculate the cross-sectional area. The sarcomere length of each myotube was adjusted to 2.5–2.6 μm. Ca2+-containing experimental solutions for force measurements ranged in calcium concentration from pCa 8.0 (no added calcium relaxing solution) to a maximal of pCa 4.3. Force developed at 25°C in each pCa solution was recorded with chart recorder 220 (Model 15–6327-57, Gould, Cleveland, OH), as well as digitized by using a multiplexed A/D converter and personal computer.
Maximum velocity of shortening was measured by using the Edman slack test (26). Myotubes were activated at pCa 4.3 until a steady-state level of isometric force was reached, and then a rapid (∼500 μsec) shortening step was impose. Steps varied in length from 4 to 14% of total fiber length; nine step lengths were applied. To determine stiffness/force ratios relative to wild-type levels, sinusoidal length oscillations (0.05% myotube length, peak to peak, at 1 kHz) were applied during maximal calcium activation. Data were saved to disk on a computer.
RESULTS
Expression of Human Cardiac TnT on a Quail Sarcomere Background.
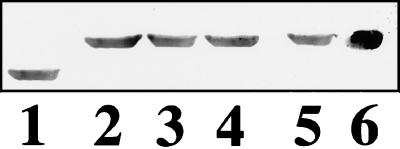
As in our previous study, myotubes derived from primary quail myocyte culture have been used to study expression of cloned human cardiac TnT cDNAs (8). However, in the present study, the CMV promoter (rather than a myosin light chain promoter) was used to drive higher levels of expression. To document the relative levels of the different TnT isoforms that were incorporated into the myofibrils, Western blotting was performed; the cardiac and endogenous quail TnTs could be distinguished by position on the blot and by differential antibody reactivity. Two different antibodies were used to probe the blots; a TnT antibody that recognized only the human cardiac TnT (Sigma product JLT-12) and an avian TnT antibody that cross-reacted with the human cardiac TnT (Serotec product T1/61). The data shown in Fig. 2 demonstrate that, complete replacement of the endogenous TnT with human cardiac TnT (wild type or mutant) was achieved, as was reported in our earlier work (8).
Figure 2.
Western blots showing relative levels of TnT in myotubes. The lanes were probed with an antibody that recognizes all vertebrate TnT isoforms (Serotec product T1/61). Lanes: 1, native TnT in control quail myotubes (control myotubes were subjected to transfection and selection protocols but with a plasmid that conferred neomycin resistance but did not contain the human cardiac TnT cDNA); 2, expressed and assembled wild-type human cardiac TnT in neomycin-selected transfected quail myotubes (note loss of endogenous TnT incorporation); 3, expressed and assembled Ile79Asn (HCM mutant) human cardiac TnT in neomycin-selected transfected quail myotubes (note loss of endogenous TnT incorporation); 4, expressed and assembled Arg92Gln (HCM mutant) human cardiac TnT in neomycin-selected transfected quail myotubes (note loss of endogenous TnT incorporation); 5, expressed and assembled ΔGlu160 (HCM mutant) human cardiac TnT in neomycin-selected transfected quail myotubes (note loss of endogenous TnT incorporation); 6, purified human cardiac TnT.
Sarcomere Assembly With Wild-Type and Mutant TnTs.
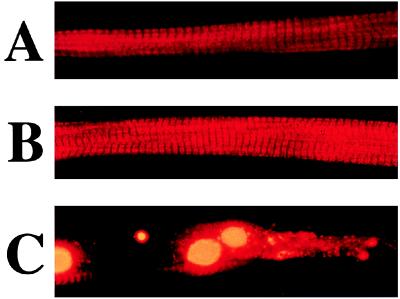
Under these conditions the ability of the transfected TnT peptides to assemble into sarcomeres was assessed by immunostaining of individual myotubes. An antibody to rabbit skeletal muscle TnT (JLT12, Sigma) did not recognize the native quail TnT sufficiently to reveal sarcomeres. Immunostaining of myotubes transfected with wild-type human cardiac TnT revealed an orderly sarcomere structure. With each of the three mutant TnTs, the majority of the myotubes had normal structure, but in each case a small percentage showed regional disruptions (Fig. 3 and Table 1).
Figure 3.
Demonstration of incorporation of wild-type and mutant TnT into sarcomeres. All myotubes were probed with an antibody that recognizes human cardiac, but not quail, TnT isoforms (Sigma product JLT-12). (A) A quail myotube demonstrating expressed (CMV promoter) and assembled human cardiac TnT from a neomycin-selected transfected culture. Note the regular sarcomeric pattern that is seen in 100% of myotubes expressing the wild-type human cardiac TnT. (B) A quail myotube demonstrating expressed (CMV promoter) and assembled mutant (Ile79Asn) human cardiac TnT from a neomycin-selected transfected culture. Note the regular sarcomeric pattern, which is seen in approximately 90% of myotubes (see Table 1) expressing the mutant cardiac TnT. (C) An example of quail myotube demonstrating expressed (CMV promoter) and assembled mutant (Ile79Asn) human cardiac TnT but with focal disruption (arrow) of the regular sarcomeric pattern. Focal disruptions are seen in approximately 10% of the myotubes expressing any of the three mutant cardiac TnTs.
Table 1.
Percentage of myotubes displaying normal or disrupted sarcomeres after CMV promoter-driven cardiac TnT expression
| % normal sarcomeres | % disrupted regions | |
|---|---|---|
| Control | 100 | 0 |
| Wild-type cardiac TnT | 100 | 0 |
| Truncated TnT mutant | 0 | 100 |
| Ile79Asn mutant | 92.5 | 7.5 |
| Arg92Gln mutant | 89 | 11 |
| ΔGlu160 mutant | 91 | 9 |
Numbers in this table are based on counts of 1,000 myotubes. The truncated TnT is described in ref. 8. It results from a splicing error that removes the C-terminal 28 amino acids and replaces them with seven incorrect amino acids.
Contractile Properties of Myotubes With Wild-Type or Mutant Cardiac TnT.
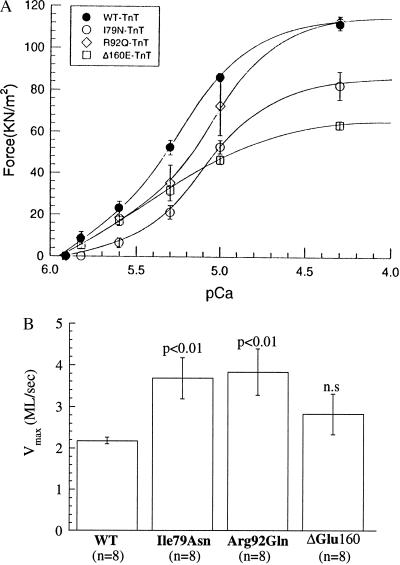
After permeabilization, individual myotubes were isolated and mounted for analysis of the contractile response to activating levels of calcium or for determination of the unloaded shortening velocity via the slack test. Maximum isometric force was not different between control myotubes transfected with vector alone (1.45 ± 0.18 kg/cm2, mean ± SEM) and myotubes transfected with wild-type human cardiac TnT (1.32 ± 0.15 kg/cm2). Thus the substitution of the human cardiac TnT for the native quail TnT did not detectably alter the maximal force of contraction. However, as shown in Fig. 4A, myotubes expressing two of the mutant cardiac TnTs (Ile79Asn and ΔGlu160) displayed a reduction of the maximal force (75% and 60% of wild-type human cardiac TnT, respectively), and the third (Arg92Gln) did not. In all cases, the stiffness/force ratio was unchanged during maximal calcium activation. Fig. 4A also demonstrates that all three mutations resulted in decreased calcium sensitivity of isometric force generation (i.e., a rightward shift of the force–pCa relationship). Interestingly, as summarized in Fig. 4B, two of the mutants resulted a doubling of the unloaded shortening velocity (Ile79Asn and Arg92Gln), and the ΔGlu160 showed a slight increase in unloaded shortening velocity that was not statistically significant.
Figure 4.
Contractile properties of myotubes expressing wild-type and mutant human cardiac TnT. (A) Force–pCa relationships for wild-type and HCM mutant cardiac TnT-containing myotubes. The corresponding Hill coefficients whereas follows: wild type, 1.94; Ile79Asn, 2.2; Arg92Gln, 1.58. For the ΔGlu160 mutation, the data at calcium concentrations less than 10−5.6 M were best fit by a Hill coefficient of 2.80, but above 10−5.6 M, the Hill coefficient was 1.44. Note that if these data are plotted relative to the maximal force for each mutant, the ΔGlu160 mutant demonstrates an apparent increase in calcium sensitivity compared with wild type, primarily due to the reduction in Hill coefficient above 10−5.6 M calcium. All other mutants show decreased calcium sensitivity, even if the data are plotted relative to maximal force. As is apparent, if the data are plotted as a function of the absolute force, all mutants show decreased calcium sensitivity compared with wild type. (B) Unloaded shortening velocities for wild-type and HCM mutant cardiac TnT-containing myotubes. The unloaded shortening velocity (Vmax) is expressed in terms of myotube lengths shortened per second (ML/sec). Student’s t tests were performed, with each mutant individually paired with wild type. The results are displayed above each bar, and the corresponding n is displayed beneath each bar.
DISCUSSION
With expression directed by a CMV promoter, the mutant human cardiac TnT replaces virtually all the native quail TnT (Fig. 2). Overexpression of wild-type cardiac TnT produced 100% normal structures. All three mutants were stably incorporated into sarcomeres (as shown by immunofluorescence with an antibody that recognizes only the human TnT; Fig. 3). However, approximately 10% of quail myotubes demonstrated disruptions or aggregates within their sarcomere structures resulting from CMV promoter-driven overexpression of either of the two missense mutants or the single codon deletion in cardiac TnT. This population of myotubes with disrupted sarcomeres was observable from the earliest time points (∼3 days after BrdUrd removal), and the percentage of these disrupted myotubes did not increase with days in culture. This suggests that the protein aggregates resulted from local high-level expression and interfered with de novo sarcomere assembly. This same phenomenon was observed to a much greater extent with the truncated cardiac TnT mutant, as documented in our earlier report (ref. 8 and Table 1). The truncated TnT tended to aggregate at much lower levels of expression, which precluded the use of the CMV promoter in that study. Thus it is likely that expression of the truncated cardiac TnT protein in human hearts could directly lead to structural disruptions. Although it is doubtful that the three mutant cardiac TnT proteins in the current study are expressed at high enough levels in human HCM patients for structural disruptions to be a primary consequence of the mutations, it cannot be ruled out that under conditions that exist in the working heart, the tendency of all of the mutant TnT proteins to aggregate may contribute to secondary structural problems associated with this type of HCM.
Contractile characterization of the mutant cardiac TnTs using myotubes with normal sarcomeric structures revealed alterations in both force production and shortening velocity due to the mutations (note that myotubes with structural abnormalities did not generate stable isometric force and could not be analyzed). Each of the mutant cardiac TnTs was associated with a shift of the force–pCa curve in the direction of decreased Ca2+ sensitivity (Fig. 4A). (However, if the forces are plotted relative to the maximal for each mutation, rather than as the absolute values used in Fig. 4A, then the ΔGlu160 mutant appears to slightly increase the calcium sensitivity relative to wild type.) The Ile79Asn and ΔGlu160 mutants reduced the maximum Ca2+-activated force (to 75% and 60% of wild-type human cardiac TnT, respectively). Consistent with the in vitro motility data of Lin et al. (19), unloaded shortening velocity was increased in myotubes expressing the Ile79Asn mutant (Fig. 4B). This increased shortening velocity also was seen for the Arg92Gln mutation and to a slight extent for the ΔGlu160 mutant.
The regulatory proteins (troponin and tropomyosin) are thought to modulate myosin cross-bridge kinetics by altering the rate at which the myosin can move from an attached non-force-generating state to a state(s) that generates force (27). In this manner the regulatory proteins are thought to control both the steady-state level of force and the rate of force generation. On the other hand, the unloaded shortening velocity during maximal calcium activation is thought to be determined solely by the kinetics of the myosin cross-bridges (28); it is limited by the rate of cross-bridge detachment from the force-generating state(s). However, the fact that the TnT mutations affect unloaded shortening velocity reveals that the regulatory proteins can accelerate cross-bridge detachment, as well as regulate the rate of force generation. Furthermore, two of the mutants show decreased force production, even at maximal levels of calcium activation. This is due to a decrease in the number of attached cross-bridges, rather than a decrease in force per cross-bridge, because the stiffness/force ratio was invariant. Thus the myosin isoforms within a striated muscle may not be the sole determinant of the maximal shortening velocity or even the cross-bridge duty cycle (the fraction of the overall actin-myosin cross-bridge cycle that a cross-bridge spends generating force). Although the current study reveals that TnT mutations can alter cross-bridge detachment rate, a clear implication is that the diversity of TnT isoforms (29), much of which is in the N-terminal region of the molecule, could function to fine tune cross-bridge kinetics, as well as calcium sensitivity.
The proposal that the regulatory proteins can alter cross-bridge detachment rate is at odds with current views on regulation, which hold that modulation of cross-bridge kinetics is via modulation of the attachment rate (27). However, for an altered attachment rate to increase the unloaded shortening velocity, the attachment rate would have to increase (30). This is inconsistent with our data, because it would lead to a longer, rather than shorter, duty cycle. The only manner in which alterations in attachment could explain our data would be if the mutations in TnT resulted in the blocking of attachment of a population of myosin heads that disproportionately contribute to drag during unloaded shortening. However, an alteration in the attachment rate for the Ile79Asn mutation is further excluded on the basis of the earlier work of Lin et al. (19). Their data revealed an increase in the in vitro motility rate (i.e., unloaded shortening) but no change in the regulated actin-myosin ATPase rate in solution and no change in the calcium sensitivity of the regulated ATPase. In these ATPase assays, there is no imposed strain on the cross-bridges, thus the duty cycle is short and subject to a different rate limiting step compared with isometric conditions in a muscle (27, 30, 31). The rate limiting step for the solution ATPase cycle is thought to be the apparent attachment rate (the rate of transition from a weakly bound non-force-generating cross-bridge state to a strongly bound force-generating state), which the data of Lin et al. (19) imply is unchanged by the Ile79Asn mutation in TnT.
The mechanism by which TnT might alter cross-bridge detachment rate is unclear, but at least in the case of the mutations described in this study it likely involves TnT–tropomyosin interactions. As shown in Fig. 5, the three mutations under investigation are in a region of TnT that interacts with tropomyosin in a Ca2+ insensitive manner (32). One could speculate that the mutations alter this interaction in a way that causes the tropomyosin either to alter the actin such that it promotes more rapid detachment of attached myosin cross-bridges or that the tropomyosin itself competes more effectively with myosin for the same sites on actin, causing more rapid detachment from actin. This is essentially the reciprocal of the accepted view of the binding of myosin cross-bridges to the thin filament leading to cooperative activation by displacing the tropomyosin from its blocking position on actin (32, 33).
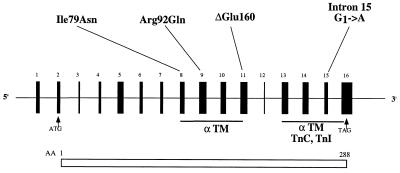
Figure 5.
Schematic of the cardiac TnT gene, indicating sites of HCM-causing mutations. The position of the three mutations investigated in this work (Ile79Asn, Arg92Gln, and ΔGlu160) are indicated, as is the splice site mutation that results in the truncated cardiac TnT that we have characterized (8). (The intron 15G→A mutation inactivates a 5′ splice donor site leading to either skipping of exon 15 or activation of a cryptic splice site.) The three mutations presented herein all lie within a region that interacts with α-tropomyosin (αTM) in a calcium insensitive manner (30). The more C-terminal region of TnT interacts in a calcium-sensitive manner with tropomyosin and interacts with the rest of the troponin complex (TnI and TnC).
In an earlier report, we concluded that a C-terminal truncation of human cardiac TnT acts as a dominant negative allele, in that it blocked calcium activation of the thin filament (8). We speculated that it was likely that all of the known cardiac TnT mutations that cause HCM act in a similar fashion to the β-cardiac myosin heavy chain mutants, providing a common mechanism of pathogenesis. Although it would appear that the cardiac TnT mutations examined in the present study do lead to reduced force production at levels of calcium activation that are germane to cardiac function, the findings suggest important differences in pathogenesis. (i) The reduction in power generation is likely more modest than that seen with myosin heavy chain mutations associated with equally poor prognosis. This may translate into a lesser stimulus for compensatory hypertrophy, which would be in keeping with the mild hypertrophy seen clinically. (ii) These findings raise the possibility of additional aspects of the pathogenesis that we have not previously recognized. We suggest that the combination of increased unloaded shortening velocity and decreased force production is indicative of an effect of the mutant cardiac TnT to shorten the cross-bridge duty cycle. Thus, in addition to decreased force output, some of the cardiac TnT mutations may increase the cost of force production (less force per ATP due to the decreased time each cross-bridge spends generating force). If this is the case, then even in the absence of hypertrophy, increased cardiac output in hearts expressing the cardiac TnT mutations could result in energy demands that cannot be met. This in turn likely would result in regional ischemia and arrhythmias, which would contribute to the high incidence of sudden death associated with these cardiac TnT mutations.
Acknowledgments
This work was supported by grants from the National Heart Lung and Blood Institute (P01-HL15835) to H.L.S. and the British Heart Foundation and Wellcome Trust to H.W.
ABBREVIATIONS
- HCM
hypertrophic cardiomyopathy
- TnT
troponin T
- CMV
cytomegalovirus
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Watkins H, Seidman J G, Seidman C E. Hum Mol Genet. 1995;4:1721–1727. doi: 10.1093/hmg/4.suppl_1.1721. [DOI] [PubMed] [Google Scholar]
- 2.Geisterfer Lowrance A A, Kass S, Tanigawa G, Vosberg H P, McKenna W, Seidman C E, Seidman J G. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 3.Poetter K, Jiang H, Hassanzadeh S, Master S R, Chang A, Dalakas M C, Rayment I, Sellers J R, Fananapazir L, Epstein N D. Nat Genet. 1996;13:63–69. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- 4.Thierfelder L, Watkins H, MacRae C, Lamas R, Vosberg H P, McKenna W J, Seidman J G, Seidman C E. Cell. 1994;77:701–712. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 5.Kimura A, Harada H, Park J E, Nishi H, Stoh M, Takahashi M, Hiroi S, Sasaoka t, Ohbuchi N, Nakamura T, et al. Nat Genet. 1997;16:379–382. doi: 10.1038/ng0897-379. [DOI] [PubMed] [Google Scholar]
- 6.Watkins H, Conner D, Thierfelder L, Jarcho J A, MacRae C, McKenna W J, Maron B J, Seidman J G, Seidman C E. Nat Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 7.Bonne G, Carrier L, Bercovici J, Cruaud C, Richard P, Hainque B, Gautel M, Labeit S, James M, Beckmann J, et al. Nat Genet. 1995;11:438–440. doi: 10.1038/ng1295-438. [DOI] [PubMed] [Google Scholar]
- 8.Watkins H, Seidman C E, Seidman J G, Feng H S, Sweeney H L. J Clin Invest. 1996;98:2456–2461. doi: 10.1172/JCI119063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vikstrom K L, Leinwand L A. Curr Opin Cell Biol. 1996;8:97–105. doi: 10.1016/s0955-0674(96)80053-6. [DOI] [PubMed] [Google Scholar]
- 10.Watkins H, Rosenzweig A, Hwang D-S, Levi T, McKenna W J, Seidman C E, Seidman J G. N Engl J Med. 1992;326:1108–1114. doi: 10.1056/NEJM199204233261703. [DOI] [PubMed] [Google Scholar]
- 11.Watkins H, McKenna W J, Thierfelder L, Suk H S, Anan R, Spirito P, Matsumori A, Moravec C, Seidman J G, Seidman C E. N Engl J Med. 1995;332:1058–1064. doi: 10.1056/NEJM199504203321603. [DOI] [PubMed] [Google Scholar]
- 12.Rayment I, Holden H M, Sellers J R, Fananapazir L, Epstein N D. Proc Natl Acad Sci USA. 1995;92:3864–3868. doi: 10.1073/pnas.92.9.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sweeney H L, Straceski A, Leinwand L A, Tikunov B, Faust L. J Biol Chem. 1994;269:1603–1605. [PubMed] [Google Scholar]
- 14.Cuda G, Fananapazir L, Zhu W S, Sellers J R, Epstein N D. J Clin Invest. 1993;91:2861–2865. doi: 10.1172/JCI116530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lankford E, Epstein N D, Fananapazir L, Sweeney H L. J Clin Invest. 1995;95:1409–1414. doi: 10.1172/JCI117795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuda G, Fananapazir L, Epstein N D, Sellers J R. J Muscle Res Cell Motil. 1997;18:275–283. doi: 10.1023/a:1018613907574. [DOI] [PubMed] [Google Scholar]
- 17.Fujita H, Sugiura S, Momomura S, Omata M, Sugi H, Sutoh K. J Clin Invest. 1997;99:1010–1015. doi: 10.1172/JCI119228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sata M, Ikebe M. J Clin Invest. 1996;98:2866–2873. doi: 10.1172/JCI119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin D, Bobkova A, Homsher E, Tobacman L S. J Clin Invest. 1996;97:2842–2848. doi: 10.1172/JCI118740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bing W, Redwood C S, Purcell I F, Esposito G, Watkins H, Marston S B. Biochem Biophys Res Commun. 1997;236:760–764. doi: 10.1006/bbrc.1997.7045. [DOI] [PubMed] [Google Scholar]
- 21.Bottinelli R, Coviello D A, Redwood C S, Pellegrino M A, Maron B J, Spirito P, Watkins H, Reggiani C. Circ Res. 1998;82:106–115. doi: 10.1161/01.res.82.1.106. [DOI] [PubMed] [Google Scholar]
- 22.Nakajima-Taniguchi C, Matsui H, Fujio Y, Nagata S, Kishimoto T, Yamauchi-Takihara K. J Mol Cell Cardiol. 1997;29:839–843. doi: 10.1006/jmcc.1996.0322. [DOI] [PubMed] [Google Scholar]
- 23.Forissier J F, Carrier L, Farza H, Bonne G, Bercovici J, Richard P, Hainque B, Townsend P J, Yacoub M H, Faure S, et al. Circulation. 1996;94:3069–3073. doi: 10.1161/01.cir.94.12.3069. [DOI] [PubMed] [Google Scholar]
- 24.Moolman J C, Corfield V A, Posen B, Ngumbela K, Seidman C E, Brink P A, Watkins H. J Am Coll Cardiol. 1997;29:549–555. doi: 10.1016/s0735-1097(96)00530-x. [DOI] [PubMed] [Google Scholar]
- 25.Sweeney H L, Feng H. Methods Cell Biol. 1997;52:275–282. [PubMed] [Google Scholar]
- 26.Edman K A P. J Physiol. 1979;291:143–159. doi: 10.1113/jphysiol.1979.sp012804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brenner B. Proc Natl Acad Sci USA. 1988;85:3265–3269. doi: 10.1073/pnas.85.9.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siemankowski R F, White H D. J Biol Chem. 1984;259:5045–5053. [PubMed] [Google Scholar]
- 29.Breitbart R E, Nguyen H T, Medford R M, Destree A T, Mahdavi V, Nadal-Ginard B. Cell. 1985;41:67–82. doi: 10.1016/0092-8674(85)90062-5. [DOI] [PubMed] [Google Scholar]
- 30.Sweeney H L, Rosenfeld S S, Brown F, Faust L, Smith J, Xing J, Stein L A, Sellers J R. J Biol Chem. 1998;273:6262–6270. doi: 10.1074/jbc.273.11.6262. [DOI] [PubMed] [Google Scholar]
- 31.Furch M, Geeves M A, Manstein D J. Biochemistry. 1998;37:6317–6326. doi: 10.1021/bi972851y. [DOI] [PubMed] [Google Scholar]
- 32.Tobacman L S. Annu Rev Physiol. 1996;58:751–792. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- 33.Lehrer S S, Geeves M A. J Mol Bio. 1998;277:1081–1089. doi: 10.1006/jmbi.1998.1654. [DOI] [PubMed] [Google Scholar]