Abstract
Misfolded proteins in the endoplasmic reticulum (ER) are often degraded in the cytosol by a process called ER-associated protein degradation (ERAD). During ERAD in S. cerevisiae, the ATPase Cdc48p associates with Der1p, a putative component of a retro-translocation channel. Cdc48p also binds a homolog of Der1p, Dfm1p, that has no known function in ERAD. Here, we show that Der1p and Dfm1p are contained in distinct complexes. While the complexes share several ERAD components, only the Dfm1p complex contains the Cdc48p cofactors Ubx1p and Ubx7p, while the Der1p complex is enriched in Ufd1p. These data suggest distinct functions for the Der1p and Dfm1p complexes.
Structured summary
MINT-6491003: Ufd1-SA (uniprotkb:P53044) physically interacts (MI:0218) with Der1-HA (uniprotkb:P38307) by anti tag coimmunoprecipitation (MI:0007)
MINT-6490940: Der1-SA (uniprotkb:P38307) physically interacts (MI:0218) with Cdc48 (uniprotkb:P25694), Usa1 (uniprotkb:Q03714), Hrd3 (uniprotkb:Q05787), Hrd1 (uniprotkb:Q08109), Ubx2 (uniprotkb:Q04228), Yos9 (uniprotkb:Q99220), Npl4 (uniprotkb:P33755) and Ufd1 (uniprotkb:P53044) by anti tag coimmunoprecipitation (MI:0007)
MINT-6490972: Dfm1-CA (uniprotkb:Q12743) physically interacts (MI:0218) with Ubx7 (uniprotkb:P38349), Ubx1 (uniprotkb:P34223), Kar2 (uniprotkb:P16474), Npl4 (uniprotkb:P33755), Yos9 (uniprotkb:Q99220), Ubx2 (uniprotkb:Q04228), Hrd1 (uniprotkb:Q08109), Hrd3 (uniprotkb:Q05787), Usa1 (uniprotkb:Q03714) and Cdc48 (uniprotkb:P25694) by anti tag coimmunoprecipitation (MI:0007)
MINT-6491016: Ufd1-SA (uniprotkb:P53044) physically interacts (MI:0218) with Dfm1-HA (uniprotkb:Q12743) by anti tag coimmunoprecipitation (MI:0007)
MINT-6491041: Ubx7-SA (uniprotkb:P38349) physically interacts (MI:0218) with Dfm1-HA (uniprotkb:Q12743) by anti tag coimmunoprecipitation (MI:0007)
MINT-6490909: Dfm1-CA (uniprotkb:Q12743) physically interacts (MI:0218) with Dfm1-HA (uniprotkb:Q12743) by anti tag coimmunoprecipitation (MI:0007)
MINT-6491029: Ubx1-SA (uniprotkb:P34223) physically interacts (MI:0218) with Dfm1-HA (uniprotkb:Q12743) by anti tag coimmunoprecipitation (MI:0007) MINT-6490896: Der1-SA (uniprotkb:P38307) physically interacts (MI:0218) with Der1-HA (uniprotkb:P38307) by anti tag coimmunoprecipitation (MI:0007)
Keywords: ER-associated degradation, Ubx proteins, Cdc48p ATPase
1. Introduction
In eukaryotic cells, misfolded soluble or membrane proteins of the endoplasmic reticulum (ER) are often transported back into the cytosol and degraded by the proteasome. This process is called ERAD, retro-translocation, or dislocation, and it involves several steps (for review, see [1]). First, the ER proteins need to be recognized as misfolded, then the proteins are retro-translocated into the cytosol, and finally they are poly-ubiquitinated and degraded by the proteasome. In yeast, ERAD substrates use distinct ubiquitin ligases, depending on where their misfolded domain is located. Proteins with misfolded lumenal or membrane domains (ERAD-L and ERAD-M substrates, respectively) use the ubiquitin ligase Hrd1p, whereas membrane proteins with misfolded cytosolic domains (ERAD-C substrates) use the ligase Doa10p [2,3]. Both Hrd1p and Doa10p associate with other membrane proteins as well as with the cytoplasmic ATPase Cdc48, which likely provides the energy to move substrates across or out of the membrane [4,5]. Similar pathways employing related components exist in higher eukaryotes [6,7].
The Hrd1p complex in yeast contains Der1p, a multi-spanning membrane protein that is required for the degradation of a variety of ERAD-L substrates[8]. The precise function of Der1p is not yet clear, but like its mammalian homologs, the Derlins, it has been implicated in the formation of a putative retro-translocation channel [9–12]. S. cerevisiae has a second Der1p-like protein, called Dfm1p (Der1-like- family-member 1) [13]. It shares 22% sequence identity with Der1p, and like Der1p, it is non-essential for the viability of yeast cells. Dfm1p is actually more related to the mammalian Derlins than Der1p itself [10]. Specifically, both Dfm1p and the Derlins posses a long C-terminal tail that interacts with Cdc48p or its mammalian homolog, the p97/VCP ATPase [9,10,14]. The mammalian Derlins can associate with one another, raising the possibility that Der1p and Dfm1p form complexes and cooperate in the same process [6,7]. In fact, the sequence similarity would suggest that Dfm1p, like Der1p, functions in ERAD, a notion that is supported by the observation that the expression of Dfm1p is upregulated by the unfolded protein response (UPR) [14,15]. In addition, a der1Δdfm1Δ double mutant triggers a stronger UPR than the der1Δ single mutant [14]. However, numerous attempts to find ERAD substrates for Dfm1p have so far failed. It thus remains unclear whether Der1p and Dfm1p function in the same pathway or have distinct roles.
Here we show that Der1p and Dfm1p are contained in distinct complexes. Nevertheless, a systematic analysis of interaction partners shows that both proteins interact with an identical set of membrane proteins, the Hrd1p-complex, and that they both associate with the Cdc48p ATPase. However, the Dfm1p complex differs from the Der1p complex in that it contains the Cdc48p cofactors Ubx1p and Ubx7p. Conversely, the Der1p complex contains higher levels of the cofactor Ufd1p. These data support the idea that Der1p and Dfm1p perform distinct cellular functions.
2. Material and Methods
Yeast strains
Chromosomal tagging of proteins was performed using standard PCR-based homologous recombination. All proteins were expressed under their endogenous promoters. The strains used are isogenic to BY4743 (MATa/α his3Δ/his3Δ leu2Δ/leuΔ ura3Δ/ura3Δ) or its MATa haploids.
Immunoprecipitation
A detailed protocol has been published previously [3]. In brief, crude membrane fractions were prepared from 50ml cultures and solubilized in 1% digitonin. The extract was incubated with either calmodulin-coupled beads (Pharmacia), IgG-coupled magnetic beads (Dynal), or S-peptide binding protein-coupled beads (Novagen). After washing, the bound proteins were eluted with SDS, separated by SDS-PAGE, and analyzed by immunblotting with antibodies to HA or with IgG, followed by incubation with peroxidase-coupled secondary antibodies.
Protein complex purification
15g of yeast cells were lysed by grinding in liquid nitrogen and membranes were solubilized in 1% digitonin. The extract was incubated with IgG-coupled magnetic beads (Dynal). After washing, proteins were eluted with SDS. The eluate was either precipitated with trichloroacetic acid or separated by SDS-PAGE, followed by Coomassie staining and excision of individual bands. In both cases, the proteins were identified by mass spectrometry.
3. Results
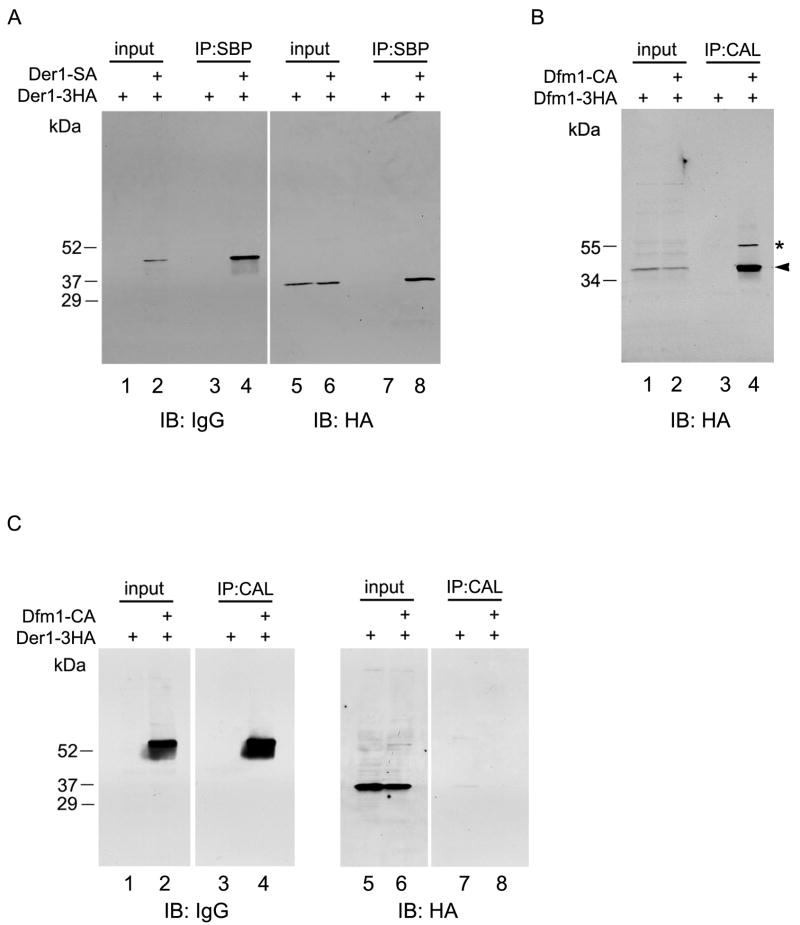
We first tested whether Der1p can self-associate, as demonstrated for its mammalian homologs. To this end, we generated two differently tagged versions of Der1p. The first construct contained at the C-terminus both S-peptide- and protein A-tags (Der1-SA), which interact with a speficic S-peptide binding protein and IgG, respectively. The second construct contained three hemagglutinin (HA) tags at the C-terminus of Der1p (Der1-3HA). As demonstrated previously, both constructs support ERAD, although they are not fully functional; nevertheless, they associate with all components of the Hrd1p complex [3]. Haploid yeast cells expressing the two constructs under their endogenous promoters were mated to obtain diploid cells expressing both proteins at the same time. The cells were broken and cell lysates were either subjected directly to SDS-PAGE (input), or the lysate was incubated with beads containing the S-peptide binding protein, and bound proteins were analyzed by SDS-PAGE (IP). In both cases, the proteins were transferred to nitrocellulose and probed with IgG or HA antibodies, followed by peroxidase-coupled secondary antibodies. As shown in Fig. 1A, lane 8, pull-down of Der1-SA co-isolated Der1-3HA. No band was seen with control cells that only expressed Der1-3HA (lane 7). These data indicate that Der1p can self-associate, consistent with results obtained with the mammalian Derlins.
Figure 1. Der1p and Dfm1p self-associate, but do not interact with each other.
(A) Yeast cells expressing an HA-tagged version of Der1p (Der1-3HA) with or without a SA-tagged version of Der1p (Der1-SA) were lysed. Samples were analyzed directly (input; 5% of total material) or after immunoprecipitation using coupled S-peptide binding protein (SBP) (IP; 95% of total material). All samples were separated by SDS-PAGE and analyzed by immunoblotting (IB) with the indicated antibodies. The arrowhead indicates Der1-3HA co-immunoprecipitated by Der1-SA. (B) As in (A), but using cells expressing tagged versions of Dfm1p (Dfm1-CA and Dfm1-3HA, respectively). The IP was performed using bead-coupled calmodulin (CAL). The arrowhead indicates Dfm1-3HA co-immunoprecipitated by Dfm1-CA. The asterisk indicates Dfm1-CA detected by IgG of the secondary antibody. (C) As in (B) but using cells expressing tagged versions of Dfm1p and Der1p (Dfm1-CA and Der1-3HA, respectively).
Next, we performed similar experiments with Dfm1p. In this case, the protein was fused at the C-terminus to a calmodulin peptide and a protein A module (Dfm1-CA) or to three HA tags (Dfm1-3HA). Cell lysates were incubated with calmodulin-coupled beads and bound proteins were analyzed by blotting with HA antibodies (Fig. 1B, lane 4). Two bands were visible, one corresponding to the co-precipitated Dfm1-3HA protein (arrow head), the other to Dfm1-CA (star), which reacts with the IgG of the secondary antibodies. No bands were seen with control cells expressing only Dfm1-3HA (lane 3). These data thus show that Dfm1p, like Der1p, can form oligomers that contain at least two Dfm1p molecules.
Finally, we tested whether Der1p interacts with Dfm1p. Dfm1-CA was co-expressed with Der1-3HA in haploid yeast cells, and pull-down experiments were performed with calmodulin-containing beads. Despite the fact that both proteins could be detected in the input fractions (Fig. 1C, lanes 2 and 6) and that Dfm1-CA was efficiently recovered in the bound fraction (lane 4), there was no co-isolation of Der1-3HA (lane 8). Thus, Der1p and Dfm1p do not appear to interact with one another.
Based on the observation that the Der1p and Dfm1p complexes appear to be distinct, we reasoned that they might contain different components. To identify interaction partners of Der1p and Dfm1p, we expressed Der1-SA and Dfm1-CA under their endogenous promoters. The yeast cells were broken and a membrane fraction was solubilized in 1% digitonin. The extract was incubated with IgG coupled to magnetic beads, and bound proteins were eluted with SDS. They were separated by SDS-PAGE and stained with Coomassie blue. A control, performed with wild type cells containing untagged proteins, showed that the major bands eluted from the beads were IgG heavy and light chains, which presumably were not covalently bound (Fig. 2, lane 1–3). However, with both Der1-SA amd Dfm1-CA a number of additional bands were seen (numbered in lanes 2 and 3). The identity of the bands was determined by mass spectrometry (Table). We also analyzed the proteins bound to the IgG beads directly by mass spectrometry after precipitation with trichloroacetic acid (Table).
Figure 2. Interaction partners of Der1p and Dfm1p.
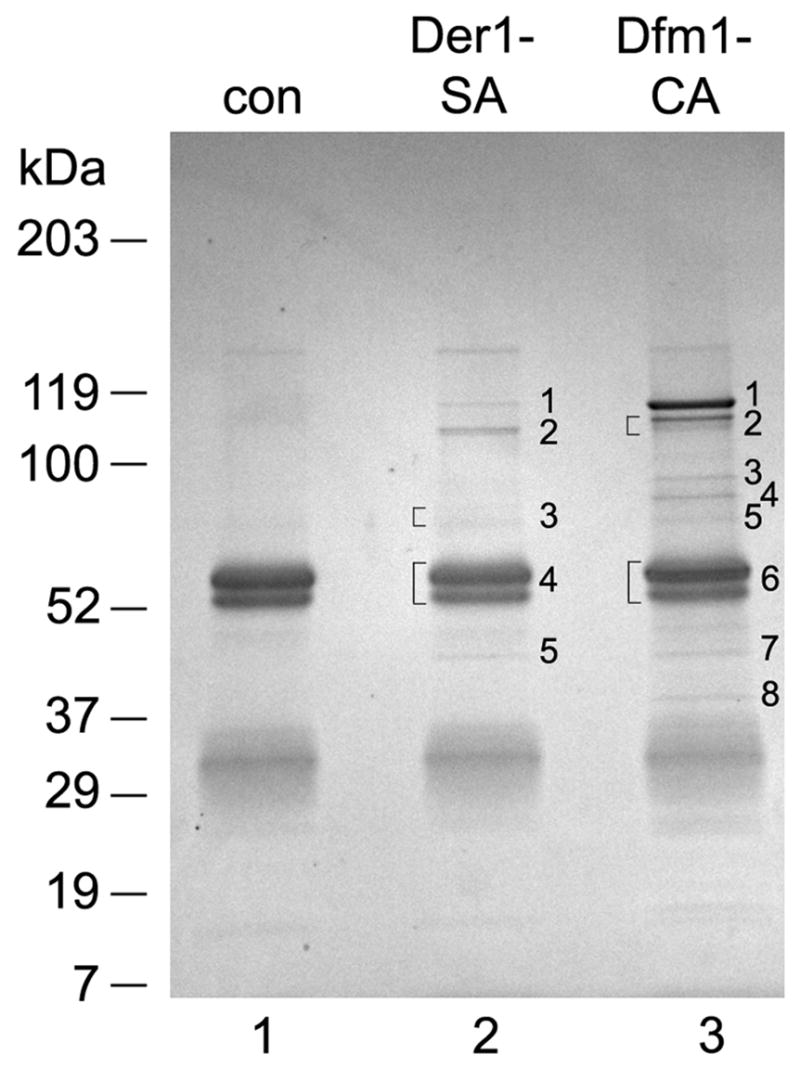
Wild-type yeast cells (con) or cells expressing either Der1-SA or Dfm1-CA were lysed and membrane fractions were solubilized with 1% digitonin. The extract was incubated with IgG-coupled magnetic beads and bound material was analyzed by SDS-PAGE and Coomassie staining. The numbered stained bands were cut out and their identity determined by mass spectrometry. Brackets indicate where closely spaced bands were cut out and analyzed together.
Table.
Summary of interacting proteins found when immunoprecipitation was performed using tagged Der1p (Der1-SA) or tagged Dfm1p (Dfm1-CA). The bound proteins were identified using mass spectrometry. We analyzed either the total protein eluate after precipitation with trichloroacetic acid (TCA), or individual protein bands after their separation in a SDS gel and staining with Coomassie blue (Ind). Numbers indicate the amount of peptides identified by mass spectromety. For multiple experiments, numbers are separated by a dash. The numbers in brackets correspond to the numbers assigned to the bands in Figure 2.
| Interacting protein | Tagged protein (Bait) | |||
|---|---|---|---|---|
| Der1-SA [5] | Dfm1-CA [6] | |||
| TCA | Ind | TCA | Ind | |
| Cdc48p | 12 | 23 [1] | 46/68 | 55 [1] |
| Usa1p | 10 | 23 [2] | 2/6 | 21 [2] |
| Hrd3p | 11 | 23 [2] | 2/9 | 22 [2] |
| Kar2 | 0 | 0 | 0/8 | 31 [3] |
| Ubx2p | 0 | 8 [3] | 0/0 | 7 [4] |
| Yos9p | 5 | 6 [3] | 2/1 | 10 [4] |
| Hrd1p | 1 | 9 [3] | 3/2 | 7 [5] |
| Npl4p | 0 | 3 [3] | 0/0 | 8 [5] |
| Ubx1p | 0 | 0 | 4/4 | 2 [6] |
| Ubx7p | 0 | 0 | 4/8 | 5 [6] |
| Ufd1p | 0 | 1 [5] | 0/0 | 0 |
All Der1p interaction partners have been identified before: Cdc48p (band 1), Usa1p and Hrd3p (band 2), Hrd1p, Ubx2p, Yos9p, and Npl4p (band 3), Der1p-TAP and Ufd1p (band 5); band 4 contained only rabbit IgG. The membrane proteins Hrd1p, Hrd3p, Usa1p, and Der1p are members of the Hrd1p complex, Npl4p and Ufd1p are cofactors of the Cdc48p ATPase, Ubx2p is a membrane-bound binding partner of Cdc48p, and Yos9p is a luminal lectin-like chaperone. For all proteins there is evidence that they participate in ERAD.
The majority of the Dfm1p components are identical to those in the Der1p complex: Cdc48p (band 1), Usa1p and Hrd3p (band 2), Yos9p and Ubx2p (band 4), and Npl4p and Hrd1p (band 5). The approximate levels of most of these proteins was about the same in the two complexes, as judged from the number of peptides identified, with the exception of Cdc48p, which was more abundant in the Dfm1p complex (see band 1 in lanes 2 and 3). Dfm1p-TAP was contained in band 6 that was dominated by IgG. In addition to these proteins, we found Kar2p (band 3) as well as Ubx1p and Ubx7p (band 6). Kar2p (BiP) is a member of the Hsp70 family of chaperones and is an abundant luminal ER protein. The detection of Ubx1p and Ubx7p is particularly interesting because both proteins are established cofactors of the Cdc48p ATPase, but have no known function in ERAD. Bands 7 and 8 contained peptides of Dfm1p and Cdc48p, respectively. These could originate from proteolysis or, in the case of Dfm1p, represent untagged Dfm1p. In contrast to the results with the Der1p complex, Ufd1p was undetectable in the Dfm1p complex. The results were the same when the samples were analyzed directly after trichloroacetic acid precipitation (Table), indicating that no major component was missed in the Coomassie-stained gel.
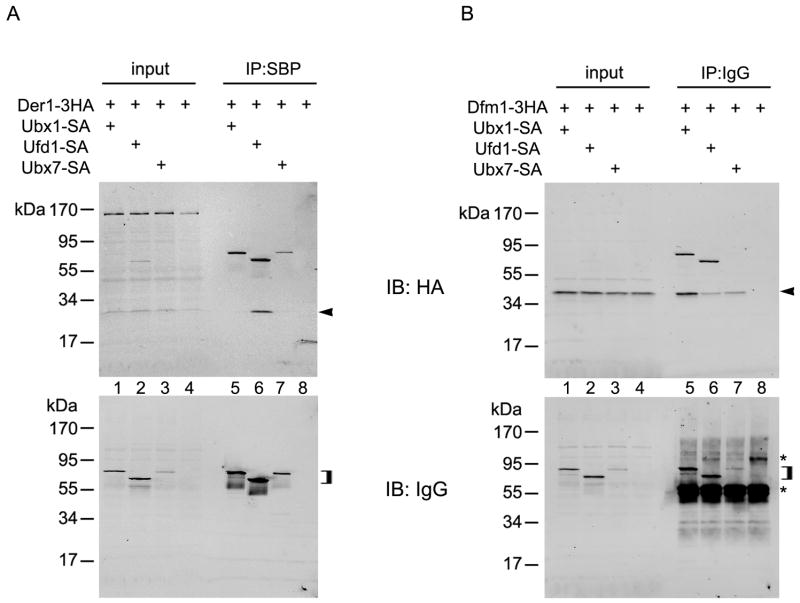
To confirm that the Der1p- and Dfm1p- complexes contain distinct cofactors of the Cdc48p ATPase, we performed pull-down experiments with tagged proteins. Der1-3HA was expressed together with Ubx1p, Ufd1p, or Ubx7p, all containing at their C-termini both S-peptide- and protein A- tags (Ubx1-SA, Ufd1-SA, Ubx7-SA). All proteins were expressed under their endogenous promoters. The expression level of Ubx7p-SA was lower than that of Ubx1p-SA or Ufd1p-SA, as determined by SDS-PAGE followed by probing with IgG (Fig. 3A, lower panel, lane 3 versus lanes 1 and 2). All SA-tagged proteins were also efficiently recovered with beads to which the S-peptide binding protein had been coupled (lanes 5–7). When probed with HA antibodies for co-precipitation of Der1-HA, a band was only seen in pull-down experiments with Ufd1-SA (Fig. 3A, upper panel, lane 6; arrow head). This band was not seen in the absence of the SA-tagged proteins (lane 8). No association of Der1-3HA was observed with either Ubx1-SA or Ubx7-SA (lanes 5 and 7) (the upper bands in lanes 5–7 result from the detection of the protein A moiety in the SA-tagged protein by the secondary antibody and serve as an additional control for the recovery of the tagged proteins). These data show that Ufd1p, but not Ubx1p or Ubx7p, is associated with the Der1p complex, in agreement with the mass spectrometry experiments. It should be noted that in the two sets of experiments, the pull-down was performed in a reciprocal manner (Ufd1-SA in Fig. 3 versus Der1-SA in Fig. 2), confirming the existence of a genuine complex.
Figure 3. Der1p and Dfm1p bind a different set of Cdc48p cofactors.
(A) Yeast cell lysates from cells expressing a tagged version of Der1p (Der1-3HA) and a tagged version of either Ubx1p (Ubx1-SA), Ufd1p (Ufd1-SA) or Ubx7p (Ubx7-SA) were analyzed either directly (input; 5% of total material) or by immunoprecipitation (IP; 95% of total material) using coupled S-peptide binding protein (SBP). The samples were separated by SDS-PAGE and analyzed by immunoblotting (IB) with the indicated antibodies. The arrowhead in the upper panel indicates Der1-3HA co-immunoprecipitated by Ufd1-SA. The square bracket in the lower panel indicates the position of the SA-tagged cofactors. (B) As in (A), except that a tagged version of Dfm1p (Dfm1-3HA) was expressed together with the SA-tagged cofactors. The immunoprecipitation was performed using IgG bound to magnetic beads (IgG). The arrowhead in the upper panel indicates Dfm1-3HA co-immunoprecipitated by Ubx1-SA, Ufd1-SA, or Ubx7-SA. The square bracket in the lower panel indicates the position of the SA-tagged cofactors. The asterisks indicate coeluted IgG and a crossreacting protein.
Similar experiments were performed with the Dfm1p complex. In this case, Dfm1-3HA was co-expressed with SA-tagged versions of Ubx1p, Ufd1p, or Ubx7p, and the SA-tagged proteins were bound to magnetic beads containing coupled IgG. When the bound proteins were probed with HA antibodies, a major Dfm1-3HA band was seen when the pull-down was performed with Ubx1-SA (Fig. 3B, upper panel, lane 5; arrow head) (the upper band results from detection of the protein A moiety in the SA-tagged protein). Smaller quantities of Dfm-3HA bands were pulled down by Ufd1-SA or Ubx7-SA (upper panel, lanes 6 and 7). Whereas Ufd1-SA was expressed at about the same level as Ubx1-SA (see lower panel, lanes 1 and 2), the level of Ubx7-SA was significantly lower (lane 3). Thus, the Dfm1p complex contains a significantly higher fraction of Ubx7p than Ufd1p. These results again confirm our mass spectrometry data, as they indicate that, in contrast to Der1p, Dfm1p is associated with Ubx1p and Ubx7p. Although small amounts of Ufd1p were associated with Dfm1p in the pull-down experiments, the amounts of Ufd1p in the Der1p complex were significantly higher (compare the ratio of the Der1-3HA band to the Ufd1-SA band in Fig. 3A, upper panel, lane 6 with the ratio of these bands in Fig. 3B, upper panel, lane 6). The low level of Ufd1p in the Dfm1p complex accounts for the lack of Ufd1p peptides in the mass spectrometry experiments.
4. Discussion
Our results show that Der1p and Dfm1p are contained in distinct complexes. The Der1p complex contains the Cdc48p cofactor Ufd1p and lacks Ubx1p and Ubx7p. Conversely, the Dfm1p complex is depleted of Ufd1p and instead contains Ubx1p and Ubx7p. Given that Npl4p forms a heterodimer with Ufd1p [16], we suspect that Npl4p is also more abundant in the Der1p complex, although it is detectable by mass spectrometry in both complexes. The mutually exclusive binding of the Ufd1p/Npl4p and Ubx1p cofactors is consistent with previous results [16,17]. The different composition of the two complexes suggests that Der1p and Dfm1p have distinct roles.
One possibility is that Dfm1p functions in a process other than protein degradation. For example, mammalian p47 (Ubx1p in yeast), appears to be involved in membrane fusion [18,19]. Other data show that Dfm1p interacts genetically with Ufe1p, an ER-SNARE [14]. Although we cannot rule out a role for Dfm1p in these processes, we did not find interacting proteins with an established role in vesicular traffic. Rather, the detection of the entire set of known ERAD components in the Dfm1p complex suggests some role in protein degradation.
In one model, the Dfm1p complex serves as a “buffer”, whose components can be activated for ERAD on demand. In this view, the Der1p complex would carry the load of retro-translocation of misfolded substrates, whereas the Dfm1p complex would sequester ERAD components in an inactive state. This idea is supported by the following observations. First, in contrast to Der1p, the deletion of Dfm1p has no effect on the degradation of a variety of misfolded ER proteins: CPY*, Sec61-2, KWW, Ste6-166, KHN, Hmg2 [13,14]. We confirmed and extended these findings to CFTR (a mammalian protein that is degraded by ERAD in yeast), Gas1* (a misfolded GPI-anchored protein), and overexpressed CPY* (a protein targeted to a post-ER compartment for destruction [20]) (our unpublished results). Dfm1p also does not appear to affect the degradation of the cytosolic substrate Ub-P-bGal (unpublished results). Second, the Der1p-specific interaction partner Ufd1p is a known ERAD component [28], while the deletion of Ubx1p or Ubx7p has no effect on the degradation of the ERAD substrates CPY* or GAS1p*, respectively (data not shown). Third, while the deletion of der1 has a synthetic growth defect with a deletion of the UPR gene ire1, an observation made for all components that are active in ERAD, no such genetic interactions was observed between ire1 and dfm1 or ubx7 (Supplementary Fig. S1; a double deletion of ubx1 and ire1 could not be constructed because the ubx1 mutant is defective in sporulation).
Interestingly, both the Dfm1p and Der1p complexes contain the two proteins involved in substrate recognition, Hrd3p and Yos9p [21–23]. This makes it unlikely that the Dfm1p complex has an entirely different set of substrates. Instead, the Dfm1p complex might recognize misfolded lumenal glycosylated ERAD-L substrates, such as CPY*, but may not be capable of initiating their retro-translocation. The reason might be that the Dfm1p complex contains a different set of Cdc48p cofactors than the Der1p complex.
Several Cdc48p partners in the Der1p- and Dfm1p- complexes interact directly with Cdc48p and modulate its activity: Ubx1p, Ubx2p, and Ubx7p through their Ubx domains [24,25], Npl4p through its Ubl domain [26], and Dfm1p, Ufd1p, and Ubx1p through a heptapeptide motif called the SHP-box [13,14] (for review see [27]). ERAD is known to require the ATPase activity of Cdc48p [28], and it is therefore intriguing that the Dfm1p complex contains Ubx1p, whose mammalian homolog inhibits the ATPase [29]. This would therefore be consistent with the idea that the protein complex containing Dfm1p, Ubx1p, and Ubx7p is inactive for ERAD. Exchanging these components for Der1p, Ufd1p, and Npl4p would result in the activation for retro-translocation. The idea that Dfm1p may be a negative regulator of ERAD has been expressed before, but was considered to be unlikely because the overexpression of Dfm1p results in the upregulation of UPR regardless of whether it contained or lacked the Cdc48p-interacting SHP box [14]. Our finding that the Dfm1p complex is associated not only with Cdc48p but with the entire set of known ERAD components suggests that the overexpression of Dfm1p may have sequestered ERAD components other than Cdc48p and thereby caused UPR. Taken together, the role for Dfm1p (and other Cdc48p binding proteins) might therefore be in regulating the activity of Cdc48p for its function in ERAD.
Supplementary Material
A yeast strain deleted for ire1 carrying a His marker was crossed with strains deleted for der1, dfm1, or ubx7, all carrying a G418 resistance marker. All strains were isogenic to BY4743. The diploids were sporulated and double-deletion mutants were isolated. Wild type cells, and single and double deletion mutants were serially diluted and grown on YPD plates at 30°C or 37°C for two days. Note that the strain carrying a double deletion of ire1 and der1 grows slower at 37°C than all the other strains.
Acknowledgments
We would like to thank L. Hicke for antibodies and A. Buchberger, D.Ng, R. Hampton, S.Michaelis, D.Wolf and T.Yoko-o for plasmids; A.M. Stanley for critical reading of the manuscript; and members of the Rapoport lab for discussions. P.C. is supported by a Jane Coffin Childs Memorial Fund for Medical Research fellowship. T.A.R. is supported by an NIH grant and is a Howard Hughes Medical Institute Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–72. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 2.Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–73. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 4.Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7:993–8. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- 5.Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- 6.Ye Y, Shibata Y, Kikkert M, van Voorden S, Wiertz E, Rapoport TA. Inaugural Article: Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A. 2005;102:14132–8. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lilley BN, Ploegh HL. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A. 2005;102:14296–301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knop M, Finger A, Braun T, Hellmuth K, Wolf DH. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. Embo J. 1996;15:753–63. [PMC free article] [PubMed] [Google Scholar]
- 9.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–7. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 10.Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–40. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- 11.Sun F, Zhang R, Gong X, Geng X, Drain PF, Frizzell RA. Derlin-1 promotes the efficient degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) and CFTR folding mutants. J Biol Chem. 2006;281:36856–63. doi: 10.1074/jbc.M607085200. [DOI] [PubMed] [Google Scholar]
- 12.Oda Y, Okada T, Yoshida H, Kaufman RJ, Nagata K, Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol. 2006;172:383–93. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hitt R, Wolf DH. Der1p, a protein required for degradation of malfolded soluble proteins of the endoplasmic reticulum: topology and Der1-like proteins. FEMS Yeast Res. 2004;4:721–9. doi: 10.1016/j.femsyr.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Sato BK, Hampton RY. Yeast Derlin Dfm1 interacts with Cdc48 and functions in ER homeostasis. Yeast. 2006;23:1053–64. doi: 10.1002/yea.1407. [DOI] [PubMed] [Google Scholar]
- 15.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–58. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 16.Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. Embo J. 2000;19:2181–92. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pye VE, Beuron F, Keetch CA, McKeown C, Robinson CV, Meyer HH, Zhang X, Freemont PS. Structural insights into the p97-Ufd1-Npl4 complex. Proc Natl Acad Sci U S A. 2007;104:467–72. doi: 10.1073/pnas.0603408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj IW. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol. 2001;3:1086–91. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- 19.Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388:75–8. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- 20.Haynes CM, Caldwell S, Cooper AA. An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER-Golgi transport. J Cell Biol. 2002;158:91–101. doi: 10.1083/jcb.200201053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8:849–54. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 22.Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–75. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 23.Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–51. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 24.Schuberth C, Richly H, Rumpf S, Buchberger A. Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep. 2004;5:818–24. doi: 10.1038/sj.embor.7400203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchberger A, Howard MJ, Proctor M, Bycroft M. The UBX domain: a widespread ubiquitin-like module. J Mol Biol. 2001;307:17–24. doi: 10.1006/jmbi.2000.4462. [DOI] [PubMed] [Google Scholar]
- 26.Bruderer RM, Brasseur C, Meyer HH. The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism. J Biol Chem. 2004;279:49609–16. doi: 10.1074/jbc.M408695200. [DOI] [PubMed] [Google Scholar]
- 27.Jentsch S, Rumpf S. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–6. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 29.Meyer HH, Kondo H, Warren G. The p47 co-factor regulates the ATPase activity of the membrane fusion protein, p97. FEBS Lett. 1998;437:255–7. doi: 10.1016/s0014-5793(98)01232-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A yeast strain deleted for ire1 carrying a His marker was crossed with strains deleted for der1, dfm1, or ubx7, all carrying a G418 resistance marker. All strains were isogenic to BY4743. The diploids were sporulated and double-deletion mutants were isolated. Wild type cells, and single and double deletion mutants were serially diluted and grown on YPD plates at 30°C or 37°C for two days. Note that the strain carrying a double deletion of ire1 and der1 grows slower at 37°C than all the other strains.