Abstract
Previously, we showed that the addition of human erythrocyte glycosphingolipids (GSLs) to nonhuman CD4+ or GSL-depleted human CD4+ cells rendered those cells susceptible to HIV-1 envelope glycoprotein-mediated cell fusion. Individual components in the GSL mixture were isolated by fractionation on a silica-gel column and incorporated into the membranes of CD4+ cells. GSL-supplemented target cells were then examined for their ability to fuse with TF228 cells expressing HIV-1LAI envelope glycoprotein. We found that one GSL fraction, fraction 3, exhibited the highest recovery of fusion after incorporation into CD4+ nonhuman and GSL-depleted HeLa-CD4 cells and that fraction 3 contained a single GSL fraction. Fraction 3 was characterized by MS, NMR spectroscopy, enzymatic analysis, and immunostaining with an antiglobotriaosylceramide (Gb3) antibody and was found to be Gal(α1→4)Gal(β1→4)Glc-Cer (Gb3). The addition of fraction 3 or Gb3 to GSL-depleted HeLa-CD4 cells recovered fusion, but the addition of galactosylceramide, glucosylceramide, the monosialoganglioside, GM3, lactosylceramide, globoside, the disialoganglioside, GD3, or α-galactosidase A-digested fraction 3 had no effect. Our findings show that the neutral GSL, Gb3, is required for CD4/CXCR4-dependent HIV-1 fusion.
HIV-1 enters permissive cells by binding to the cellular receptor, CD4 (1), followed by gp120-gp41-mediated fusion of the viral and target cell membranes (2, 3). In addition to CD4, several members of the chemokine-receptor family have been shown to act as necessary coreceptors for HIV-1 fusion and infection (4, 5). Chemokine-receptor usage varies depending on the viral strain and the target cells used and is the primary determinant of viral tropism (6–8). Although the discovery of chemokine receptors has revolutionized our thinking about the way HIV-1, HIV-2, and the simian immunodeficiency virus gain entry into the cell (9), some crucial pieces of the puzzle are missing. Certain HIV-1 strains have been shown to infect CD4+ cells in the absence of known chemokine receptors (10, 11). Moreover, a panel of HIV-1 and HIV-2 strains, whose infection of CXCR4+ CD4+ RD rhabdomyosarcoma cells was inhibited by a mouse anti-CXCR4 monoclonal antibody (12G5), resisted 12G5 inhibition on T cell lines. These observations suggest that CXCR4 could be processed or presented differently depending on the cell type, allowing some strains to evade 12G5 inhibition. Therefore, it is conceivable that other cofactors, possibly glycosphingolipids (GSLs), are involved, enabling individual HIV strains to choose between alternate modes of entry into cells.
The GSL hypothesis is based on a number of observations, including recovery of fusion of nonsusceptible cells after transfer of protease- and heat-resistant components from human erythrocytes (12, 13), physicochemical studies on the interactions of the gp120 V3 loop with defined GSLs (14–17), and inhibition of HIV-1 infection in vitro after treatment of cells with several inhibitors of GSL biosynthesis (18–20).
Recently, we have shown that incorporation of GSLs isolated from human (but not bovine) erythrocytes into the membranes of fusion-incompetent CD4+ cells complements subsequent HIV-1 fusion (20). In this study, we have followed up on this observation by isolating individual components from the human erythrocyte GSL mixture by using silica-gel column chromatography and testing the fractions for HIV-1LAI-gp120-gp41-mediated fusion activity after incorporation into CD4+ nonhuman cells and GSL-depleted HeLa-CD4 cells (HeLa-CD4/GSL−). We have identified a neutral GSL fraction, fraction 3 (Fr. 3), in the crude GSL mixture, which conferred the highest HIV-1 fusion activity when incorporated into nonsusceptible cells. Structural analysis of this fusion-active fraction showed its chemical structure to be Gal(α1→4)Gal(β1→4)Glc-Cer (Gb3).
MATERIALS AND METHODS
Materials.
Fluorescent probes were obtained from Molecular Probes, and tissue culture media were obtained from GIBCO/BRL. Phospholipids were purchased from Avanti Polar Lipids, and 1-phenyl-2-hexadecanoylamino-3-morpholino-1-propanol (PPMP), GSLs, neutral GSL Qualmix standards, and rabbit anti-asialo-GM2 antibody were from Matreya (Pleasant Gap, PA). Other reagents were from Sigma. Mouse anti-galactosylceramide mAb was from Boehringer Mannheim. Rat anti-Gb3 mAb (38.13) was generated as described (21).
Fractionation of Human Erythrocyte GSLs.
Total GSLs were extracted from human erythrocytes as described (20). The GSL mixture was fractionated further on a silica-gel column by using a CHCl3/MeOH solvent mixture of increasing polarity. The solvents were removed under vacuum, and the components of individual GSL fractions were detected by chromatography on silica-gel TLC. Bial’s reagent, which detects sugars, GSLs, sulfolipids, and gangliosides, was used to detect the GSLs on the TLC plates.
Acetylation of Fr. 3.
The GSL was acetylated with acetic anhydride/pyridine in a 1:2 ratio (vol/vol) for 24 h at room temperature. The reagents were evaporated under reduced pressure. The residue was taken up in water and extracted three times with ethyl acetate. The combined organic extracts were washed with water and brine and dried by filtration through a pad of sodium sulfate. The crude peracetate was purified on a silica-gel column with a gradient of CH3OH (0–3%) CH2Cl2.
Electrospray Liquid-Chromatography/MS Analysis.
A Hewlett–Packard 5989B mass spectrometer with an Analytica (Branford, CT) electrospray source was used to analyze GSL samples with the same parameters described (22). Samples and known standards were injected onto a C-18 HPLC column (Phenomenex, Belmont, CA; 5 μm; 2.0 × 150 mm) and separated by using a mobile phase consisting of water/MeOH with 0.5% NaOH/hexane changing linearly from 12:88:0 to 0:98:2 (vol/vol) in 7 min at the flow rate of 0.3 ml/min, after holding at the initial composition for 3 min. The final mobile-phase composition was maintained for 15 min.
NMR Spectroscopy.
NMR spectroscopy was performed with a Bruker AMX spectrometer at 500 MHz (Billerica, MA). We collected 1H one-dimensional spectra at different temperatures from 288 K to 323 K controlled by a Eurotherm variable temperature unit with an accuracy of ±0.1 K. The 90° pulse width was 10.85 ms, and the sweep width was 5,000 Hz. The spectra were processed with Gaussian multiplication (lb = −1, gb = 0.1). Two-dimensional total correlation spectroscopy (TOCSY) was performed with the standard pulse sequence (23), and the data were collected with a sweep width of 4,466 Hz in both dimensions and with a spin-lock mixing time of 65 ms.
High-Performance TLC (HPTLC) Immunostaining of Fr. 3.
GSLs were separated on glass-backed HPTLC plates (Analtech) by using CHCl3/MeOH/H2O (75:21.5:3.5, vol/vol) as the mobile phase. The samples (5 μg per lane) were run in duplicate sets. Plates were air dried and split into equal halves. A portion was sprayed with Bial’s reagent, and the other part was stabilized mechanically by a 2-min bath in 0.1% polyisobutyl methacrylate in n-hexane. The dried plates were sprayed gently with PBS and immediately immersed into Tris/BSA buffer to block nonspecific binding for 30 min at room temperature. The plates were then treated with appropriate dilutions of anti-GSL antibodies for 1 h at room temperature. Antibody binding was detected with an alkaline phosphatase detection kit (Boehringer Mannheim).
Enzymatic Analysis of Fr. 3.
Dried aliquots of Fr. 3 and authentic GSLs (20 μg) were digested by incubating for 2 h with an excess of (7,000 units) α-galactosidase A [αGalA; prepared as described in ref. 24 in 80 mM sodium acetate (pH 4.1)/0.25% Nonidet P-40/0.25% sodium taurocholate]. After Folch partition (25), the lower phase was dried and redissolved in CHCl3/MeOH (2:1), and an aliquot was subjected to HPTLC. Spots were visualized by spraying with 0.1% 5-hydroxy-1-tetralone in 80% sulfuric acid (26) and scanned on a Storm 860 fluorescent scanner (Molecular Dynamics) with a blue fluorescent filter set (450-nm excitation filter; 520-nm emission filter) and a photomultiplier tube (voltage of 900 V).
Cells.
GP4F, TF228, and HeLa-CD4 cells were obtained and grown as described (20). GP4F cells were infected with the recombinant vaccinia vCB3 to express human CD4. To reduce surface GSLs from HeLa-CD4 cells, the cells were grown in the presence of the GSL-synthesis inhibitor PPMP (10 μM) for at least 7 days as described (20).
Addition of GSLs to CD4+ Cells.
GSLs were added to CD4+ cells as described (20). Briefly, liposomes containing egg phosphatidylcholine/egg phosphatidylethanolamine/GSLs (3:1.5:1, wt/wt; 0.9 mg/ml total lipid) were prepared in PBS without Ca2+ and Mg2+ by extrusion through a 0.2-μm filter by using an extruder (Lipex Biomembranes, Vancouver, BC). HeLa-CD4 cells on microwells (5 × 104 per dish) were infected with PR/8 influenza virus (A/PR8/34/H1N1) overnight at 37°C. Target cells were treated with 5 μg/ml trypsin for 5 min at room temperature to convert the hemagglutinin precursor to its fusogenic form. Liposomes (1 ml) were allowed to bind to hemagglutinin-expressing cells. Liposome–cell fusion was induced by brief exposure (60 s) of the cells to pH 5.1, followed by incubation in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. The modified cells were then used as targets in the cell–cell fusion assays described below.
HIV-1 Envelope Glycoprotein-Mediated Cell–Cell Fusion.
To monitor cell–cell fusion, target cells were labeled with the cytoplasmic dye 5- and 6-{[(4-chloromethyl)benzoyl]amino}tetramethylrhodamine (CMTMR; 10 μM, excitation/emission 541/565 nm) for 2–4 h at 37°C (13). Cells expressing gp120-gp41LAI (TF228 cells) were labeled either with the fluorescent membrane probe DiO (3,3′-dioctadecyloxacarbocyanine perchlorate; excitation/emission 484/501) or aqueous probe calcein-AM (excitation/emission 496/517 nm) as described (13). Fluorescently labeled TF228 cells were cocultured with CMTMR-labeled target cells for 2–4 h at 37°C. For assays in which the HeLa-CD4 cells were treated with PPMP, all incubation media contained the inhibitor.
After fusion, the cells were examined with an Olympus IX70 inverted microscope (New Hyde Park, NY) with the U-MNG filter cube for the rhodamine probes and the U-MNIBA filter cube for the other probes as described (20). Image analysis for dye mixing was performed with metamorph image analysis software (Universal Imaging, West Chester, PA). The total number of cells testing positive for each dye was scored. Bright-field images were used to distinguish false–positives where labeled cells were lying over one another without actual fusion. The extent of fusion was calculated as percentage of fusion = 100 × number of cells positive for both dyes/number of cells positive for CMTMR.
RESULTS
Identification of a Fusion-Active Fraction.
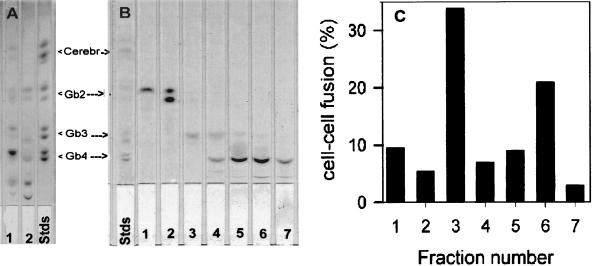
To determine the fusion-active GSL(s) in the human erythrocyte GSL mixture (Fig. 1A, lane 1), we isolated individual components by using silica-gel column chromatography. We collected seven GSL fractions from the crude GSL mixture. The relative mobility of these GSL fractions on silica-gel TLC plates is shown in Fig. 1B. Liposomes, containing one of these fractions, were prepared and incorporated into the membranes of fusion-incompetent cells. Fig. 1C shows fusion activity with GSL-supplemented CD4+ GP4F cells. Fr. 3-enriched target cells showed the highest recovery of fusion, and fraction 6 also showed higher recovery than other fractions. Addition of fractions 1, 2, 4, 5, and 7 did not result in significant fusion. As can be seen in Fig. 1B, Fr. 3 is one of the minor components in the crude GSL mixture. Resorcinol spray analysis of the TLC (27) indicated that, of the seven fractions collected, only fractions 5 and 6 contained acidic GSLs (data not shown). Furthermore, Fr. 3 was not present in the bovine GSL mixture (Fig. 1A, lane 2), consistent with our previous observation that addition of GSLs from bovine erythrocyte membranes did not rescue gp120-gp41-mediated fusion (20).
Figure 1.
Recovery of fusion after addition of various GSL fractions isolated from the crude human erythrocyte mixture. (A) TLC analysis of erythrocyte GSLs. GSLs were isolated as described in Materials and Methods. Total lipid (25–50 μg) was spotted on a 5 × 20-cm silica-gel TLC plate. The plate was developed in CHCl3/MeOH/H2O (65:25:4, vol/vol). At the end of the run, the plate was air dried, sprayed with Bial’s reagent, heated to develop the spots, scanned with a Hewlett–Packard 4P scanner, and photographed. GSLs isolated from human erythrocytes are in lane 1, and GSLs isolated from bovine erythrocytes are in lane 2; GSL standards are in lane 3. (B) TLC of various GSL fractions purified by silica-gel column. The crude GSL mixture from human erythrocytes (shown in A, lane 1) was fractionated by silica-gel column chromatography. GSLs in the fractions were analyzed by chromatography on silica-gel TLC as in A. Lanes 1–7 are fraction numbers. (C) Recovery of fusion of CD4+ nonhuman cells by GSL fractions. GP4F cells were plated on microwells and infected with vCB3 to express human CD4 on the cell surface. Liposomes containing different GSL fractions were incorporated into the membranes of the target cells and the cells were labeled with CMTMR. DiO-labeled TF228 cells were cocultured with GSL-supplemented target cells for 4–6 h at 37°C. Images were collected, and fusion was calculated as described in Materials and Methods. Data presented here are normalized to the recovery of fusion by the crude human GSL fraction.
Structural Analysis of Fr. 3.
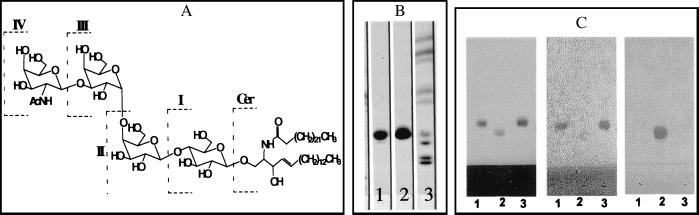
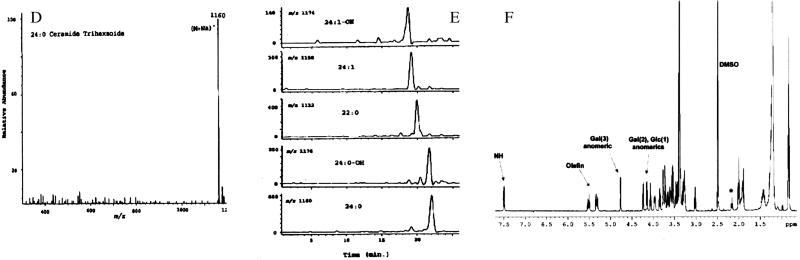
Silica-gel TLC analysis of Fr. 3 (Fig. 2B, lane 1) showed a single component (>99% pure) with relative mobility very similar to that of Gb3 (Fig. 2B, lane 2). Neither Fr. 3 nor Gb3 reacted with resorcinol spray reagent, indicating that Fr. 3 is a neutral GSL (data not shown). The chemical structure of GSLs of the “globo” series is shown in Fig. 2A. A comparison of electrospray liquid-chromatography/MS analysis of Fr. 3 with known standards indicated that this fraction contained mainly ceramide trihexosides. They were detected as natriated molecules (M+Na+) as is shown for 24:0-trihexoside as an example in Fig. 2D. Fig. 2E shows mass ion chromatograms of Fr. 3 GSL containing 24:0, 22:0, or 24:1 fatty acyl moieties and their hydroxy derivatives. Presence of these fatty acids was also confirmed by the gas chromatographic analysis after transmethylation (data not shown). NMR analysis corroborated the data obtained from enzymatic hydrolysis (see below) and MS. Fig. 2F shows the 1H spectrum of Fr. 3 in 98:2 DMSO-d6/D2O; the relevant peaks are labeled with arrows. Only one amide proton was observed (ceramide), and no acetate peaks were present, confirming the absence of amino sugars in the head group. In addition, there were no indications of the presence of neuraminic acid units in the molecule in the proton spectrum. There were three anomeric protons, and their spin systems were traced by total correlation spectroscopy (data not shown). Coupling constants measured for the anomeric proton doublets indicated two β-linked (7.6 and 7.7 Hz) and one α-linked (3.6 Hz) saccharides. Data for 13C also showed the presence of three anomeric carbons and the one-bond 1JCH coupling constants (measured in an heteronuclear multiple-quantum correlation spectrum) consistent with the stereochemical assignments of the anomeric centers. Exhaustive acetylation of Fr. 3 (see Materials and Methods) and purification yielded a compound with 11 acetate methyl groups present in the 1H NMR spectrum. This number of free hydroxyl groups in the molecule was consistent with a trihexose-containing ceramide glycoside. In addition, comparison of the NMR data obtained for Fr. 3 with that of an authentic sample of ceramide trihexoside (Fig. 2B, lane 2, top spot) and comparison of the NMR data obtained for Fr. 3 with the reported chemical shifts and coupling constants of synthetic Gb3 showed the two to be virtually identical. A minor set of peaks were observed in the fatty acid region of the spectrum, and these were attributed to a small percentage of material in which the length of the hydrocarbon chain attached at the ceramide amino group varied (see MS analysis, Fig. 2E). To define the carbohydrate moieties of Fr. 3 further, we examined its interaction with an anti-Gb3 antibody (38.13; ref. 21) that is known to recognize the terminal Galα1→4Galβ motif (28). Fig. 2C shows immunostaining of Fr. 3, Gb3 and asialo-GM2 on HPTLC plates with 38.13 and an anti-asialo-GM2 antibody. The 38.13 antibody reacted with Fr. 3 and Gb3 to similar extents but not with asialo-GM2, which contains N-acetylgalactosamine (β1→4) as the terminal sugar group (Fig. 2C Center). Conversely, the asialo-GM2 antibody reacted only with asialo-GM2 (Fig. 2C Right). These results show that the polar head group of Fr. 3 bears Galα1→4Galβ sugar linkages.
Figure 2.
(A) Chemical structure of globo-series GSLs. Cer, ceramide; I, glucosylceramide; II, lactosyl ceramide {Gb2; [Gal(β1→4)Glc-Cer]}; III, Gb3 [Gal(α1→4)Gal(β1→4)Glc-Cer]; and IV, globoside [Gb4;GalNAc(β1→3)Gal(α1→4)Gal(β1→4)Glc-Cer]. (B) TLC of Fr. 3. GSLs were chromatographed on silica-gel TLC plates (as shown in Fig. 1A) and sprayed with orcinol. Lane 1, Fr. 3; lane 2, Gb3 (top spot); lane 3, the same neutral GSL standards as shown in Fig. 1 A and B. (C) HPTLC immunostaining. GSLs (5 μg) were chromatographed on silica-gel HPTLC plates. The plates were air dried and sprayed with orcinol (Left), incubated with 38.13 antibody (Center), or incubated with asialo-GM2 antibody (Right). Lane 1, Fr. 3; lane 2, asialo-GM2; lane 3, Gb3. (D) Mass spectrum of 24:0 Gb3 detected from Fr. 3. (E) Mass ion chromatogram of various molecular species of Gb3 present in Fr. 3. (F) 1H NMR proton spectrum of Fr. 3 in 98:2 DMSO-d6/D2O at 298 K. Relevant peaks are labeled with arrows. Numbers in parentheses refer to the position of the particular sugar in the trisaccharide with glucose attached to the ceramide at position 1. The peak marked with an asterisk is thought to arise from minor variations in the length of the ceramide fatty-acid chains.
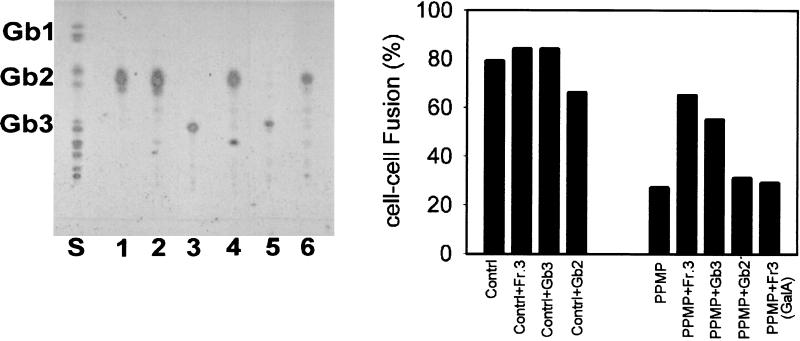
To confirm the exact nature of carbohydrate motifs of Fr. 3 further, we performed enzymatic analysis of Fr. 3 by using αGalA, which is specific for the hydrolysis of the (α1→4)-galactosyl linkage found in Gb3. Fig. 3 Left shows that treatment of Fr. 3 with αGalA resulted in the formation of a product comigrating with ceramide dihexoside, which we suggest represents Gb2. Before digestion, Fr. 3 migrated coincident with Gb3, used as positive control in this assay system. The αGalA preparation was tested for α- and β-glycosidase activity against synthetic substrates and shown to be reactive for only α-galactosyl-containing substrates. αGalA did not cleave sugar groups from Gb2, confirming the specificity of the enzyme. Fr. 3 was efficiently hydrolyzed by ceramide glycanase, resulting in the formation of ceramide and a trihexoside, confirming that the sugar attached to the ceramide in Fr. 3 was glucose attached by β-linkage (data not shown). Based on our structural analysis, we conclude that Fr. 3 isolated from a human erythrocyte GSL mixture is a ceramide trihexoside that possesses a polar head group identical to the well characterized Gb3.
Figure 3.
Enzymatic digestion of Fr. 3. (Left) Fr. 3 was digested with αGalA in parallel with standard GSLs, and the results were analyzed by HPTLC. Lane S, mixed GSL standards; lane 1, Gb2 with no enzyme; lane 2, Gb2 + αGalA; lane 3, Gb3 with no enzyme; lane 4, Gb3 + αGalA; lane 5, Fr. 3 with no enzyme; and lane 6, Fr. 3 + αGalA. (Right) Fusion activity after addition of αGalA-digested Fr. 3. Digested lipid was extracted, incorporated into liposomes, and transferred to HeLa-CD4/GSL−. Fusion activity was monitored as described for Fig. 1C. Controls represent untreated Hela-CD4 cells. Gb2 was added as an additional control.
Fusion Activity of HeLa-CD4/GSL− supplemented with GSLs.
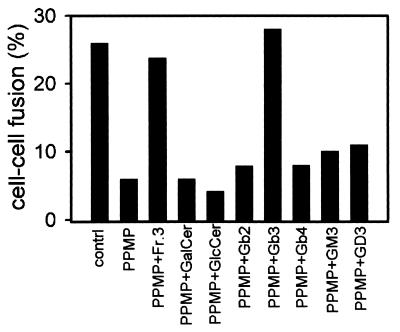
HeLa-CD4/GSL− exhibit reduced susceptibility to HIV-1 env-mediated cell fusion (20) when cultured in the presence of PPMP, a competitive inhibitor of ceramide:UDP-glucosyltransferase (29). HeLa-CD4/GSL− were supplemented with a variety of well defined GSLs, and their fusion with HIV-1LAI env-expressing cells was monitored. Fig. 3 Right shows recovery of fusion activity after addition of either Fr. 3 or Gb3 to HeLa-CD4/GSL−. Under identical conditions, addition of neither αGalA-treated-Fr. 3 nor Gb2, a precursor of Gb3, to the target cells rescued fusion activity, indicating that the Galα1→4 motif is critical. Gb4, the major neutral GSL in the human erythrocyte membrane (30) arises from Gb3 with addition of N-acetylgalactosamine to the terminal galactose residue of Gb3 in a β1→3 linkage (see Fig. 2A). Incorporation of Gb4 into the plasma membranes of HeLa-CD4/GSL− did not result in recovery of fusion with TF228 cells (Fig. 4). This observation suggests that the terminal α-galactose residue of Fr. 3/Gb3 must be at the terminal position for HIV-1 env-mediated fusion. It has been shown that the disialoganglioside, GD3, Gb2, and the monosialoganglioside GM3, specifically interact with HIV-1 gp120 (41), and recent studies have shown that human GM3 (but not bovine GM3) interacts preferentially to form a trimolecular complex GM3-CD4-gp120 (16, 17). Fig. 4 shows that addition of none of these GSLs to HeLa-CD4/GSL− promoted HIV-1 env-mediated fusion.
Figure 4.
Recovery of fusion activity by addition of various GSLs. The lipids were incorporated into liposomes and transferred to HeLa-CD4/GSL−, and fusion activity was monitored as described in Fig. 1C. Controls represent untreated Hela-CD4 cells. Data are from one of three similar experiments. GD3 is NeuNAcα2→8NeuNAcα2→3Galβ1→4Glcβ1→1Cer.
DISCUSSION
We found that CD4+ cell lines, which are not susceptible to HIV-1 env-mediated fusion, became fusogenic targets after their complementation with Fr. 3, a minor component in the crude GSL mixture extracted from human erythrocytes. Fraction 6 also exhibited fusion-complementation activity, although to a lesser extent than Fr. 3. Because Fr. 3 contained only a single neutral GSL component as shown in TLC and because this GSL was also present at a low level in fraction 6, we applied a variety of biophysical, biochemical, and immunological methods to elucidate its structure. MS and NMR spectroscopy of Fr. 3 indicated the presence of three sugar molecules and a ceramide backbone, indicating that Fr. 3 is a GSL of the trihexosyl-ceramide type. Cleavage of Fr. 3 by αGalA and the specific binding of the anti-Gb3 mAb 38.13 to Fr. 3 confirm that the terminal sugar moiety of Fr. 3 is Galα1→4Galβ1. Taken together, these data establish Fr. 3 as a neutral GSL containing the oligosaccharide sequence Galα1→4Galβ1→4Glc, identical to Gb3. Addition of Fr. 3 or pure Gb3 (but not αGalA-digested Fr. 3 or Gb2) to HeLa-CD4/GSL− complemented subsequent fusion with HIV-1 env-expressing cells, indicating the importance of the Galα1→4Galβ1 motif. Furthermore, the inability of Gb4 to rescue fusion under identical conditions shows that the Galα1→4 linkage in the polar-head group of the GSL must be the terminal linkage required for HIV-1 fusion.
On certain Burkitt lymphoma cell lines, Gb3 has been identified as a GSL antigen, CD77 (21), which seems to be involved in signal transduction leading to apoptosis (31, 32). It serves as the cell-surface receptor for Shiga toxin and Shiga-like toxin (Verotoxin; ref. 33) and is known to play a role in the affinity of the α interferon receptors (34). Gb3 is also closely linked to Fabry disease, in which patients lack α-galactosidase, causing a block in the catabolic pathway of this molecule, which results in accumulation of Gb3 (35).
Although HIV-1 preferentially infects the cells of the monocyte/macrophage lineage and T lymphocytes, previous reports indicate that Langerhans cells (36), dendritic cells (37), glial cells (38), some colonic cells (15), and B lymphocytes (39) are also infected by HIV-1. The presence of Gb3 on B cell lines (21) and human gut epithelial cells (40) suggests that it might play a role in HIV-1 infection in these cells. Because some cultured intestinal CD4− cell lines support galactosylceramide-dependent HIV-1 entry (14), we hypothesize that galactosylceramide promotes binding to gp120 and that Gb3 is involved in fusion in these cells. The CD4-transfected CXCR4− human astroglioma cell line U87-CD4, which expresses a high level of Gb3 as indicated by staining with the 38.13 antibody (data not shown), is not susceptible to HIV-1LAI infection, indicating that Gb3 is not able to act in the absence of CXCR4 for this viral strain. However, it is possible that Gb3 can serve as a coreceptor for other viral strains that enter U87-CD4 cells (10, 11).
Gb3 is present at very low levels in CD4+ lymphocytes (41) and is undetectable by 38.13 antibody staining on SupT1 cells (data not shown). Inhibition of HIV-1 envelope glycoprotein-mediated cell fusion after treatment of SupT1 cells with PPMP (20) indicates that Gb3 may still may be involved at these low levels. Treating HeLa cells with PPMP did not alter the levels of cell-surface-expressed CD4 and CXCR4 as monitored by flow cytometry and fluorescence microscopy of specific mAb-stained cells (data not shown). The reduced fusion susceptibility of CD4+ CXCR4+ cells lacking Gb3 indicates that the role of the GSL might be to arrange the receptors in a proper disposition to interact with the HIV-1 envelope glycoprotein. The nature of these specific interactions remains to be explored.
Acknowledgments
We thank Dr. Zdenka L. Jonak for the TF228.1.16 cell line, Dr. John Silver for HeLa cell lines, Dr. Judith White for GP4F cells, Drs. Chris Broder and Edward Berger for the vaccinia recombinant, Dr. David Roberts for expert advice on the characterization of GSLs, Mr. Patrick Rose for testing various cells lines for expression of Gb3, Dr. Kathy Wehrly for advice on the handling of U87CD4 cells, and members of the Blumenthal lab for insightful comments. U87-CD4 cells were provided by the National Institute of Allergy and Infectious Diseases AIDS Research and Reference Reagent Program. This work was supported by the National Institutes of Health Intramural AIDS Targeted Antiviral Program.
ABBREVIATIONS
- CMTMR
5- and 6-{[(4-chloromethyl)benzoyl]amino}tetramethylrhodamine
- DiO
3,3′-dioctadecyloxacarbocyanine perchlorate
- Fr. 3
fraction 3
- αGalA
α-galactosidase A
- Gb2
lactosylceramide [Gal(β1→4)Glc-Cer]
- Gb3
globotriaosylceramide [Gal(α1→4)Gal(β1→4)Glc-Cer]
- Gb4
globoside [GalNAc(β1→3)Gal(α1→4)Gal(β1→4)Glc-Cer]
- Glc-Cer
glucosylceramide
- GSL
glycosphingolipid
- HeLa-CD4/GSL−
GSL-depleted HeLa-CD4 cells
- HPTLC
high-performance thin layer chromatography, PPMP, 1-phenyl-2-hexadecanoylamino-3-morpholino-1-propanol
References
- 1.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 2.Moore J P, Jameson B A, Weiss R A, Sattentau Q J. In: Viral Fusion Mechanisms. Bentz J, editor. Boca Raton, FL: CRC; 1993. pp. 233–289. [Google Scholar]
- 3.Blumenthal R, Dimitrov D S. In: Handbook of Physiology. Hoffman J F, Jamieson J C, editors. New York: Oxford Univ. Press; 1997. pp. 563–603. [Google Scholar]
- 4.Berger, E. A. (1997) AIDS 11, Suppl. A, S3–S16. [PubMed]
- 5.Broder C C, Dimitrov D S. Pathobiology. 1996;64:171–179. doi: 10.1159/000164032. [DOI] [PubMed] [Google Scholar]
- 6.Moore J P, Trkola A, Dragic T. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 7.Doms R W, Peiper S C. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 8.Littman D R. Cell. 1998;93:677–680. doi: 10.1016/s0092-8674(00)81429-4. [DOI] [PubMed] [Google Scholar]
- 9.Dimitrov D S. Cell. 1997;91:721–730. doi: 10.1016/s0092-8674(00)80460-2. [DOI] [PubMed] [Google Scholar]
- 10.McKnight A, Wilkinson D, Simmons G, Talbot S, Picard L, Ahuja M, Marsh M, Hoxie J A, Clapham P R. J Virol. 1997;71:1692–1696. doi: 10.1128/jvi.71.2.1692-1696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjorndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyo E M. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dragic T, Picard L, Alizon M. J Virol. 1995;69:1013–1018. doi: 10.1128/jvi.69.2.1013-1018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puri A, Morris S J, Jones P, Ryan M, Blumenthal R. Virology. 1996;219:262–267. doi: 10.1006/viro.1996.0244. [DOI] [PubMed] [Google Scholar]
- 14.Yahi N, Baghdiguian S, Moreau H, Fantini J. J Virol. 1992;66:4848–4854. doi: 10.1128/jvi.66.8.4848-4854.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delezay O, Koch N, Yahi N, Hammache D, Tourres C, Tamalet C, Fantini J. AIDS. 1997;11:1311–1318. doi: 10.1097/00002030-199711000-00004. [DOI] [PubMed] [Google Scholar]
- 16.Hammache D, Pieroni G, Yahi N, Delezay O, Koch N, Lafont H, Tamalet C, Fantini J. J Biol Chem. 1998;273:7967–7971. doi: 10.1074/jbc.273.14.7967. [DOI] [PubMed] [Google Scholar]
- 17.Hammache D, Yahi N, Pieroni G, Ariasi F, Tamalet C, Fantini J. Biochem Biophys Res Commun. 1998;246:117–122. doi: 10.1006/bbrc.1998.8531. [DOI] [PubMed] [Google Scholar]
- 18.Picard L, Dragic T, Wiels J, Alizon M. Perspect Drug Discovery Des. 1996;5:143–153. [Google Scholar]
- 19.Mizrachi Y, Lev M, Harish Z, Sundaram S K, Rubinstein A. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;11:137–141. doi: 10.1097/00042560-199602010-00004. [DOI] [PubMed] [Google Scholar]
- 20.Puri A, Hug P, Munoz-Barroso I, Blumenthal R. Biochem Biophys Res Commun. 1998;242:219–225. doi: 10.1006/bbrc.1997.7941. [DOI] [PubMed] [Google Scholar]
- 21.Wiels J, Fellous M, Tursz T. Proc Natl Acad Sci USA. 1981;78:6485–6488. doi: 10.1073/pnas.78.10.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H Y, Wang T-C, Ma Y-C. Anal Chem. 1994;66:3977–3982. doi: 10.1021/ac00094a020. [DOI] [PubMed] [Google Scholar]
- 23.Bax A, Griffey R H, Hawkins L B. J Magn Reson. 1983;55:301–315. [Google Scholar]
- 24.Kusiak J W, Quirk J M, Brady R O. J Biol Chem. 1978;253:184–190. [PubMed] [Google Scholar]
- 25.Folch J, Lees M, Stanley G H S. J Biol Chem. 1957;225:497–509. [PubMed] [Google Scholar]
- 26.Watanabe K, Mizutsa M. J Lipid Res. 1998;36:1848–1855. [PubMed] [Google Scholar]
- 27.Kundu S K. Methods Enzymol. 1981;72:185–204. doi: 10.1016/s0076-6879(81)72012-3. [DOI] [PubMed] [Google Scholar]
- 28.Nudelman E, Kannagi R, Hakomori S, Parsons M, Lipinski M, Wiels J, Fellous M, Tursz T. Science. 1983;220:509–511. doi: 10.1126/science.6836295. [DOI] [PubMed] [Google Scholar]
- 29.Abe A, Inokuchi J, Jimbo M, Shimeno H, Nagamatsu A, Shayman J A, Shukla G S, Radin N S. J Biochem (Tokyo) 1992;111:191–196. doi: 10.1093/oxfordjournals.jbchem.a123736. [DOI] [PubMed] [Google Scholar]
- 30.Yamakawa T, Nishimura S, Kamimura M. Jpn J Exp Med. 1965;35:201–207. [PubMed] [Google Scholar]
- 31.Taga S, Carlier K, Mishal Z, Capoulade C, Mangeney M, Lecluse Y, Coulaud D, Tetaud C, Pritchard L L, Tursz T, et al. Blood. 1997;90:2757–2767. [PubMed] [Google Scholar]
- 32.Khine A A, Firtel M, Lingwood C A. J Cell Physiol. 1998;176:281–292. doi: 10.1002/(SICI)1097-4652(199808)176:2<281::AID-JCP6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 33.Lingwood C A. Trends Microbiol. 1996;4:147–153. doi: 10.1016/0966-842x(96)10017-2. [DOI] [PubMed] [Google Scholar]
- 34.Ghislain J, Lingwood C A, Fish E N. J Immunol. 1994;153:3655–3663. [PubMed] [Google Scholar]
- 35.Brady R O, Gal A E, Bradley R M, Martensson E, Warshsaw A L, Laster L. N Engl J Med. 1997;276:1163–1167. doi: 10.1056/NEJM196705252762101. [DOI] [PubMed] [Google Scholar]
- 36.Zambruno G, Giannetti A, Bertazzoni U, Girolomoni G. Immunol Today. 1995;16:520–524. doi: 10.1016/0167-5699(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 37.Cameron P, Pope M, Granelli-Piperno A, Steinman R M. J Leukocyte Biol. 1996;59:158–171. doi: 10.1002/jlb.59.2.158. [DOI] [PubMed] [Google Scholar]
- 38.Ensoli F, Wang H, Fiorelli V, Zeichner S L, De Cristofaro M R, Luzi G, Thiele C J. J Neurovirol. 1997;3:290–298. doi: 10.3109/13550289709029470. [DOI] [PubMed] [Google Scholar]
- 39.Fritsch L, Marechal V, Schneider V, Barthet C, Rozenbaum W, Moisan-Coppey M, Coppey J, Nicolas J C. Virology. 1998;244:542–551. doi: 10.1006/viro.1998.9120. [DOI] [PubMed] [Google Scholar]
- 40.Jacewicz M S, Acheson D W, Mobassaleh M, Donohue-Rolfe A, Balasubramanian K A, Keusch G T. J Clin Invest. 1995;96:1328–1335. doi: 10.1172/JCI118168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delezay O, Hammache D, Fantini J, Yahi N. Biochemistry. 1996;35:15663–15671. doi: 10.1021/bi961205g. [DOI] [PubMed] [Google Scholar]