Abstract
It has long been assumed that HIV-1 evolution is best described by deterministic evolutionary models because of the large population size. Recently, however, it was suggested that the effective population size (Ne) may be rather small, thereby allowing chance to influence evolution, a situation best described by a stochastic evolutionary model. To gain experimental evidence supporting one of the evolutionary models, we investigated whether the development of resistance to the protease inhibitor ritonavir affected the evolution of the env gene. Sequential serum samples from five patients treated with ritonavir were used for analysis of the protease gene and the V3 domain of the env gene. Multiple reverse transcription–PCR products were cloned, sequenced, and used to construct phylogenetic trees and to calculate the genetic variation and Ne. Genotypic resistance to ritonavir developed in all five patients, but each patient displayed a unique combination of mutations, indicating a stochastic element in the development of ritonavir resistance. Furthermore, development of resistance induced clear bottleneck effects in the env gene. The mean intrasample genetic variation, which ranged from 1.2% to 5.7% before treatment, decreased significantly (P < 0.025) during treatment. In agreement with these findings, Ne was estimated to be very small (500–15,000) compared with the total HIV-1 RNA copy number. This study combines three independent observations, strong population bottlenecking, small Ne, and selection of different combinations of protease-resistance mutations, all of which indicate that HIV-1 evolution is best described by a stochastic evolutionary model.
HIV-1 is characterized by a very high degree of genetic diversity (1–3). Even in a single infected individual, the virus can be best described as a population of distinct, but closely related, genetic variants, referred to as a quasispecies (4, 5). A deterministic evolutionary model of HIV-1 has been proposed to describe HIV-1 evolution. This model assumes a large population size of HIV-1 (6–9) in which competition between constantly generated viral variants with slight fitness differences determines the exact frequency of mutants (10). According to this model, evolution will be inevitable and, in principle, predictable. Recently, however, Leigh Brown argued that chance may play a significant role in the evolution of HIV-1, because the effective population size (Ne) in an infected individual may be small (11, 12). Ne is defined as an ideal population that is drawn randomly from the total population and that has the same statistical characteristics as the total population. In our case, Ne can be described as the average number of virus variants that produce infectious progeny. A stochastic evolutionary model would take into account the impact of chance on the evolution of HIV-1.
To investigate whether HIV-1 evolution is best described by a deterministic or a stochastic evolutionary model, we have studied how a strong selective pressure on the viral protease affects genetic variation and evolution of a distant, unrelated gene, i.e., coselection on the env gene. If the population size of HIV-1 would be infinite (deterministic model) protease-inhibitor treatment would be expected to have no effect on the genetic variation in the env gene, whereas such effects would be expected if evolution of HIV-1 follows the stochastic model. For this reason, we sequenced part of the env gene from multiple clones obtained from samples drawn at the start of protease-inhibitor treatment and at a time point when genotypic protease resistance had developed and the serum HIV-1 RNA levels had rebounded to near baseline levels. We found that development of genotypic resistance induced a strong bottleneck effect in the distant env gene, as evidenced both by phylogenetic analysis and by a significant reduction in the genetic variation in the HIV-1 RNA population. In addition, estimates showed that Ne is small. Together, these data indicate that stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy.
MATERIALS AND METHODS
Study Population.
We selected five patients from a population of patients receiving ritonavir monotherapy (13). Patients 125, 127, and 134 were antiviral-treatment naive, whereas patient 129 had been treated with 500 mg/day zidovudine for 22 months followed by addition of 1.5 mg/day zalcitabine for 10 months and subsequently 0.75 mg/day zalcitabine for 11 months. Patient 224 had received 600 mg/day zidovudine for 69 months followed by 300 mg/day zidovudine and 2.25 mg/day zalcitabine for 3 months. After a washout period of 3 weeks, ritonavir therapy was initiated. Patient 224 was treated with 1000 mg/day of ritonavir; patients 129, 125, and 127 were treated with 800 mg/day of ritonavir; and patient 134 was treated with 600 mg/day ritonavir.
CD4 counts in peripheral blood were determined (14). Serum was collected at the start of therapy and at regular intervals during the study. Serum from patients 125 and 127, who were naive to antiviral treatment, was also collected before the start of ritonavir therapy.
Isolation and Quantitation of Viral RNA.
RNA was extracted from 100 μl of serum (15) and was quantified by using a prototype Roche assay (16).
Amplification and sequence analysis of the viral protease.
After viral-RNA isolation, an equivalent of 10 μl of serum was used to reverse transcribe and amplify the protease gene (nucleotides 2252–2548). A one-tube reverse transcription–PCR procedure was performed, essentially as described by Nijhuis et al. (17), with 1 mM MgCl2, 10 pmol of primer 5′prot-1 (5′-AGG CTA ATT TTT TAG GGA AGA TCT GGC CTT CC-3′, nucleotides 2077–2108; Pharmacia), and primer 3′prot-1 (5′-GCA AAT ACT GGA GTA TTG TAT GGA TTT TCA GG-3′, nucleotides 2733–2702; Pharmacia). After this first reverse transcription–PCR, the amount of amplified product was increased further in a nested amplification by using 12 pmol of primer 5′prot-2 (5′-TCA GAG CAG ACC AGA GCC AAC AGC CCC A-3′, nucleotides 2135–2162), 11 pmol of primer 3′prot-2 (5′-AAT GCT TTT ATT TTT TCT TCT GTC AAT GGC-3′, nucleotides 2649–2620), and 2 mM MgCl2.
Direct sequencing of the protease gene was performed with the Thermo sequenase fluorescent-labeled primer cycle sequencing kit (Amersham) by using the fluorescent-labeled oligonucleotides JA187 (5′-AAC TTT TGG GCC ATC CAT TC-3′, nucleotides 2611–2592) and JA186 (5′-AGA GCC AAC AGC CCC ACC AG-3′, nucleotides 2147–2166) and an ALF sequencer (Pharmacia). The amplified viral protease genes obtained from patients 129 and 224 were also cloned and sequenced as described above.
Amplification and sequence analysis of the viral envelope (V3 region).
After viral RNA isolation, an equivalent of 4 μl of serum was used to reverse transcribe and amplify the V3 region by using the XL RNA PCR kit (Perkin–Elmer). These reactions were performed essentially as described by the manufacturer by using oligonucleotide JA170 [5′-GTG ATG TAT T(A/G)C A(A/G)T AGA AAA ATT C-3′, nucleotides 7388–7364] to enable cDNA synthesis. Then, the cDNA was amplified by using oligonucleotides JA170 and JA167 [5′-TAT C(C/T)T TTG AGC CAA TTC C(C/T)A TAC A-3′, nucleotides 6846–6869]. The amount of amplified product was increased further in a nested PCR by using the Expand high-fidelity PCR system (Boehringer Mannheim). These amplification reactions were performed essentially as described by the manufacturer by using oligonucleotides JA168 [5′-ACA ATG (C/T)AC ACA TGG AAT TA(A/G) GCC A-3′, nucleotides 6958–6982] and JA169 [5′-AGA AAA ATT C(C/T)C CTC (C/T)AC AAT TAA A-3′, nucleotides 7373–7349].
The effectiveness of RNA extraction and cDNA synthesis was checked by limiting-dilution PCR, which showed that the number of successfully extracted and reverse-transcribed RNA molecules that were added to each PCR exceeded 25 in most cases and in no case was lower than 5. To reduce the risk of sampling artifacts further, the products from two to four independent PCRs were pooled before cloning by using the Ligator kit (R & D systems). The V3 region of these clones was sequenced with the Thermo sequenase fluorescent-labeled primer cycle sequencing kit (Amersham) by using the fluorescent-labeled oligonucleotides JA168 and JA169 and an ALF sequencer.
Calculation of Genetic Distances and Phylogenetic Analysis.
The sequences were aligned manually with the sequence editor in the genetic data environment software package (18). Amino acid translations were made in the genetic data environment. Genetic distances were calculated with mega (19) under the Tamura–Nei model and Γ-distributed substitution rates with the α-parameter set to 0.38 according to Leitner et al. (20). The distances were also used to construct phylogenetic trees by using the neighbor-joining method as implemented in the neighbor program in phylip (21). The distances and trees were calculated from gap-stripped alignments. The length of the alignment differed slightly between patients and ranged from 318 to 348 nucleotides. To visualize and edit the trees graphically, the program treetool (version 1.0) in the genetic data environment was used. Prototype sequences were obtained from the Los Alamos database. In addition, sequences from 114 Dutch homosexual men and 32 Dutch intravenous drug users, kindly provided by Carla Kuiken (Los Alamos), were used in some analyses.
The sequences have been deposited in GenBank under accession numbers AF100001–AF100140.
Calculation of Ne.
The program fluctuate (22) was used to estimate Ne at each time point from each patient. A starting tree was calculated with the program neighbor in the phylip package (21), under the unweighted pair-group method of averages setting. fluctuate then calculated the parameter Theta (θ) by a maximum likelihood method. The model with which the calculations were completed assumed a variable population size, empirical nucleotide frequencies, a transition/transversion ratio of 1.42, and a single rate over sites. The mutation rate per generation (μ) was assumed to be 3.4 × 10−5 (23). For a haploid organism, such as HIV, Theta is defined as 2 × Ne × the mutation rate per generation (θ = 2Neμ).
RESULTS
Analysis of Viral Dynamics and Evolution of Genotypic Protease Resistance.
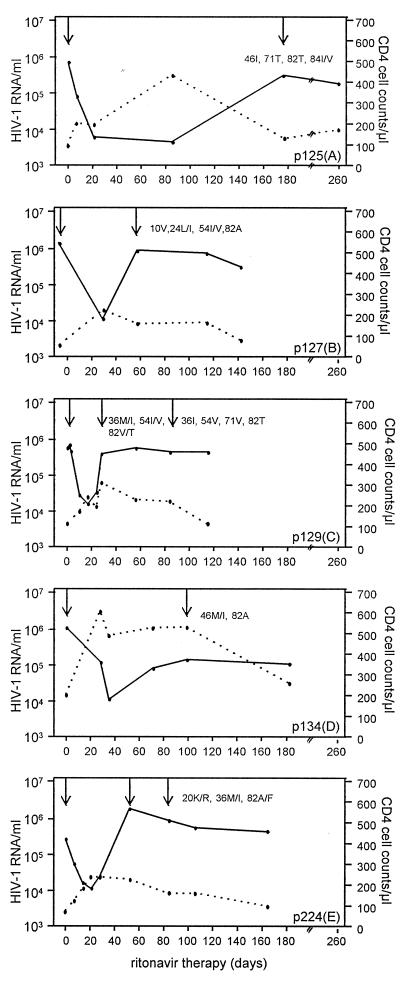
Responses in serum HIV-1 RNA levels and CD4 cell counts for each of the five patients treated with ritonavir is depicted in Fig. 1. The five patients had serum HIV-1 RNA levels, in terms of log10 of copies per ml, between 5.4 and 6.1 copies per ml at the start of therapy. Sequence analysis of the protease gene at the start of therapy showed no amino acid changes associated with ritonavir resistance.
Figure 1.
Serum HIV-1 RNA levels (solid lines) and CD4 cell counts (dotted lines) from start of ritonavir therapy in patients 125(A), 127(B), 129(C), 134(D), and 224(E). The times for detection of protease amino acid changes associated with selection during ritonavir treatment are indicated.
During treatment, an initial decline of 1.4–2.2 log10 units in serum HIV-1 RNA was observed (Fig. 1). Despite this reduction, all patients harbored between 3.7 and 4.2 log10 units of HIV-1 RNA copies per ml. The initial decrease in RNA load was mirrored by an increase in CD4 cells. After this initial phase, the serum HIV-1 RNA levels increased despite continued ritonavir treatment and then remained relatively stable during the observation period. In all patients, the increase in HIV-1 RNA was associated with the appearance in serum of viral variants displaying known ritonavir mutations (refs. 24–26; Fig. 1). Despite the fact that ritonavir resistance developed in all five patients, none displayed the same combination of protease mutations, suggesting a stochastic element in the evolution of ritonavir resistance.
Bottleneck Effect in the env V3 Region After Development of Ritonavir Resistance.
Next we wanted to study whether the selection of particular HIV-1 protease variants harboring amino acid changes associated with ritonavir therapy had measurable effects on other regions of the HIV-1 genome (i.e., the effect of coselection). For this reason, we sequenced the C2/V3 region of the env gene from multiple clones obtained from samples drawn at the start of treatment and at a time point when genotypic ritonavir resistance had developed and the serum HIV-1 RNA levels had rebounded to near baseline levels (Fig. 2).
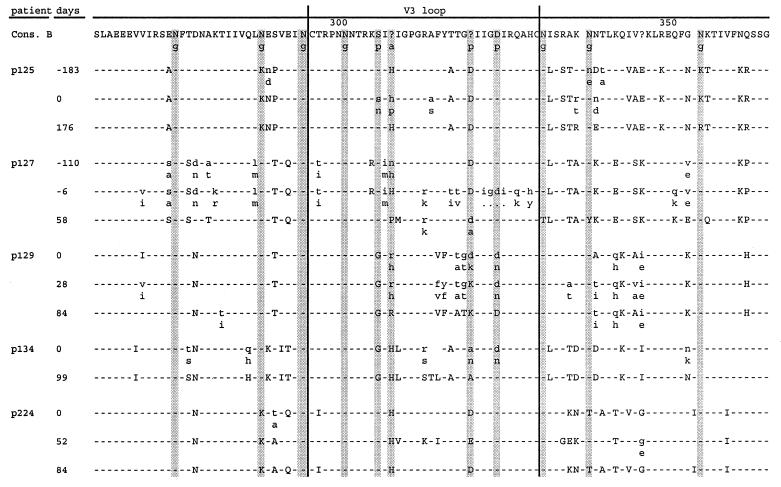
Figure 2.
Amino acid sequence of part of the HIV-1 envelope protein in patients 125, 127, 129, 134, and 224 from start of ritonavir therapy as compared with the sequence of HIV-1 consensus B. Pretherapy sequences are listed for the antiviral-therapy naive patients 125 and 127. Uppercase letters refer to an amino acid change that dominated the population, whereas heterogeneous amino acid positions are indicated with lowercase letters. Particular positions known to be involved in glycosylation (g), antigenicity (a), and phenotype (p) are indicated in gray.
To calculate the intrasample genetic distances in samples obtained before and after development of genotypic ritonavir resistance, 10 individual clones in each sample were analyzed (Table 1). The mean intrasample distances before treatment ranged from 1.2% to 5.7%. After development of genotypic resistance, the mean intrasample distances had decreased significantly (P < 0.025, Friedman ANOVA). In three patients, 125, 134, and 224, the mean intrasample distances decreased strongly (from 2.7% to 0.4%; from 2.9% to 0.4%; and from 1.2% to 0.1%, respectively). In the other two patients, 127 and 129, mean intrasample distances were only moderately reduced compared with pretreatment values (from 4.8% to 2.2% and from 5.7% to 3.6%, respectively). Additional samples obtained from patients 129 and 224 soon after development of genotypic resistance showed a similar trend toward reduced intrasample variation compared with pretreatment samples. Sequences obtained from samples collected approximately half a year before the start of ritonavir treatment were also available for patients 125 and 127, who were both naive to antiviral therapy. These two samples had very similar intrasample genetic distances compared with the corresponding pretreatment samples (2.2–2.7% and 5.4–4.8%, respectively), indicating that intrasample variation is relatively stable in the absence of a deliberately imposed selection pressure, at least over these relatively short time periods. Collectively, these data indicate that the development of genotypic ritonavir resistance represents a strong genetic bottleneck for HIV-1. Thus, selection of HIV-1 variants with mutations in the protease gene conferring ritonavir resistance greatly affects the entire genetic configuration of the virus population and leads to greatly reduced intrapatient genetic variation.
Table 1.
Mean intrasample variation in the HIV-1 population before, at start, and during ritonavir therapy
| Patient | Mean intrasample variation, % (SD, %)
|
|||
|---|---|---|---|---|
| Before therapy | Day 0 | Early in therapy | Late in therapy | |
| 125 | 2.2 (1.0) | 2.7 (1.0) | − | 0.4 (0.3) |
| 127 | 5.4 (3.2) | 4.8 (2.4) | − | 2.2 (2.2) |
| 129 | − | 5.7 (3.8) | 3.9 (3.1) | 3.6 (2.8) |
| 134 | − | 2.9 (1.2) | − | 0.4 (0.3) |
| 224 | − | 1.2 (0.6) | 0.7 (0.6) | 0.1 (0.1) |
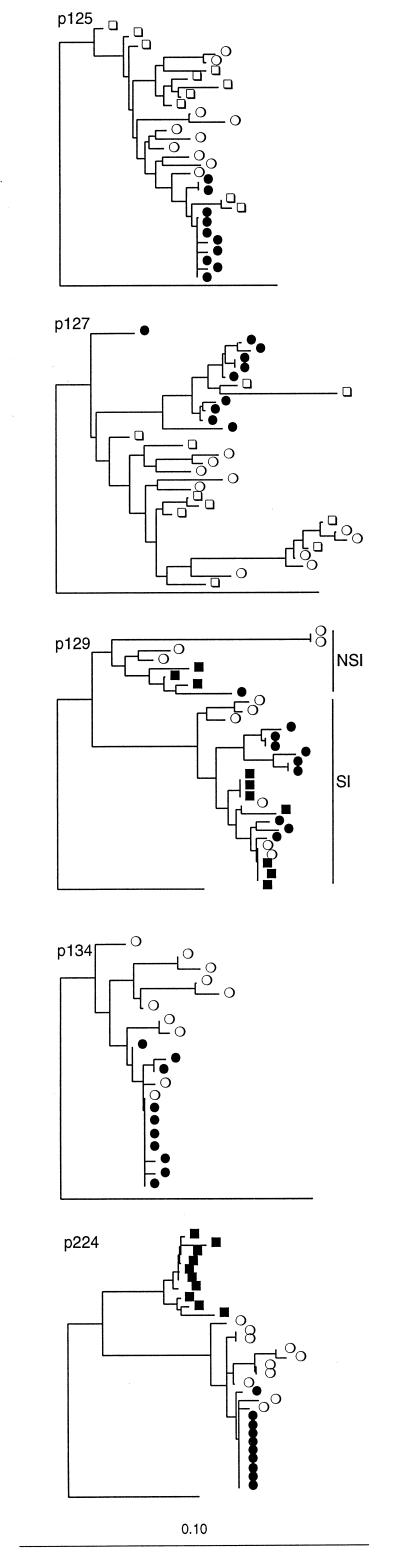
The genetic distance estimates were also used to construct phylogenetic trees. In a first analysis all clones from all patients were included in one tree together with reference sequences (not shown). This tree showed that all clones clustered in a patient-specific manner. Fig. 3 shows the V3 trees for each patient. Overall, we found that the phylogenetic tree also gave evidence for strong bottleneck effects on the V3 domain as a result of ritonavir treatment. However, some interesting individual differences could be discerned. The tree for patient 125 showed that sequences obtained from both pretreatment samples were separated by relatively long branches, which is consistent with the comparably large intrasample distances. The posttreatment sequences formed a very tight cluster that seemed to have been drawn directly from the pretreatment population. The tree derived from the sequences from patient 127 showed a similar picture, except that the pretreatment population was more diverse and formed four distinct clusters. Again, the posttreatment sequences formed a much tighter cluster that seemed to have been derived from V3-sequence variants that were relatively rare in the pretreatment population. The tree for patient 129, the only patient harboring env genes encoding both SI viruses that use the CXCR4 coreceptor and NSI viruses that use the CCR5 coreceptor, was quite distinct from that of the other patients, harboring only NSI-virus isolates. The tree displays two clearly separated sequence clusters that both contained sequences from all three time points analyzed. Interestingly, the sequences in these two clusters displayed either SI or NSI genotypes, as indicated by the amino acid variations at position 320 in the envelope protein (Fig. 2; refs. 27 and 28). The proportion of variants with SI genotype seemed to increase over time. Clearly, both SI and NSI variants passed the bottleneck imposed by the protease inhibitor, which in part explains the comparably large genetic distances observed after development of genotypic resistance in this patient. Patient 134 was similar to patient 125 in that the posttreatment population formed a tight cluster that seemed to be derived from the more heterogeneous pretreatment population. Finally, the tree derived from the sequences obtained from patient 224 showed a dramatically different picture. Before treatment, the population was relatively heterogeneous. Initiation of treatment was followed by a dramatic shift in the virus population such that a distinct group of sequences completely dominated the population 52 days after the start of ritonavir treatment. By day 84, a new and equally dramatic shift had occurred, whereby the population was dominated by a homogeneous cluster of sequences closely related to the pretreatment variants. To exclude the possibility that day 52 sequences were not authentic and that the results could be explained by sample mix-up or contamination, we reanalyzed the sequences from patient 244 together with the V3 sequences from 114 Dutch homosexual men and 32 Dutch intravenous drug users (kindly provided by Carla Kuiken). In this phylogenetic analysis, the sequences from patient 224 formed a monophyletic cluster clearly separated from the remaining 146 Dutch control sequences (data not shown), indicating strongly that the day 52 sequences originated from patient 224 and that the ritonavir treatment had induced a dramatic, but temporary, genetic change in the V3 domain.
Figure 3.
Phylogenetic trees constructed by using the neighbor-joining method and V3 nucleotide sequences from patients 125, 127, 129, 134, and 224. □, sequences obtained from samples drawn 100–200 days before start of ritonavir treatment; ○, sequences obtained at start ■, sequences obtained early after start; and •, sequences obtained later after start. “NSI” and “SI” denote sequences from patient 129 with nonsyncytium-inducing and syncytium-inducing phenotypes, respectively. The trees were rooted against the database sequence JRCSF, and the length of branch leading to the root has been shortened by a factor of 2.
Small Ne.
As described above, the analysis of genetic distances and evolutionary relationships gave evidence for a genetic bottleneck imposed by ritonavir. These findings suggest a small Ne. We calculated Ne for each sample (Table 2). This calculation showed that Ne ranged from 500 to 15,000. The values derived from the samples obtained after the start of treatment should be interpreted with some caution, because the small intrasample genetic distance in these samples gives very little information about the population structure. These low values for Ne are in perfect agreement with our other analyses and explain the strong bottleneck effect induced by ritonavir.
Table 2.
Ne before, at start, and during ritonavir therapy
| Patient |
Ne
|
|||
|---|---|---|---|---|
| Before therapy | Day 0 | Early in therapy | Late in therapy | |
| 125 | 5300 | 7000 | − | 7700 |
| 127 | 4500 | 2300 | − | 500 |
| 129 | − | 1100 | 450 | 800 |
| 134 | − | 2100 | − | 7500 |
| 224 | − | 1300 | 16000 | − |
DISCUSSION
In this study, we investigated whether HIV-1 evolution is best described by deterministic or stochastic evolutionary models by analyzing how the development of resistance to a protease inhibitor, ritonavir, affects evolution of an unrelated gene, env. We show that development of ritonavir resistance induces strong bottleneck effects in the env gene, as evidenced by significant reductions in genetic diversity. This result, together with the finding that Ne is small, indicates that evolution of HIV-1 is influenced strongly by stochastic processes.
We made three independent observations that indicated that stochastic processes strongly influence evolution of HIV-1 during suboptimal protease-inhibitor therapy. First, we showed that development of genotypic ritonavir resistance induced strong bottleneck effects in a distant gene segment, env, as evidenced both by phylogenetic analysis and by a significant reduction in the intragenetic variation of the viral RNA population. Analysis of the genotypic variation in two antiviral-therapy naive patients before the start of ritonavir therapy indicated that normally, in the absence of a deliberately imposed selection pressure, genetic variation is relatively stable, at least over these short time periods. In contrast, protease-inhibitor therapy induced a reduction in the genetic variation in the viral RNA population in all five patients. The largest reductions were observed in four patients harboring only NSI variants, whereas in patient 129, who harbored both NSI and SI variants, the genetic variation was less reduced. In this particular patient, both variants passed the bottleneck imposed by the protease inhibitor, which may explain the comparably large genetic variation after the development of genotypic resistance.
Our findings are in agreement with recent observations published by Delwart et al. (49) who also reported changes in the genetic composition of the plasma virus population during treatment with a protease inhibitor. In their studies, which used DNA-heteroduplex tracking rather than RNA sequencing, the alterations seemed to be transient. It should be stressed that it is unlikely that sampling artifacts influenced our genetic-distance estimates, because we obtained samples when the virus load had rebounded to near baseline levels, and we controlled the efficiency of RNA extraction and cDNA synthesis by limiting dilution. In contrast to the evolutionary bottlenecking observed during protease-inhibitor therapy, no consistent impact on the evolution of the env gene has been observed during zidovudine treatment; Leigh Brown et al. (30, 31) reported no impact on evolution of the env gene during zidovudine therapy, whereas a transient effect was described by Zhang et al. (29). The fact that zidovudine is a weaker inhibitor of HIV-1 replication than ritonavir might explain this seemingly contradictory observation. Another explanation might be that Leigh Brown et al. (30, 31) examined the proviral DNA compartment, which evolves more slowly and lags behind the plasma RNA compartment examined in our study (7, 32, 33).
The observation that development of genotypic resistance to a protease inhibitor induces strong bottleneck effects in the viral population implies that the size of the replicating viral population is relatively small. However, HIV-1 RNA determinations showed that all five patients harbored at least 104 RNA copies per ml of serum, which in terms of body volume would indicate an almost infinite size of the virus population. These findings imply that only a fraction of the HIV-1 RNA molecules effectively contribute to the production of infectious progeny (Ne). To test this hypothesis, we calculated Ne in all five patients and showed that Ne is many orders of magnitude smaller than would be expected based on the HIV-1 RNA copy number. This observation of a relatively small Ne is in agreement with our results that show genetic bottlenecking and is the second indication that stochastic processes influence the evolution of HIV-1. Furthermore, our estimates of Ne are nicely in agreement with recent estimations of Ne (12).
Several factors are likely to contribute to the fact that Ne is very small compared with the total HIV-1 RNA number. It has been shown that the majority of virus particles are noninfectious (HIV-1 RNA exceeds the infectious virus titer 104- to 105-fold; refs. 6 and 9), probably because deleterious mutations are induced by the error-prone reverse transcriptase enzyme (23) or because “mechanical” errors result in misassembly (34). The number of infectious virus particles may be reduced further by immunological inactivation (i.e., neutralization) or physiological inactivation of potentially infectious particles. The number and availability of target cells may be limiting, thereby reducing Ne (35–37). In addition, a successfully infected cell must be activated to produce new virus particles (38, 39), and activated T cells may be killed by cytotoxic T cells before they release virus. Finally, Ne may be reduced, because generation shifts are not synchronized, i.e., the generations overlap.
In addition to strong bottlenecking and a small Ne, a third indication that stochastic processes strongly influence HIV-1 evolution was that different combinations of HIV-1 protease mutations evolved in each patient. This observation is in agreement with other studies showing that the mutational pathways leading to resistance to other protease inhibitors (40) or to zidovudine have considerable interindividual differences (31, 40, 41). However, it should be mentioned that these interindividual differences in mutational pathways could also be influenced by factors other than stochastic processes such as differences in the backbone sequence of the virus or differences in drug levels.
It is interesting to discuss our findings in relation to recombination, which is an important characteristic of retrovirus replication. At first glance, our data seem to argue against rapid recombination. Only in patient 224 is it possible that recombination caused the removal of the divergent env sequences seen after the development of resistance. That we do not observe recombination in all patients may be explained by the fact that for recombination to take place there must be several virus variants present, and the strong bottleneck induced by the protease inhibitor may have created a situation where there was essentially nothing with which to recombine.
An important implication of stochastic evolution is the effect on the fitness of the population. It has been postulated that because of the heterogeneity of a viral population, any genome sampled at random is likely to harbor deleterious mutations relative to the consensus, thus generating progeny with decreased fitness. Moreover, during stochastic evolution in which chance plays an important role, it is possible that the population will be dominated by the first variant to appear, even though others may be more fit. These hypotheses are in agreement with an important concept in population genetics known as “Muller’s ratchet.” Muller’s ratchet predicts that when mutation rates are high and a significant proportion of mutations are deleterious, a kind of irreversible ratchet mechanism will decrease the mean fitness of small populations of asexual organisms. Several in vitro investigations have shown that Muller’s ratchet operates on RNA viruses and that it can dramatically reduce fitness (1, 42–45). Because we observed strong bottlenecking and a small Ne, we anticipate Muller’s ratchet to be also effective in vivo in our patients. This expectation is supported by publications describing viruses harboring ritonavir-resistant proteases and displaying a reduced fitness (46–48).
Our study combines three independent observations: strong population bottlenecking, small Ne, and selection of different combinations of protease mutations, all indicating that HIV-1 evolution is best described by stochastic evolutionary models.
Acknowledgments
We thank Prof. Danner and Dr. Borleffs for providing serum samples and clinical information on the patients. We also thank Dr. Kuiken for providing sequences from 114 Dutch homosexual men and 32 Dutch intravenous drug users. Financial support was provided by the Dutch Health Research Council, The Hague, The Netherlands (Project W0-94-148/94063).
ABBREVIATIONS
- NSI
non-syncytium-inducing
- SI
syncytium-inducing
Footnotes
References
- 1.Domingo E, Escarmis C, Sevilla N, Moya A, Elena S F, Quer J, Novella I S, Holland J J. FASEB J. 1996;10:859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 2.Coffin J M. Curr Top Microbiol Immunol. 1992;176:143–164. doi: 10.1007/978-3-642-77011-1_10. [DOI] [PubMed] [Google Scholar]
- 3.Domingo E, Holland J J. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 4.Eigen M. Steps Toward Life. Oxford: Oxford Univ. Press; 1992. [Google Scholar]
- 5.Eigen M. Gene. 1993;135:37–47. doi: 10.1016/0378-1119(93)90047-7. [DOI] [PubMed] [Google Scholar]
- 6.Piatak M, Jr, Saag M S, Yang L C, Clark S J, Kappes J C, Luk K C, Hahn B H, Shaw G M, Lifson J D. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 7.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, et al. Nature (London) 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 8.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Nature (London) 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 9.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 10.Coffin J M. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 11.Leigh Brown A J, Richman D D. Nat Med. 1997;3:268–271. doi: 10.1038/nm0397-268. [DOI] [PubMed] [Google Scholar]
- 12.Leigh Brown A J. Proc Natl Acad Sci USA. 1997;94:1862–1865. doi: 10.1073/pnas.94.5.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danner S A, Carr A, Leonard J M, Lehman L, Gudiol F, Gonzales J, Raventos A, Rubio R, Bouza E, Pintado V, et al. New Engl J Med. 1995;333:1528–1533. doi: 10.1056/NEJM199512073332303. [DOI] [PubMed] [Google Scholar]
- 14.Roos M T, Miedema F, Eeftinck Schattenkerk J K, de Wolf F, Goudsmit J, Lange J M, Danner S A, Out T A, Schellekens P T. Neth J Med. 1989;34:132–141. [PubMed] [Google Scholar]
- 15.Boom R, Sol C J, Salimans M M, Jansen C L, Wertheim-van Dillen P M, van der Noordaa J. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nijhuis M, Boucher C A B, Schuurman R. BioTechniques. 1995;19:178–180. [PubMed] [Google Scholar]
- 18.Smith S. gde, Genetic Data Environment. Ann Harbor, MI: Millipore Imaging Systems; 1992. , Version 2.2. [Google Scholar]
- 19.Kumar S, Tamura K, Nei M. mega, Molecular Evolutionary Genetic Analysis. Pennsylvania State Univ., University Park, PA: Institute of Molecular Evolutionary Genetics; 1993. , Version 1.01. [Google Scholar]
- 20.Leitner T, Kumar S, Albert J. J Virol. 1997;71:4761–4770. doi: 10.1128/jvi.71.6.4761-4770.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenstein J. phylip, Phylogenetic Inference Package. Seattle: Univ. of Washington; 1991. , Version 3.4. [Google Scholar]
- 22.Kuhner M K, Yamato J, Felsenstein J. fluctuate, Metropolis-Hastings Markov Chain Monte Carlo Genealogy Sampler. Seattle: Univ. of Washington; 1997. , Version 1.1 (beta). [Google Scholar]
- 23.Mansky L M, Temin H M. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruiz L, Nijhuis M, Boucher C, Puig T, Bonjoch A, Martinez-Picado J, Marfil S, de Jong D, Burger D, Arno A, et al. J Acquired Immune Defic Syndr Hum Retrovirol. 1998;19:19–28. doi: 10.1097/00042560-199809010-00003. [DOI] [PubMed] [Google Scholar]
- 25.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P J, Mo H M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, et al. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 26.Markowitz M, Mo H, Kempf D J, Norbeck D W, Bhat T N, Erickson J W, Ho D D. J Virol. 1995;69:701–706. doi: 10.1128/jvi.69.2.701-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Jong J J, Goudsmit J, Keulen W, Klaver B, Krone W J, Tersmette M, de Ronde A. J Virol. 1992;66:757–765. doi: 10.1128/jvi.66.2.757-765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y M, Dawson S C, Landsman D, Lane H C, Salzman N P. J Virol. 1994;68:425–432. doi: 10.1128/jvi.68.1.425-432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leigh Brown A J, Cleland A. AIDS. 1996;10:1067–1073. [PubMed] [Google Scholar]
- 31.Cleland A, Watson H G, Robertson P, Ludlam C A, Leigh Brown A J. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;12:6–18. doi: 10.1097/00042560-199605010-00002. [DOI] [PubMed] [Google Scholar]
- 32.Kozal M J, Shafer R W, Winters M A, Katzenstein D A, Merigan T C. J Infect Dis. 1993;167:526–532. doi: 10.1093/infdis/167.3.526. [DOI] [PubMed] [Google Scholar]
- 33.Kroodsma K L, Kozal M J, Hamed K A, Winters M A, Merigan T C. J Infect Dis. 1994;170:1292–1295. doi: 10.1093/infdis/170.5.1292. [DOI] [PubMed] [Google Scholar]
- 34.Layne S P, Merges M J, Dembo M, Spouge J L, Conley S R, Moore J P, Raina J L, Renz H, Gelderblom H R, Nara P L. Virology. 1992;189:695–714. doi: 10.1016/0042-6822(92)90593-e. [DOI] [PubMed] [Google Scholar]
- 35.de Jong M D, Veenstra J, Stilianakis N I, Schuurman R, Lange J M A, de Boer R J, Boucher C A B. Proc Natl Acad Sci USA. 1996;93:5501–5506. doi: 10.1073/pnas.93.11.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Boer R J, Boucher C A B. Proc R Soc London Ser B. 1996;263:899–905. doi: 10.1098/rspb.1996.0133. [DOI] [PubMed] [Google Scholar]
- 37.de Jong M D, de Boer R J, de Wolf F, Foudraine N, Boucher C A B, Goudsmit J, Lange J M A. AIDS. 1997;11:F79–F84. doi: 10.1097/00002030-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 38.Wong J K, Hezareh M, Günthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 39.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, et al. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 40.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, et al. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boucher C A, O’Sullivan E, Mulder J W, Ramautarsing C, Kellam P, Darby G, Lange J M, Goudsmit J, Larder B A. J Infect Dis. 1992;165:105–110. doi: 10.1093/infdis/165.1.105. [DOI] [PubMed] [Google Scholar]
- 42.Duarte E, Clarke D, Moya A, Domingo E, Holland J. Proc Natl Acad Sci USA. 1992;89:6015–6019. doi: 10.1073/pnas.89.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Novella I S, Elena S F, Moya A, Domingo E, Holland J J. J Virol. 1995;69:2869–2872. doi: 10.1128/jvi.69.5.2869-2872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke D K, Duarte E A, Moya A, Elena S F, Domingo E, Holland J. J Virol. 1993;67:222–228. doi: 10.1128/jvi.67.1.222-228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chao L. Nature (London) 1990;348:454–455. doi: 10.1038/348454a0. [DOI] [PubMed] [Google Scholar]
- 46.Zennou V, Mammano F, Paulos S, Mathez D, Clavel F. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nijhuis M, Schuurman R, de Jong D, Schipper P, Boucher C. Antiviral Ther. 1997;92:61–62. (abstr.). [Google Scholar]
- 49.Delwart E L, Pan H, Neumann A, Markowitz M. J Virol. 1998;72:2416–2421. doi: 10.1128/jvi.72.3.2416-2421.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]