Abstract
Vibrio cholerae, the etiologic agent of the diarrheal disease cholera, is a Gram-negative bacterium that belongs to the γ subdivision of the family Proteobacteriaceae. The physical map of the genome has been reported, and the genome has been described as a single 3.2-Mb chromosome [Majumder, R., et al. (1996) J. Bacteriol. 178, 1105–1112]. By using pulsed-field gel electrophoresis of genomic DNA immobilized in agarose plugs and digested with the restriction enzymes I-CeuI, SfiI, and NotI, we have also constructed the physical map of V. cholerae. Our analysis estimates the size of the genome at 4.0 Mb, 25% larger than the physical map reported by others. Our most notable finding is, however, that the V. cholerae chromosome appears to be not the single chromosome reported but two unique and separate circular megareplicons.
A comprehensive Vibrio cholerae genetic map has been unattainable because of the relative paucity of efficient genetic exchange systems for this organism. Conjugation in V. cholerae is possible and is mediated by the P factor, but unlike the F factor of Escherichia coli, the P factor cannot integrate into the chromosome, and hence cannot induce Hfr donors. Although many genes have been mapped relative to one another, the distance within these gene clusters is small relative to the entire genome; thus, a physical map is needed to determine the positions of genes on the chromosome.
Pulsed-field gel electrophoresis (PFGE) has been used to assess genomic diversity, to identify the location of specific genes, to determine the size of the genome, to detect gross chromosome alterations, and to document the existence of multiple replicons in a bacterium. Previously, the existence of multiple chromosomes or permanent megareplicons in the family Proteobacteriaceae has been primarily limited to members of the α subdivision [the exception being Burkholderia cepacia (1), a member of the β subdivision]. Members of the α group demonstrated to have two-chromosome genomes include Brucella melitensis (2), Rhodobacter sphaeroides (3), Agrobacterium tumefaciens (4), and Rhizobium meliloti (5). We now report that V. cholerae contains not the single circular chromosome previously reported (6), but two chromosomes.
MATERIALS AND METHODS
Bacterial Strains.
V. cholerae 395, a classical Ogawa strain, and V. cholerae E7946, an El Tor Ogawa strain were from collections at the Center for Vaccine Development and have previously been demonstrated to produce cholera in volunteers (7). V. cholerae AI1837, an O139 Bengal strain, was isolated from a Bangladeshi patient with severe cholera in early 1993 (a gift of M. J. Albert, International Centre for Diarrheal Disease Research, Bangladesh) and also has been shown to produce cholera in volunteers (8). V. cholerae 392, a non-O1 environmental isolate, was from collections at the Center for Vaccine Development. V. cholerae S-21, a non-O1 clinical isolate and Vibrio mimicus strain 2031 were a gift of J. Powell, Division of Hospital Epidemiology, University of Maryland, Baltimore.
Pulsed-Field Gel Electrophoresis.
Chromosomal DNA was prepared from the bacterial strains by using the technique of Trucksis et al. (9) with minor modification. The bacteria were grown in Luria–Bertani (LB) broth at 37°C to an optical density at 600 nm of 0.9. Cells were washed according to previously described protocols (9) and mixed with an equal volume of 2.0% inbed agarose (New England Biolabs) and cast in a mold. The individual agarose inserts were treated as previously described (9) except that lysozyme (5 mg/ml) was substituted for lysostaphin. After processing, the inserts were incubated for at least 6 hr with restriction enzyme I-CeuI or NotI at 37°C or with SfiI at 50°C. Electrophoresis was performed with either a CHEF-DR II or III system (Bio-Rad) in 0.5× TBE buffer (45 mM Tris/45 mM boric acid/1 mM EDTA, pH 8.3) at 4–10°C. Different electrophoresis conditions used were based on the size of the DNA fragments to be resolved. Electrophoresis parameters are given in each figure legend. After electrophoresis, gels were stained with ethidium bromide. The molecular weights of restriction fragments were determined by comparison with λ DNA concatamers, Saccharomyces cerevisiae chromosome, and low-range pulsed-field gel markers (New England Biolabs) as molecular weight markers.
Southern Blotting.
Southern blots of restriction endonuclease-digested DNAs were transferred to Magnagraph nylon membrane (Micron Separations, Westboro, MA). The genes were mapped by using either restriction fragments isolated from plasmids containing the cloned genes or with oligonucleotide probes derived from published gene sequences (Table 1). DNA fragment probes were labeled with [α-32P]dCTP by using the random-primed DNA labeling kit (Amersham) according to the instructions of the manufacturer. Hybridizations were performed at 37°C in 25% formamide buffer. The following oligonucleotide gene probes were used: pcd4, 5′-ACTGAAGGCTCTCATCACAGGCATG-3′; ompW, 5′-TTGCCTAGCCGTACTTGCAGCCCTA-3′; mutL, 5′-ATGACTGCGACCGAGAAGATAGAAG-3′; ompV, 5′-TCTAGGCACTTATCTGACTGGCAGC-3′; and hlyT, 5′-GCTATCTATCACAGTGATGCCGTGT-3′. The oligonucleotides were radiolabeled with [γ-32P]dATP by using a 5′ end labeling protocol (10). The rpoS gene probe was a 600-bp fragment amplified from E. coli strain NM522 (11) genomic DNA using primers T1 (5′-GCGCGTCGCGCACTGCGTGG-3′) and T2 (5′-CTGGCCAGCACTTCACGCTG-3′) derived from the published E. coli DNA sequence (12). PCR with Taq polymerase (2 units) was performed in 50 μl containing chromosomal DNA template (50 ng), primers (2 μM each), dNTPs (200 μM), MgCl2 (2 mM), (NH4)2SO4 (17 mM), dimethyl sulfoxide (5 μl), and 2-mercaptoethanol (0.07%). The amplification cycle was denaturation at 94°C for 1 min, annealing at 50°C for 2 min, and extension at 72°C for 2 min. The amplification cycle was repeated 29 times and followed by final extension at 72°C for 10 min.
Table 1.
Gene probes used for V. cholerae gene mapping
| Gene | Map location | Function | Source (ref.) |
|---|---|---|---|
| ctxA† | I-1/2, S-B/T | Cholera toxin A subunit | J. B. Kaper (22) |
| dam | I-4, S-I | DNA adenine methylase | J. Das (23) |
| epsE | I-4, S-A | Extracellular secretion protein | M. Sandkvist (24) |
| exbB† | I-2, S-J | High affinity iron transport system | S. M. Payne (25) |
| dnaE | I-1, S-S | Alpha subunit of DNA polymerase III | A. Franco (26) |
| fur† | I-2, S-C | Ferrichrome uptake, regulation of iron uptake | S. B. Calderwood (27) |
| hap† | I-2, S-J | Hemagglutinin protease | C. C. Hase (28) |
| hlyA† | I-2, S-E | Hemolysin | J. B. Kaper (29) |
| hlyT | I-3, S-D | Hemolysin | Oligonucleotide (30) |
| hutA | I-1, S-D | Heme transport receptor | S. M. Payne (25) |
| irgB | I-3, S-P | Iron-regulated virulence protein | S. B. Calderwood (31) |
| malEFG† | I-2, S-J | Maltose transport | H. Lang (32) |
| malQ† | I-2, S-M | Maltose metabolism | H. Lang (32) |
| mshA | I-3, S-V | Mannose-sensitive hemagglutinin pilin | R. K. Taylor (33) |
| mutL | I-6, S-A | DNA repair | Oligonucleotide (34) |
| nanH | I-1, S-H | Neuraminidase | J. B. Kaper (35) |
| ompS† | I-2, S-M | Growth-phase-dependent maltoporin | H. Lang (36) |
| ompT | I-1, S-H | Outer membrane protein | J. B. Kaper (unpublished data) |
| ompU | I-3, S-D | Outer membrane protein | J. B. Kaper (37) |
| ompV | I-1, S-K | Outer membrane protein | Oligonucleotide (38) |
| ompW† | I-2, S-J | Outer membrane protein | Oligonucleotide (39) |
| pcd4† | I-2, S-M | Confers resistance to bacteriophage | Oligonucleotide (40) |
| recA | I-3, S-P | Homologous recombination | J. B. Kaper (41) |
| rfaD | I-5, S-A | Core oligosaccharide biosynthesis | J. B. Kaper (42) |
| rpoS | I-3, S-P | Alternate sigma factor | M. Trucksis, E. coli probe (43) |
| tagA | I-1, S-B | Lipoprotein | D. K. Karaolis (44) |
| tcpA | I-1, S-B | Toxin-coregulated pilus | R. K. Taylor (45) |
| tcpG/dsbA | I-7, S-A | Periplasmic thiol:disulfide interchange protein | R. K. Taylor (46) |
| toxR | I-1, S-R | Transcription-activating transmembrane DNA-binding protein | J. B. Kaper (47) |
| vibA | I-1, S-B | Vibriobactin siderophore synthesis | S. M. Payne (48) |
| viuA | I-1, S-S | Vibriobactin receptor | S. B. Calderwood (49) |
There are two copies of CTXø in the genome. I, I-CeuI band; S, SfiI fragment.
Genes located on replicon II.
Isolation of SfiI or I-CeuI Linking Fragments.
To isolate SfiI-linking fragments of V. cholerae genomic DNA, a comparison was made of the PFGE pattern of V. cholerae 395 genomic DNA digested with NotI alone or NotI plus SfiI. NotI fragments that disappeared in the double-digestion products identified those NotI fragments having SfiI site(s). The NotI fragments identified as containing SfiI sites were excised from agarose gels, digested to completion with HindIII (which cuts V. cholerae DNA frequently), and labeled with [α-32P]dCTP as above. These linking probes were then used in Southern hybridizations of SfiI-digested V. cholerae 395 genomic DNA. In a similar manner, a comparison was made of the PFGE pattern of V. cholerae 395 genomic DNA digested with SfiI alone or I-CeuI plus SfiI. SfiI fragments that disappeared in the double-digestion products identified those SfiI fragments having I-CeuI site(s). These I-CeuI linking fragments were used in a Southern hybridization as was done for the SfiI linking fragments.
RESULTS
Genome Size.
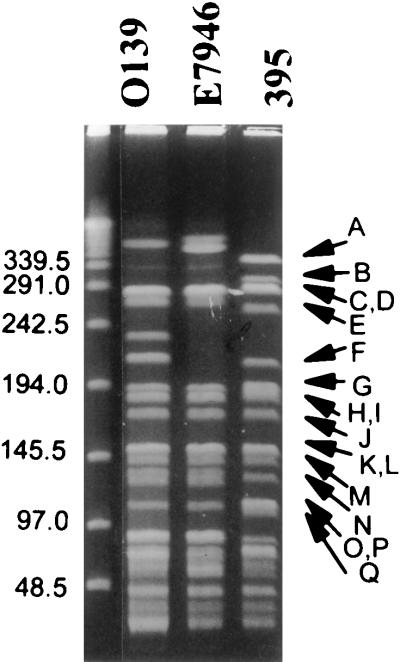
The size of the V. cholerae 395 genome was estimated by compilation of the fragment sizes resulting from SfiI restriction enzyme digestion (Table 2). Several bands resulting from SfiI digestion had an increased staining intensity relative to other bands. These were determined to be doublets after they were excised from the gel and digested with a second restriction enzyme, NotI; the summation of the resulting fragments was found to be twice that of the original-sized SfiI fragment. The SfiI digestion yielded 33 fragments from 364 to 4 kb in size, which were designated A thru GG (Fig. 1). The sum of the fragments yielded a genome size estimated to be 3,988 kb. The I-CeuI-digested genomic DNA produced 10 fragments discernible following ethidium bromide staining of PFGE (Fig. 2). The fragments ranged in size from 1,579 to 5 kb, and were designated 1 to 10.
Table 2.
Sizes of the SfiI(fragment)- and I-CeuI(band)-generated restriction fragments from classical V. cholerae 395
| Fragment | Size, kb | Band | Size, kb |
|---|---|---|---|
| A | 364 | 1 | 1378 |
| B | 302 | 2 | 1579 |
| C | 287 | 3 | 330 |
| D | 287 | 4 | 256 |
| E | 259 | 5 | 173 |
| F | 210 | 6 | 119 |
| G | 198 | 7 | 75 |
| H | 189 | 8 | 67 |
| I | 187 | 9 | 6 |
| J | 172 | 10 | 5 |
| K | 150 | ||
| L | 150 | ||
| M | 143 | ||
| N | 138 | ||
| O | 113 | ||
| P | 113 | ||
| Q | 104 | ||
| R | 84 | ||
| S | 75 | ||
| T | 71 | ||
| U | 71 | ||
| V | 65 | ||
| W | 58 | ||
| X | 46 | ||
| Y | 40 | ||
| Z | 31 | ||
| AA | 22 | ||
| BB | 21 | ||
| CC | 12 | ||
| DD | 9 | ||
| EE | 8 | ||
| FF | 5 | ||
| GG | 4 | ||
| Total | 3988 |
The sizes of bands one and two were determined by summation of their SfiI fragments. The mobility of band 2 in the I-CeuI digestion is aberrant relative to the other fragments because of the covalently closed circular nature of this species.
Figure 1.
PFGE of SfiI-digested V. cholerae DNA. λ concatamers (lane 1), O139 V. cholerae strain AI1837 (lane 2), El Tor V. cholerae strain E7946 (lane 3), and classical V. cholerae strain 395 (lane 4). The V. cholerae strain 395 fragments are identified to the right of lane 4. Pulse ramps were 13 to 28 sec for 60 hr at 180 V in 1.8% agarose.
Figure 2.
PFGE of I-CeuI-digested V. cholerae DNA. Digested fragments were separated in different agarose concentrations to permit separation of both large and small fragments. (A) O139 V. cholerae strain AI1837 (lane 1), El Tor V. cholerae strain E7946 (lane 2), classical V. cholerae strain 395 (lane 3), and yeast-chromosome pulsed-field gel marker (lane 4). The first three bands are identified to the left of lane 1. Pulse ramps were 60 to 90 sec for 24 hr at 200 V in 1.0% agarose. Band 2 is circular, and so the molecular weight cannot be determined by comparison with linear molecular-weight markers. (B) O139 V. cholerae strain AI1837 (lane 1), El Tor V. cholerae strain E7946 (lane 2), and classical V. cholerae strain 395 (lane 3). Identification of the migration of λ concatamers is shown to the left of lane 1. Fragments 3 to 10 are identified on the right of lane 3. Pulse ramps were 13 to 28 sec for 60 hr at 180 V in 1.8% agarose.
The V. cholerae Genome Contains Two Replicons.
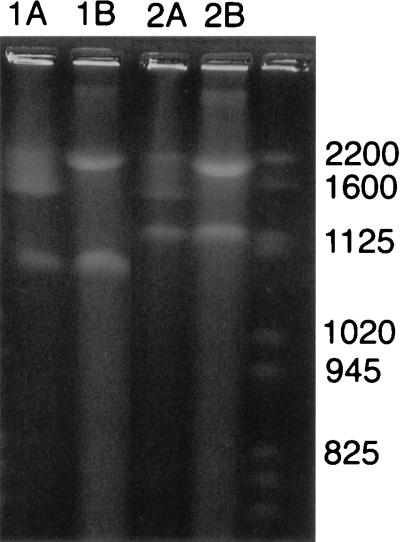
An interesting observation was noted when intact V. cholerae DNA was electrophoresed. For this experiment, total undigested genomic DNA was electrophoresed side-by-side with I-CeuI-digested genomic DNA (Fig. 3). The undigested genomic DNA appeared as two megabase-sized fragments. The top fragment (replicon I) appeared as a diffuse smear on the gel, suggesting that it was chromosomal DNA, and the lower fragment (replicon II) appeared as a more distinct band. The I-CeuI-digested genomic DNA appeared as 10 bands. Band 2 in the I-CeuI digestion appeared the same as the smaller band in the undigested DNA (Fig. 3). In our preparations of I-CeuI-digested DNA, band 2 always appeared to stain with less intensity than the other bands, suggesting a difference in ethidium bromide uptake that would occur if band 2 was covalently closed circular rather than linear DNA (Fig. 2A). When total genomic DNA was subjected to I-SceI digestion, the result was two bands (data not shown). The migration of the smaller band was the same as band 2 of the I-CeuI-digested DNA and the smaller band in the undigested DNA. These results are consistent with the second band in all three preparations representing a circular replicon that is uncut by I-CeuI or I-SceI.
Figure 3.
PFGE of undigested versus I-CeuI-digested V. cholerae DNA. O139 V. cholerae strain AI1837 (lane 1) and classical V. cholerae strain 395 (lane 2). Lanes marked A are undigested and lanes marked B are I-CeuI-digested genomic DNA. Yeast-chromosome molecular-weight markers are in the far right lane. Pulse ramps were 60 to 90 sec for 60 hr at 200 V in 1.3% agarose.
PFGE of undigested genomic DNA prepared from other strains of V. cholerae, an El Tor biotype strain E7946 (data not shown), a prototypic O139 serotype strain AI1837 (Fig. 3), and non-O1 strains 392 and S-21 (data not shown) also revealed two bands migrating about the same distance as the two replicons of the V. cholerae classical biotype strain 395 (Fig. 3). Examination of V. mimicus confirmed the presence of two bands following PFGE of undigested genomic DNA. Thus, these members of the genus Vibrio, and most likely all Vibrio species, have two unique replicons.
The Circular Map of the 2.4-Mb and 1.6-Mb Replicons.
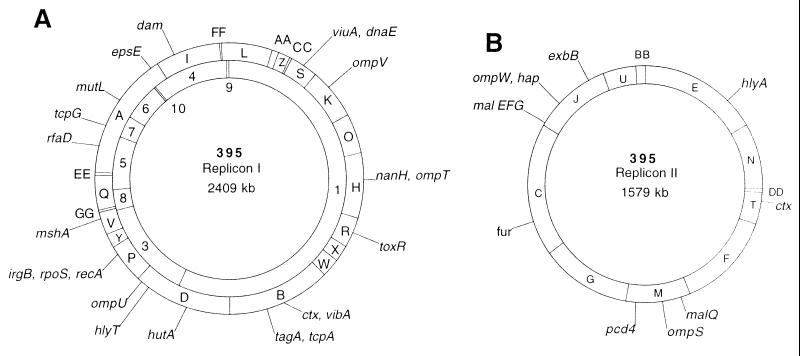
To order the SfiI fragments within the two largest bands separated in the I-CeuI digest, bands 1 and 2 were excised from the agarose gel and digested with a second restriction enzyme, SfiI. The digests were electrophoresed in parallel and with an SfiI digest of total genomic DNA. I-CeuI band 1 yielded 13 discernible SfiI fragments, of which 11 corresponded to fragments present in the total genomic DNA SfiI digest and two were new bands not present in the total genomic DNA SfiI digest and therefore represent I-CeuI–SfiI fragments. These two bands represent part of the SfiI fragments (L and D) present at the ends of the large I-CeuI fragment. The intact SfiI bands contained in band 1 of the I-CeuI digest were B, H, K, O, R, S, W, X, Z, AA, and CC.
I-CeuI band 2 excised from the agarose gel and digested with SfiI produced 10 discernible fragments, all of which corresponded to fragments present in the total genomic DNA SfiI digest. These corresponded to SfiI bands C, E, F, G, J, M, N, T, U, and BB. The summation of the SfiI fragments gave a size of 1579 kb for the second replicon. The linkage map of these SfiI bands was established by using NotI fragments encoding an SfiI site(s). Linkage confirmed the unique and separate circular nature of the second replicon.
A comparison was made of the PFGE pattern of V. cholerae 395 genomic DNA digested with SfiI or I-CeuI and SfiI. SfiI fragments that disappeared in the double-digestion identified those SfiI fragments having I-CeuI site(s). These became I-CeuI linking fragments and were used as probes in a Southern hybridization of I-CeuI-digested genomic DNA. Each probe (an SfiI fragment containing an I-CeuI site) hybridized with two of the I-CeuI genomic DNA fragments except for SfiI fragment A, which hybridized with I-CeuI bands 4, 5, 6, 7, and 9 (data not shown). The order of these I-CeuI bands was determined by using NotI linking fragments (see below) and the hybridization signals of specific gene probes (Table 1). None of the I-CeuI linking fragments hybridized with I-CeuI band 2. The analysis of this data allowed the determination of two circular maps of all 10 I-CeuI bands (Fig. 4). The exact position of the smallest I-CeuI band (band 10) could not be determined. This band may represent the recently described cryptic plasmid linked to the CTX prophage (13) and thus may be extrachromosomal.
Figure 4.
The physical map of the two replicons of classical V. cholerae strain 395. The circles represent the I-CeuI (Inner) and SfiI (Outer) maps. The order of genes within each SfiI fragment are arbitrary. (A) Replicon I. (B) Replicon II.
To construct the SfiI linkage map, a comparison was made of the PFGE pattern of V. cholerae 395 genomic DNA digested with NotI alone or SfiI plus NotI. NotI fragments that contained SfiI site(s) were used as probes in a Southern hybridization of SfiI-digested genomic DNA.
Positioning of V. cholerae Genes on the Physical Map.
The relative chromosomal locations of 44 genes (including 9 rrn operons) were determined on the physical map of V. cholerae 395 genome by hybridization using homologous or heterologous genes or synthetic oligonucleotides as probes (Table 1). The genes were positioned arbitrarily on fragments to which they hybridized and do not reflect the true order of genes within each chromosomal fragment. The I-CeuI sites were taken as the positions of the rrn operons.
A copy of the V. cholerae CTX φ element (14) encoding cep, orfU, ace, zot, and ctxAB was found on each of the replicons. This placement is consistent with previous reports that the two copies of ctxAB are widely separated in the classical biotypes of V. cholerae (15). The toxR virulence gene regulator and the V. cholerae pathogenicity island (16) containing the tcp operon, as well as all of the rrn operons, were located on the larger replicon.
DISCUSSION
Our suspicion that the V. cholerae chromosome may exist as two separate replicons was based on the observation that when undigested genomic DNA was subjected to electrophoresis, two megabase-sized fragments were visible. In addition, we were unable to convincingly link the I-CeuI fragments into a single circular chromosome. The final clue came from the observation that immobilized genomic DNA subjected to pulsed-field gel electrophoresis after digestion with another rarely cutting restriction enzyme, I-SceI, produced two fragments, the smaller of which appeared exactly like one of two megabase-sized fragments produced by I-CeuI digestion. This fragment in both digestions always appeared to stain lighter than the other bands. We now have confirmed (by linkage of SfiI fragments contained in this band) that this fragment was not cut by either I-SceI or I-CeuI; the presumed reason it did not stain well was because it was constrained in its uptake of ethidium bromide by its covalently closed circular nature, i.e., it was a separate circular megareplicon.
This study demonstrates multiple replicons in V. cholerae, an enteric bacterial pathogen with a complex genomic structure. Proteobacteriaceae consists of four subdivisions, δ/ɛ, α, γ, and β in the Woese classification based on 16S rRNA sequences (17). Previously, bacteria in the Proteobacteriaceae shown to have a multiple-replicon genome (except for Burkholderia cepacia) were all members of the α group, which also contains bacteria that have a single chromosome. Similarly, the γ subdivision, which includes the genus Vibrio, contains bacteria that also have a single chromosome, such as Pseudomonas aeruginosa and E. coli. Thus,there appears to be heterogeneity in genomic structure within the subdivisions. Other members of the γ subdivision should be examined to determine whether the genus Vibrio is an exception or whether this complex genomic structure is more prevalent.
An important question is whether the two replicons of V. cholerae represent two chromosomes. The definition of a chromosome versus a megaplasmid is the subject of some debate (18, 19). Most investigators suggest that the presence of essential or housekeeping genes defines a chromosome. These could include 16S rRNA genes, metabolically essential genes (such as those encoding proteins involved in metabolism of amino acids), or heat-shock proteins (such as groE or dnaK). Other properties suggested as necessary to define a chromosome include a size representing some significant percentage of the total genome, non-self-transmissibility, or the observation that the replicon is consistently present in strains. We would conclude that the second replicon represents a chromosome for the following reasons. First, the second replicon represents 40% of the entire genome by size. Second, all three V. cholerae strains examined, representing a classical, an El Tor, and an O139 serotype, contain both replicons. Third, both replicons are present in a fixed stoichiometry relative to each other. Finally, each replicon contains unique genes, the only duplication seen so far being the presence of the CTX φ element in both replicons of the classical biotype.
The finding that the V. cholerae chromosome consists of two unique and circular replicons could account for the difficulty in constructing a comprehensive genetic map of V. cholerae using traditional chromosome mobilization techniques. In one study (20), most chromosomal markers could be assigned to one of three linkage groups, but some markers showed little or no linkage. In a later study (21), a circular map was presented, but 85% of the markers clustered into ≈25% of the map, and the remaining three markers were positioned at widely scattered intervals throughout the rest of the map.
V. cholerae has presented us with many surprises in the 1990s. Recent reports have described truly novel developments, such as the spread of cholera into South America in 1991 and the emergence of V. cholerae O139 in 1992, as well as novel explanations for long-standing phenomena, such as the discovery of the CTXφ bacteriophage, which uses the toxin-coregulated pilus (TCP) as a receptor. The recognition that the V. cholerae genome contains two chromosomes is further evidence that despite more than a century of study, this species still presents surprises and challenges to us.
Acknowledgments
We thank Jim Galen for expert DNA sequence analysis and Kristen Ziglari, HongRong Cai, and Tim Conn for technical assistance. We are grateful to the many colleagues who generously provided cloned genes. This work was supported by Public Health Service Grant AI35717 (M.T.) and AI19716 (J.B.K.) from the National Institutes of Health.
ABBREVIATION
- PFGE
pulsed-field gel electrophoresis
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Cheng H-P, Lessie T G. J Bacteriol. 1994;176:4034–4042. doi: 10.1128/jb.176.13.4034-4042.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michaux S, Paillisson J, Carles-Nurit M-J, Bourg G, Allardet-Servent A, Ramuz M. J Bacteriol. 1993;175:701–705. doi: 10.1128/jb.175.3.701-705.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suwanto A, Kaplan S. J Bacteriol. 1989;171:5850–5859. doi: 10.1128/jb.171.11.5850-5859.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allardet-Servent A, Michaux-Charachon S, Jumas-Bilak E, Karayan L, Ramuz M. J Bacteriol. 1993;175:7869–7874. doi: 10.1128/jb.175.24.7869-7874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sobral B W S, Honeycutt R J, Atherly A G, McClelland M. J Bacteriol. 1991;173:5173–5180. doi: 10.1128/jb.173.16.5173-5180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majumder R, Sengupta S, Khetawat G, Bhadra R K, Roychoudhury S, Das J. J Bacteriol. 1996;178:1105–1112. doi: 10.1128/jb.178.4.1105-1112.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine M M, Black R E, Clements M L, Nalin D R, Cisneros L, Finkelstein R A. In: Volunteer Studies in Development of Vaccines Against Cholera and Enterotoxigenic Escherichia coli: A Review. Holme T, Holmgren J, Merson M H, Mollby R, editors. Amsterdam: Elsevier; 1981. pp. 443–459. [Google Scholar]
- 8.Morris J G, Jr, Losonsky G A, Johnson J A, Tacket C O, Nataro J P, Panigrahi P, Levine M M. J Infect Dis. 1995;171:903–908. doi: 10.1093/infdis/171.4.903. [DOI] [PubMed] [Google Scholar]
- 9.Trucksis M, Wolfson J S, Hooper D C. J Bacteriol. 1991;173:5854–5860. doi: 10.1128/jb.173.18.5854-5860.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 11.Mead D A, Skorupa E S, Kemper B. Nucleic Acids Res. 1985;13:1103–1118. doi: 10.1093/nar/13.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulvey M R, Loewen P C. Nucleic Acids Res. 1989;17:9979–9991. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubin E J, Lin W, Mekalanos J J, Waldor M K. Mol Microbiol. 1998;28:1247–1254. doi: 10.1046/j.1365-2958.1998.00889.x. [DOI] [PubMed] [Google Scholar]
- 14.Waldor M K, Mekalanos J J. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 15.Sporecke I, Castro D, Mekalanos J J. J Bacteriol. 1984;157:253–261. doi: 10.1128/jb.157.1.253-261.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. Proc Natl Acad SciUSA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsen G J, Woese C R, Overbeek R. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolsto A-B. Mol Microbiol. 1997;24:241–248. doi: 10.1046/j.1365-2958.1997.3501715.x. [DOI] [PubMed] [Google Scholar]
- 19.Suwanto A, Kaplan S. J Bacteriol. 1992;174:1135–1145. doi: 10.1128/jb.174.4.1135-1145.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker C, Gauthier D, Tate A, Richardson K, Romig W R. Genetics. 1979;91:191–214. doi: 10.1093/genetics/91.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newland J W, Green B A, Holmes R K. Infect Immun. 1984;45:428–432. doi: 10.1128/iai.45.2.428-432.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lockman H, Kaper J B. J Biol Chem. 1983;258:13722–13726. [PubMed] [Google Scholar]
- 23.Bandyopadhyay R, Das J. Gene. 1994;140:67–71. doi: 10.1016/0378-1119(94)90732-3. [DOI] [PubMed] [Google Scholar]
- 24.Overbye L J, Sandkvist M, Bagdasarian M. Gene. 1993;132:101–106. doi: 10.1016/0378-1119(93)90520-d. [DOI] [PubMed] [Google Scholar]
- 25.Henderson D P, Payne S M. Mol Microbiol. 1993;7:461–469. doi: 10.1111/j.1365-2958.1993.tb01137.x. [DOI] [PubMed] [Google Scholar]
- 26.Franco A A, Yeh P E, Johnson J A, Barry E M, Guerra H, Maurer R, Morris J G., Jr Gene. 1996;175:281–283. doi: 10.1016/0378-1119(96)00155-2. [DOI] [PubMed] [Google Scholar]
- 27.Litwin C M, Boyko S A, Calderwood S B. J Bacteriol. 1992;174:1897–1903. doi: 10.1128/jb.174.6.1897-1903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hase C C, Finkelstein R A. J Bacteriol. 1991;173:3311–3317. doi: 10.1128/jb.173.11.3311-3317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldberg S L, Murphy J R. J Bacteriol. 1984;160:239–244. doi: 10.1128/jb.160.1.239-244.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams S G, Attridge S R, Manning P A. Mol Microbiol. 1993;9:751–760. doi: 10.1111/j.1365-2958.1993.tb01735.x. [DOI] [PubMed] [Google Scholar]
- 31.Goldberg M B, Boyko S A, Calderwood S B. Proc Natl Acad Sci USA. 1991;88:1125–1129. doi: 10.1073/pnas.88.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang H, Jonson G, Holmgren J, Palva E T. Infect Immun. 1994;62:4781–4788. doi: 10.1128/iai.62.11.4781-4788.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marsh J W, Marsh W, Sun D X, Taylor R K. Infect Immun. 1996;64:460–465. doi: 10.1128/iai.64.2.460-465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bera T K, Ghosh S K, Das J. Nucleic Acids Res. 1989;17:6241–6251. doi: 10.1093/nar/17.15.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galen J E, Vimr E R, Lawrisuk L, Kaper J B. In: Cloning, Sequencing, and Expression of the Gene, nanH, for Vibrio cholerae Neuraminidase. Sack R B, Zinnaka Y, editors. Tokyo: KTK Scientific Publishers; 1990. pp. 143–153. [Google Scholar]
- 36.Lang H, Palva E T. Mol Microbiol. 1993;10:891–901. doi: 10.1111/j.1365-2958.1993.tb00960.x. [DOI] [PubMed] [Google Scholar]
- 37.Sperandio V, Girón J A, Silveira W D, Kaper J B. Infect Immun. 1995;63:4433–4438. doi: 10.1128/iai.63.11.4433-4438.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pohlner J, Meyer T F, Jalajakumari M B, Manning P A. Mol Gen Genet. 1986;205:494–500. doi: 10.1007/BF00338088. [DOI] [PubMed] [Google Scholar]
- 39.Jalajakumari M B, Manning P A. Nucleic Acids Res. 1990;18:2180. doi: 10.1093/nar/18.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biswas S K, Chowdhury R, Das J. J Bacteriol. 1992;174:6221–6229. doi: 10.1128/jb.174.19.6221-6229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldberg I, Mekalanos J J. J Bacteriol. 1986;165:715–722. doi: 10.1128/jb.165.3.715-722.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comstock L E, Maneval D, Panigrahi P, Joseph A, Levine M M, Kaper J B, Morris J G, Johnson J A. Infect Immun. 1995;63:317–323. doi: 10.1128/iai.63.1.317-323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiratsu K, Yamamoto K, Makino K. Genes Genet Syst. 1997;72:115–118. doi: 10.1266/ggs.72.115. [DOI] [PubMed] [Google Scholar]
- 44.Harkey C W, Everiss K D, Peterson K M. Gene. 1995;153:81–84. doi: 10.1016/0378-1119(94)00811-6. [DOI] [PubMed] [Google Scholar]
- 45.Rhine J A, Taylor R K. Mol Microbiol. 1994;13:1013–1020. doi: 10.1111/j.1365-2958.1994.tb00492.x. [DOI] [PubMed] [Google Scholar]
- 46.Peek J A, Taylor R K. Proc Natl Acad Sci USA. 1992;89:6210–6214. doi: 10.1073/pnas.89.13.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller V L, Mekalanos J J. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wyckoff E E, Stoebner J A, Reed K E, Payne S M. J Bacteriol. 1997;179:7055–7062. doi: 10.1128/jb.179.22.7055-7062.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butterton J R, Stoebner J A, Payne S M, Calderwood S B. J Bacteriol. 1992;174:3729–3738. doi: 10.1128/jb.174.11.3729-3738.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]