Abstract
The HIV-1 envelope protein gp120 induces apoptosis in hippocampal neurons. Because chemokine receptors act as cellular receptors for HIV-1, we examined rat hippocampal neurons for the presence of functional chemokine receptors. Fura-2-based Ca imaging showed that numerous chemokines, including SDF-1α, RANTES, and fractalkine, affect neuronal Ca signaling, suggesting that hippocampal neurons possess a wide variety of chemokine receptors. Chemokines also blocked the frequency of spontaneous glutamatergic excitatory postsynaptic currents recorded from these neurons and reduced voltage-dependent Ca currents in the same neurons. Reverse transcription–PCR demonstrated the expression of CCR1, CCR4, CCR5, CCR9/10, CXCR2, CXCR4, and CX3CR1, as well as the chemokine fractalkine in these neurons. Both fractalkine and macrophage-derived chemokine (MDC) produced a time-dependent activation of extracellular response kinases (ERK)-1/2, whereas no activation of c-JUN NH2-terminal protein kinase (JNK)/stress-activated protein kinase, or p38 was evident. Furthermore, these two chemokines, as well as SDF-1α, activated the Ca- and cAMP-dependent transcription factor CREB. Several chemokines were able also to block gp120-induced apoptosis of hippocampal neurons, both in the presence and absence of the glial feeder layer. These data suggest that chemokine receptors may directly mediate gp120 neurotoxicity.
AIDS is associated with deficits of both the immune and nervous systems. Infection with HIV-1 or related viruses produces a spectrum of cognitive and motor problems, together with neuronal loss in different parts of the brain, including the hippocampus (1, 2). However, as HIV-1 is not thought to replicate in neurons, the molecular basis for AIDS-associated neurodegeneration is unclear. Overexpression of the HIV-1 envelope protein gp120 in central glial cells (3), as well as treatment of human or rat neurons with gp120 in culture (4–7), produces apoptotic neuronal death. To explain this neuronal death, it has been suggested that gp120 causes the release of neurotoxic agents from brain macrophages and microglia, although another possibility is that gp120 is directly toxic to neurons (8). We have shown previously that gp120 induces cell death of hippocampal neurons both in the presence and absence of glia (4).
Recent studies have demonstrated that a variety of chemokine receptors act as cellular receptors for the HIV coat protein gp120, either together with or in the absence of CD4 (9–12). Chemokines (chemotactic cytokines) are a family of related proteins that have been shown to be essential for information transfer and signaling between different types of immune cells (13). Chemokine structure and effects indicate that they fall into four families, typified by the cysteine residues in their sequences (13, 14). In α-chemokines, such as SDF-1α, one amino acid separates the first two cysteines (CXC), whereas in β-chemokines, such as RANTES, the first two cysteines are next to one another (CC). Two other emerging “families” of chemokines may be typified by fractalkine, in which the first two cysteines are separated by three amino acid residues (CXXXC), and lymphotactin, which has only two cysteines in total. Chemokines exert their effects by activating a family of G-protein-coupled receptors. α-Chemokines activate the receptors CXCR1–5, β-chemokines activate the receptors CCR1–10, and fractalkine activates the receptor CX3CR1. However, there are many “orphan” chemokine receptors in the literature that await the identification of their appropriate ligand(s). It was shown that for infection of macrophages by M-tropic (i.e., macrophage-selective) strains of HIV-1, CD4 and the chemokine receptor CCR5 were required, and that the infection of T lymphocytes by T-tropic (i.e., T lymphocyte-selective) strains of HIV-1 used the CXCR4 receptor (9, 13–17). The precise determinants of HIV-1 tropism in vivo are clearly very complex and continue to be defined. Thus, some strains of HIV-1 exhibit dual tropism, and others can also use chemokine receptors in addition to CCR5 and CXCR4. To date, at least 10 chemokine receptors, including CCR3, CCR2b, CCR8, and CX3CR1, as well as the orphan chemokine receptors GPR15 (BOB), STRL33 (BONZO), GPR1, and the viral encoded receptor US28, have been shown to act as coreceptors for HIV-1 infection under some circumstances (18). These receptors may be very important in certain instances, such as HIV-1 effects in the brain, where CCR3 appears to be a major coreceptor for HIV-1 infection of microglia (19).
Chemokine receptors are expressed in normal brain and in brains from AIDS patients (20, 21), as well as in human fetal neurons in culture (12). Homologous chemokine receptors are present in both rat and mouse (22–24). Recent evidence indicates that these receptors are involved in rat cerebellar development, namely in the formation of the neuronal granule layer (25). However, the function of chemokine receptors in the different brain regions and their interaction with the HIV-1 envelope protein are still open questions, and their role in the central nervous system is largely unknown.
We now demonstrate that hippocampal neurons possess a wide variety of functional chemokine receptors. Activation of these receptors produces effects on neuronal signal transduction, synaptic transmission, and neuronal survival, including inhibition of gp120-induced neurotoxicity. Neuronal chemokine receptors may act as a direct conduit for gp120-induced neurodegeneration.
METHODS
Neuronal Cultures.
Primary hippocampal neurons were obtained from the hippocampi of E17–18 rat embryos, as previously described (4, 26). The neurons were plated onto either 15-mm glass coverslips (for survival experiments, fura-2 video imaging, electrophysiology, and immunocytochemistry) or 35-mm tissue culture dishes (for Western blot analyses and RNA preparation) and cultured in DMEM defined medium containing N2.1 supplement (Life Technologies, Grand Island, NY). A feeder layer of secondary astrocytes facing the neurons was used to support their growth and differentiation. Astroglia contamination of neuronal cultures was lower than 5%, as assessed by glial fibrillary acidic protein immunostaining. Virtually pure neuronal cultures were obtained when cells were plated at very low density (i.e., on glass coverslips). Microglial contamination of neuronal cultures was negligible (≪1%), whereas in the glial feeder layer microglia were <3%. Nevertheless, for some experiments, both neuronal and astrocyte cultures were further purified by immunopanning. Briefly, an antimouse affinity-purified polyclonal antibody was incubated on polystyrene dishes for 12 hr at 4°C in 50 mM Tris buffer (pH 9.5). Dishes were then incubated with a monoclonal antibody (Accurate, Westbury, NY) against the β-2 integrins CD11b/c, constitutively expressed on microglial surfaces, in 0.2% BSA for 2 hr at 4°C. Neuronal cell suspensions and the secondary astroglia obtained after a 6-hr shaking (200 rpm at 37°C), were incubated for 30 min on anti-CD11b/c-coated dishes and plated subsequently on coverslips or culture dishes, respectively. Microglial contamination before and after immunopanning was assessed by immunofluorescence staining. Cytosine arabinofuranoside (10 μM) was added to the cultures 24–48 hr after plating to halt nonneuronal cell proliferation.
Sources of Chemokines and gp120.
Human and mouse MDC, human MCP-1, and human thymus and activation-regulated chemokine (TARC) (chemically synthesized or recombinant proteins) were obtained as previously described (27, 28). Human RANTES, murine macrophage inflammatory protein (MIP)-1α, human SDF-1α, human interleukin (IL)-8, and human soluble fractalkine were all recombinant proteins from R & D Systems. Lyophilized proteins were reconstituted (100 μg/ml) in 0.1% BSA/PBS and aliquots stored at −20°C. Recombinant HIV-1IIIB gp120 and SIVmac251 gp120 were purchased from Intracel, Issaquah, WA, diluted (100 μg/ml and 300 μg/ml, respectively) in 0.1% BSA/PBS and stored at −70°C. Working solutions (100×) were prepared just before starting treatments; 0.1%BSA/PBS (10 μl/ml) was used as a control. The preparations of recombinant gp120 were purified by immunoaffinity chromatography to >95% purity. We did not observe neurotoxicity in experiments with denatured gp120 (100°C 10 min; neuronal survival = 81 + 3%, mean + SEM). Moreover, the addition of an anti-gp120IIIB (Intracel, 4 μg/ml) to the gp120 preparation inhibited the gp120 neurotoxicity by 45%. The absence of mycoplasmal contamination in gp120 preparations was assessed by Hoechst 33342 staining (Molecular Probes).
Fura-2 Video Imaging.
Neurons (7 to 16 days in culture) were loaded with 2 μM fura-2 acetoxymethylester (Molecular Probes) for 30 min at room temperature, washed and then incubated for an additional 30 min. Coverslips were superfused constantly with a balanced salt solution of the following composition (in mM): 159 NaCl/5 KCl/2 CaCl2/0.4 MgSO4/0.5 MgCl2/0.64 KH2PO4/3 NaHCO3/0.2% BSA (320–330 mOsm/kg, pH 7.35). All measurements were made at room temperature. Each cell in the image was analyzed independently for each time point in the captured sequence. Cells loaded with fura-2 were used to obtain Rmin and Rmax and to calculate the calibration curve as described previously (4).
Electrophysiology.
Whole-cell patch clamp experiments were performed on 10-to 16-day-old neurons (29). The cells were perfused continually with a solution containing 140 mM NaCl/10 mM Hepes/10 mM glucose/2 mM CaCl2/1 mM MgCl2/5 mM KCl. Patch pipettes (3–5 MΩ) were filled with an internal solution containing 100 mM KCl/1 mM MgCl2/10 mM Hepes/10 mM 1,2-bis(2-aminophenoxy)ethane-N,N, N′,N′-tetraacetate acid/3.6 mM Mg-ATP/0.1 mM GTP/14 mM creatine phosphate/50 units per ml creatine phosphokinase, which was adjusted to pH 7.2 with KOH. Once the whole-cell configuration was obtained, the cell was held at −60 mV and allowed to dialyze for at least 5 min before events were acquired continuously by using pclamp6.0.3 (Fetchex program; Axon Instruments, Foster City, CA) software. Activity was recorded for a control period of 3 min before application of the drug via the perfusate. An equal period was recorded in the presence of the drug and washout was followed for up to 18 mins after drug application. For Ca2+ current experiments, Ba2+ was used as the charge carrier, and hence the bathing medium contained 151 mM tetraethylammonium Cl/10 mM Hepes/10 mM glucose/5 mM BaCl2/1 mM MgCl2, with the internal pipette solution remaining unchanged. Ba2+ currents were obtained by means of a depolarizing step (90 mV/200 ms/0.033 Hz) from the holding potential (−80 mV). Drugs were applied by means of the perfusate for a period of 2 min following an initial stable baseline period.
Reverse Transcription–PCR.
Total RNA was extracted from 7- to 8-day-old neurons by using Trizol (Life Technologies) reagent after removal of the glial feeder layers and washing with Hanks’ balanced salt solution or PBS. First, strand cDNA was generated from 1 μg total RNA by using SuperScriptII (Life Technologies) and was primed by oligo(dT) oligonucleotide. cDNA was treated subsequently with 10 units of RNase-H (Life Technologies). Reactions were diluted 50% with sterile water, and 2 μl of cDNA was used for each PCR reaction. PCR reactions by using Taq polymerase were primed by oligonucleotides made according to published rat cDNA sequences (CCR10, CX3CR1/V28, CXCR1, CXCR2), mouse sequences (CCR1, CXCR4, CCR5, fractalkine), or human/mouse degenerate oligonucleotides (CCR2, CCR3, CCR4, CCR6, CCR7, CCR8, CD11b), and reactions with water rather than cDNA were run in parallel as controls. PCR products were gel purified and cloned into the pT7-Blue vector (Novagen) or pGEM-T-EZ vector (Promega). Sequences from at least three clones of each receptor were obtained from both strands, or both strands of the PCR product were directly sequenced, by automated DNA sequencing (ABI 370) to confirm the identity of the PCR product. The following oligonucleotides were used for amplification of chemokine receptors: CCR1 (TTTTAAGGCCCAGTGGGAGTT CACTCACCG and TGGTATAGCCACATGCCTTTGAAACAGCTGC); CCR2 (CT(G/A)TC(C/A)ACATC TC(G/A)TTCTC(G/T)(G/A)TTTA and CCCAAAGACCCAC TCATTTGCAGC); CCR3 (CTGCTAC(T/A)CAGGAATCAT(T/C)AAAAC and GTTCTTTCCA(G/T)(C/T)TTCT CAC(T/C)AGGAAG); CCR4 (GGCAAGGACC(C/T)TGAC(C/T)TATGGGGTCATCAC and GTGCAGTCCTGAAGGACTTC(A/T)AGCTCCACCAG); CCR5 (TACC AGATCTCAGAAAGAAGGTTTTCATTA and GCGTTTGACCATGTGTTTTCGG AAGAACACT); CCR6 (GAGCCCATCA(G/C)GTGGAAGCTGCTG and GGCAG CA(A/G)TGCAGGAAAGCCAGGAC); CCR7 (CGTGCTGGTGGTGGCTCTCC TTGTC and CACAGGACAGCTTGCTGATGAGAAG); CCR8 (GTGGT(G/C)TCT GGC(C/T)TTTATTACATTGG and ACATCCAT CCAAGATGT GCA); CCR9/10 (CGAGACATGCCCACCATCGCTTCTCCC and ATGCCAGGCCACAGAGATGG CCC); CX3CR1 (CCTCACCATGCCTACCTCCTTCCCGGAATTG and CGTGAG CTTGCACATGGCGTTGTGGAGGC); CXCR1 (ATGGCCGAGGCTGAGTATTT CATCTGGATTG and GATCACTCTCTGGTAATTCTCAGAAGTTTA); CXCR2 (ATGGGAGAAATCAGGGTGGATAATTTCAGC and GAGGGGCCACAGTTTAAGTAAACAGTCTTA); CD11b (CTGGCTACATTGGGAAAAACTGTG AG and CTCCTTCTCAAAGCGCCTGTACTCCC); CXCR4 (ATGGAAATATACACTTCG GATAACTACTC and TTAGCTGGAGTGAAAACTT GAGGATTCTG). In the case of CCR2, CCR3, CCR6, CCR7, CCR8, CXCR1, and CD11b, other primer pairs were also used in PCR reactions and failed to amplify the corresponding cDNAs.
Western Blot Analyses and Immunocytochemistry.
After removal of glial cell feeder layers, neurons were washed with balanced salt solution (see above) and left in the incubator for 4 hr before treatments with chemokines were started. At the times indicated, cells were scraped from the dish, centrifuged, and the resulting pellets were resuspended in lysis buffer (30). Cells were kept at 4°C for 15 min with constant agitation, lysed by using a syringe, and then centrifuged (14,000 rpm, 5 min). Supernatants were stored on ice and pellets further processed to extract nuclear proteins by using a hyperosmotic buffer (30). The protein concentration in the total lysate was determined by bicinchoninic acid protein assay (Pierce). Cell lysates were resolved by SDS-polyacrylamide (10%) gel electrophoresis and then transferred to a nitrocellulose membrane (Amersham). Antibodies selectively recognizing the active forms of ERK1/2 (Promega, 1:5000), c-JUN NH2-terminal protein kinase (JNK)1/2 (Promega, 1:2500), p38/high osmolarity glycerol response (Promega, 1:2000), and CREB (Upstate Biotechnology, Lake Placid, NY; 1:2000) were used for immunoblotting. Digitized images of the films obtained by enhanced chemiluminescence (Pierce) were used for densitometric analysis with un-scan it software (Silk Scientific, Orem, UT). For immunocytochemistry, antibody against the phosphorylated form of CREB (1:500) was used on neurons exposed to chemokines in the absence of glia.
Neuronal Death.
Neurons were plated on glass coverslips (1–2 × 104 cells/cm2) and cultured as described above. Fluorescein diacetate (10 μg/ml), propidium iodide (10 μg/ml), and Hoechst 33342 (5 μg/ml) were used to stain viable and dead cells and to evaluate differences between normal and apoptotic neurons (4). Alive and dead neurons in each microscopic field were counted, and the percentage of apoptotic neurons was calculated. (i) Apoptosis induced by glia deprivation: coverslips with neurons were moved to 12 × multiwells containing their conditioned medium plus the different chemokines. Neuronal viability was evaluated after 24–48 hr. (ii) Apoptosis induced by gp120: treatments with gp120 and/or chemokines in the presence of glia were started on the seventh day of culture and were carried on for 3–4 days (4). Cultures were pretreated briefly (20–40 min) with the different chemokines before the addition of gp120. For experiments in the absence of glia, neurons were grown facing the glial feeder layer for the first 6 days of culture and were moved to dishes containing their conditioned medium when experimental treatments started. Cell survival was evaluated 24 hr after the addition of gp120 and/or chemokines. Although in these conditions there was a higher background of cell death, because of the glial deprivation, gp120 neurotoxicity, as well as the protective actions of chemokines, was still evident. One-way ANOVA, followed by the Newman-Keuls multiple comparison procedure, was used for statistical analysis of the neuronal survival experiments.
RESULTS AND DISCUSSION
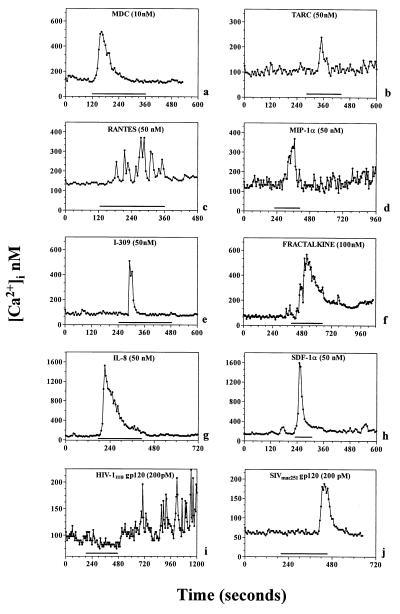
We examined hippocampal neurons in culture for the presence of functional chemokine receptors by using Ca imaging and electrophysiological and molecular biological paradigms. We used a bilaminar culture system in which pure populations of hippocampal pyramidal neurons were cultured on coverslips in close juxtaposition to an astrocyte feeder layer (4), although it is important to note that these experiments (Figs. 1–4) were carried out in the absence of the glial feeder layer. We observed two effects of chemokines on neuronal Ca signaling, depending on whether neurons were synaptically coupled. Some cultured hippocampal pyramidal neurons were characterized by stable baseline [Ca]i levels that did not exhibit spontaneous fluctuations. In these neurons, chemokines produced increases in [Ca]i observed in the neuronal soma, while superfusion of cells with normal buffer produced no change in [Ca]i. Depending on the cell under investigation, chemokines (0.1 − 100 nM) produced either [Ca]i oscillations or single [Ca]i “spikes” (Fig. 1). Effects in this concentration range were observed with a variety of chemokines thought to activate a wide spectrum of CC, CXC, and CX3C chemokine receptors (9, 27, 31). These included human (h) or murine (m) MDC (which activates CCR4), RANTES (CCR5/CCR1/CCR3-preferring), MIP-1α (CCR5/CCR1-preferring), SDF-1α (which activates CXCR4), soluble (s) fractalkine (CX3CR1-preferring), IL-8 (CXCR1/CXCR2-preferring), TARC (CCR4-preferring) and I-309 (CCR8-preferring). On the other hand, MCP-1 (CCR2-preferring) proved to be ineffective. As previously reported (32), gp120 also increased [Ca]i in neurons (Fig. 1). It is likely that the effects of chemokines were produced directly on neurons rather than through the secondary release of substances from any contaminating microglia in the neuronal cultures. Thus, we were unable to detect microglia in the neuronal layer using staining with anti-CD11b/c (33) and estimate that, if present, contamination was ≪1%. Furthermore, similar effects of chemokines were obtained on neuronal cultures that were selectively depleted of any microglia by immunopanning by using anti-CD11b/c.
Figure 1.
[Ca2+]i transients evoked by chemokines and gp120s in hippocampal neurons. Chemokines that activate different CCRs (MDC, TARC, RANTES, MIP-1α, and I-309), CXCRs (IL-8 and SDF-1α), as well as CX3CR1 (fractalkine) receptors were used for the times indicated by the bars (2–4 min). Representative traces from single neurons are shown in each panel. A total of 85 cells of 167 showed [Ca2+]i increases when stimulated by hMDC (32 of 62 by mMDC). Nine of 34 neurons were responsive to TARC, 14 of 21 to RANTES, 12 of 19 to MIP-1α, 12 of 42 to I-309, 23 of 58 to fractalkine, 18 of 43 to IL-8, 21 of 44 to SDF-1α, 26 of 55 to HIV-1IIIB gp120, and 9 of 23 to SIVmac251 gp120.
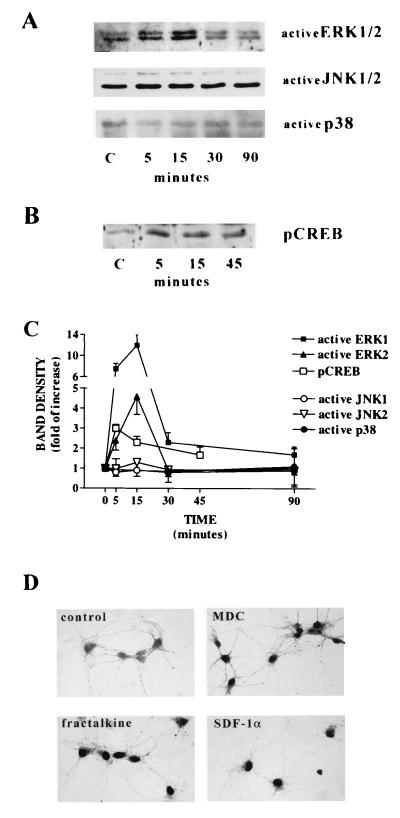
Figure 4.
Effect of chemokines on ERK1/2, JNK1/2, p38, and CREB in hippocampal neuronal cultures. Western blots of total cell lysates (A) and of nuclear extracts (B) were obtained from pure neuronal cultures treated with 100 nM fractalkine after a 4-hr preincubation in balanced salt solution in the absence of glia. Antibodies selectively recognizing the activated form of ERK1/2, JNK1/2, p38, and CREB were used. Similar results were obtained with hMDC (10 nM; not shown). (C) Densitometric analysis of the effect of fractalkine (100 nM) on ERK1/2, JNK1/2, p38, and CREB. Each data point represents the average of two to three similar experiments. (D) Nuclear localization of activated CREB (pCREB) in neurons treated with 10 nM hMDC, 100 nM fractalkine, and 100 nM SDF-1α (10 min in the absence of glial feeder layer).
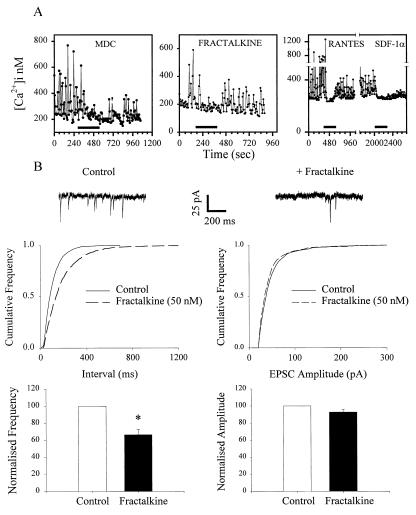
A second type of neuron observed in the cultures exhibited large spontaneous oscillations in [Ca]i. We have previously shown that these neurons are synaptically coupled to each other and that the [Ca]i oscillations, which are blocked by glutamate receptor antagonists or tetrodotoxin, are due to ongoing excitatory transmission between neurons (26). When chemokines (h and mMDC, RANTES, MIP-1α, SDF-1α, and soluble fractalkine) were added to these cells, the amplitude of the spontaneous [Ca]i oscillations decreased (Fig. 2A) any chemokine-induced [Ca]i increase being obscured by the rapidly oscillating nature of [Ca]i under basal conditions in these cells. In several experiments, we tested a second chemokine on a neuron after washout of the first agonist and reversal of its response. In such cases, we often observed a response to the second chemokine as well, indicating that individual neurons may possess more than one chemokine receptor type (Fig. 2A). These results suggest that, like several other types of G-protein-coupled receptors, activation of chemokine receptors can inhibit excitatory transmission between hippocampal neurons (34). We tested this possibility further electrophysiologically. The two chemokines we examined, mMDC and soluble fractalkine, were able to block the frequency, but not the amplitude, of spontaneous glutamatergic excitatory postsynaptic currents recorded from these neurons (Fig. 2B). Such observations are consistent with the effects of chemokines on spontaneous glutamate receptor-mediated [Ca]i transients described above and show that presynaptic chemokine receptors can regulate the release of glutamate at these synapses (29). Although the precise mechanism by which chemokines inhibit glutamate release is unclear, we did observe that fractalkine (100 nM) inhibited the voltage-dependent Ca current recorded from these hippocampal neurons (inhibition vs. control = 20 ± 5.9%, n = 5). On the other hand, mMDC (100nM, n = 7) did not block Ca currents, suggesting that, as in the case of other G-protein-coupled receptors, chemokines may produce presynaptic inhibition by diverse mechanisms (34).
Figure 2.
Inhibition of spontaneous synaptic activity by chemokines. (A) [Ca2+]i oscillations were blocked by hMDC and mMDC, fractalkine, RANTES, and SDF-1α in different neurons. In total, 18 of 37 oscillating neurons were responsive to mMDC (10 nM), 19 of 20 to fractalkine (100 nM), 7 of 7 to RANTES (50 nM), and 17 of 17 to SDF-1α (50 nM). (B) Fractalkine (50 nM) reduced the frequency, but not the amplitude, of excitatory postsynaptic currents recorded from hippocampal neurons (7 of 9 neurons). Similar results were obtained with mMDC (data not shown; 3 of 7 neurons). ∗ = P < 0.001 paired Student’s t test.
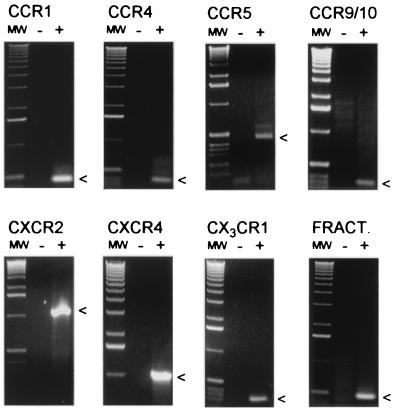
The conclusion that hippocampal neurons contain several types of chemokine receptors was further investigated by using a molecular biological approach. A series of reverse-transcription–PCR reactions were carried out by using primers specific for rat chemokine receptor sequences when available. When published rat sequences were not available, amplification with multiple primer pairs was attempted, including degenerate primers made according to published human and rat sequences and primers made according to mouse sequences. cDNA prepared from hippocampal neuron cell cultures was used as a template for all reactions, and the PCR products were subsequently cloned and sequenced. In this manner, we determined that CCR1, CCR4, CCR5, CCR9/10, CXCR2, CXCR4, CX3CR1 (V28), as well as the chemokine fractalkine (35), are expressed in rat hippocampal neurons (Fig. 3). We were unable to demonstrate expression of CCR2, CCR3, CCR6, CCR7, CCR8, CXCR1, or the β2-integrin CD11b under the same conditions. The lack of detectable CCR3 or CD11b expression is consistent with a lack of microglia (19, 33). In summary, therefore, the data clearly show that hippocampal neurons possess a wide variety of chemokine receptors, and activation of these receptors can lead directly to rapid changes in neuronal Ca signaling and synaptic transmission.
Figure 3.
Reverse-transcription–PCR reaction products from RNA extracted from pure hippocampal neuronal cultures were run on 1.2% agarose gels. Arrows indicate expected PCR product sizes. Lanes labeled (-) represent water controls.
We also found that chemokines activated other signaling pathways in hippocampal neurons. Addition of MDC or soluble fractalkine produced clear time-dependent activation of the MAP kinase-signaling pathway. For example, as can be seen in Fig. 4, fractalkine and hMDC produced a time-dependent activation of ERKs-1 and 2, whereas no activation of JNK/stress-activated protein kinase or p38 was evident. Furthermore, we also showed that several chemokines we tested activated the Ca and cyclic AMP-dependent transcription factor CREB (Fig. 4). gp120 may interact with chemokine receptors in a fundamentally different fashion from the chemokines themselves, producing different patterns of signaling (7, 36–38). For example, it has been reported recently that gp120 activates both ERK and JNK kinases in cultured neurons (7).
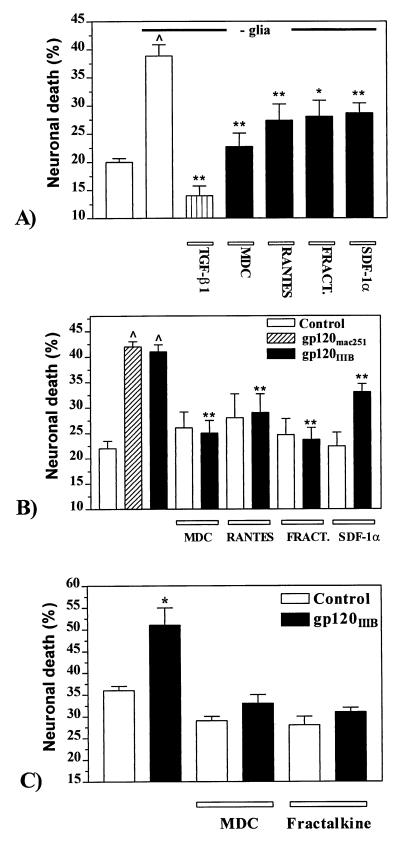
Consistent with these patterns of signaling produced by chemokines, we found that several of them could act as survival-promoting factors for hippocampal neurons (39, 40). It has been observed previously that, on removal of their glial cell feeder layer, hippocampal neurons die by apoptotic mechanisms over the next few days (41). Addition of hMDC, RANTES, SDF-1α, or soluble fractalkine to the culture medium slowed the rate of death considerably. Fig. 5A illustrates this effect and, as a comparison, also shows the effect of transforming growth factor (TGF)-β1, a cytokine that we have previously demonstrated promotes survival under these same circumstances (41). We and others have demonstrated previously that hippocampal neurons also die by apoptosis following the addition of the HIV-1 envelope protein gp120 to the culture medium, and that these effects could be inhibited by TGF-β1 (4–6, 8). As shown in Fig. 5B, neuronal death was produced by both HIV-1IIIB gp120 (T-tropic) and SIVmac251 gp120 (M-tropic). The same chemokines that were shown to be effective in promoting neuronal survival following glial deprivation were also able to block gp120IIIB-induced apoptosis of hippocampal neurons (Fig. 5B). On the other hand, MCP-1 was ineffective, consistent with the observed lack of [Ca]i response for this chemokine. Under the conditions used, SDF-1α was somewhat less effective than hMDC, soluble fractalkine, or RANTES. However, we also observed that if we increased the concentration of SDF-1α to 100 nM and the preincubation time to 4 hr before addition of gp120IIIB, the degree of protection produced was higher and thus equaled that observed with the other chemokines. Neurotoxic effects of gp120IIIB and inhibitory effects of chemokines were also observed when both the neuronal cultures and astrocyte feeder layers were depleted selectively of microglia by using immunopanning, indicating that the observed effects were not mediated by microglia. Indeed, we have shown that gp120 can produce apoptotic neuronal death in the complete absence of the glia feeder layer (4), and that death under these circumstances could also be inhibited by chemokine treatment (Fig. 5C). Thus, it appears that gp120 can be directly toxic to cultured hippocampal neurons, although it remains to be seen whether this is also true in vivo.
Figure 5.
Effect of chemokines on neuronal survival. (A) Apoptosis induced by removal of the glial cell feeder layer was reduced by hMDC or mMDC (10 nM), RANTES (10 nM), fractalkine (100 nM), SDF-1α (50 nM), and TGF-β1 (5 ng/ml). Data are expressed as mean ± SEM of the percent of dead cells from five different cultures. In each experiment, two to three coverslips per treatment were evaluated and neurons were counted from 10–12 fields of each coverslip. The neurons analyzed for each data point were: 3409 (control with glia), 2828 (control without glia), 591 (TGF-β1), 1948 (MDC), 2376 (RANTES), 2095 (fractalkine), and 1499 (SDF-1α). ^ = P < 0.0001 vs. control with glia; ∗∗ = P < 0.0001 and ∗ = P < 0.001 vs. control without glia. (B) Neurotoxicity induced by HIV-1IIIB gp120 and SIVmac251 gp120 in hippocampal cultures. gp120IIIB-induced neurotoxicity was inhibited by different types of chemokines. Chemokines (at the same concentrations reported above) and/or gp120 (200 pM) were added to the neuronal cultures at 7 days in culture and apoptotic cells counted after 3–4 days. The mean ± SEM of neuronal death from 12 separate experiments is reported and three to five coverslips (10–12 fields per coverslip) were counted from each experiment. The total number of cells counted for each treatment was: 6,145 (control), 6,812 (gp120IIIB), 1,778 (gp120mac251), 2,154 (hMDC), 2,615 (hMDC+gp120IIIB), 2,367 (RANTES), 2,998 (RANTES+gp120IIIB), 1,872 (fractalkine), 2,449 (fractalkine + gp120IIIB), 3,125 (SDF-1α), and 3,750 (SDF-1α+gp120IIIB). ^ = P < 0.0001 vs. control; ∗∗ = P < 0.0001 vs. gp120IIIB alone. (C) HIV-1IIIB gp120-induced neurotoxicity in neuronal cultures in the absence of glia. Neurons were treated for 24 hr with gp120 (200 pM) and/or hMDC (100 nM) or fractalkine (100 nM). Total number of cells counted from three different experiments: control = 2932, gp120 = 2740; MDC = 1,086, MDC + gp120 = 1,068, fractalkine = 1,300, fractalkine + gp120 = 1,631; ∗ = P < 0.01 vs. control.
It is interesting to note that both gp120s produced [Ca]i signals in hippocampal neurons (Fig. 1, traces i and j). It is likely that the gp120s produced their effects following binding to neuronal chemokine receptors. For example, if we down-regulated the response to SDF-1α (42) by pretreating neurons with this chemokine, the response to gp120IIIB was also absent, although RANTES was still effective (data not shown).
There are several conclusions suggested by the results of the present experiments. The first is that central neurons possess a variety of functional chemokine receptors that can regulate [Ca]i and other signaling pathways. Activation of these receptors can produce rapid regulation of excitatory synaptic transmission and longer-term effects on neuronal survival. The expression of some chemokines and their cognate receptors, such as fractalkine and CX3CR1, has been observed previously by Northern hybridization of brain tissue (35, 43). Expression of other chemokine and receptor pairs, such as MDC and CCR4, has not been noted in brain, perhaps because expression is limited to particular subregions such as the hippocampus. Additional chemokines and chemokine receptors may prove to be relevant in neuronal function, since numerous orphan sequences have been identified from brain cDNA libraries in the Expressed Sequence Tag database (unpublished observations). The only receptor currently identified for I-309 is CCR8, which was not amplified from our neuronal cultures. The response seen for I-309 may therefore be due to a novel neuronal receptor yet to be identified. The receptors present on neurons include CCR5 and CXCR4, which are thought to be the normal chemokine receptors for gp120 binding to macrophages and lymphocytes. Thus, it seems likely that gp120-induced neuronal death observed in culture and in association with the AIDS dementia syndrome may occur by a direct mechanism rather than, or in addition to, the indirect release of neurotoxins from nonneuronal cells (8). Binding of gp120 to neurons might initiate several events, including interference with the normal trophic effects of chemokines. These events may be directly neurotoxic or could enhance the sensitivity of neurons to other factors such as glutamate receptor activation (4, 8). The mechanism by which chemokines inhibit gp120 neurotoxicity may include competition at chemokine receptor binding sites, as is thought to occur with cells in the immune system (9). However, the fact that several types of chemokines show protective effects also indicates a fundamentally different mechanism of action as well, probably mediated at the intracellular signaling level downstream of gp120 binding (Fig. 4). Precedent for such a mechanism may be the simultaneous protective effects of hMDC on the infectivity of both M and T tropic HIV strains (44). Thus, agonists at several types of chemokine receptors may find therapeutic utility in combating the cognitive and motor changes associated with AIDS.
Acknowledgments
We thank Chris P. Mauer, Dongjun Ren, and Umar M. Shakur for helpful technical assistance. T.J.B. was supported by the Wellcome Prize Traveling Research Fellowship, and O.M., A.F., A.A.S., and R.J.M. were supported by the National Institutes of Health.
ABBREVIATIONS
- IL
interleukin
- TGF
transforming growth factor
- MDC
macrophage-derived chemokine
- ERK
extracellular response kinase
- TARC
thymus and activation-regulated chemokine
- JNK
c-JUN NH2-terminal protein kinase
- MIP
macrophage inflammatory protein
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Price R W, Brew B, Sidtis J, Rosenbaum M, Scheck A C, Cleary P. Science. 1988;239:586–592. doi: 10.1126/science.3277272. [DOI] [PubMed] [Google Scholar]
- 2.Masliah E, Ge N, Achim C L, Hansen L A, Wiley C A. J Neuropathol Exp Neurol. 1992;51:585–593. doi: 10.1097/00005072-199211000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Toggas S M, Masliah E, Rockenstein E M, Rall G F, Abraham C R, Mucke L. Nature (London) 1994;367:188–193. doi: 10.1038/367188a0. [DOI] [PubMed] [Google Scholar]
- 4.Meucci O, Miller R J. J Neurosci. 1996;16:4080–4088. doi: 10.1523/JNEUROSCI.16-13-04080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenneman D E, Westbrook G L, Fitzgerald S P, Ennist D L, Elkins K L, Ruff M R, Pert C B. Nature (London) 1988;335:639–642. doi: 10.1038/335639a0. [DOI] [PubMed] [Google Scholar]
- 6.Muller W E G, Schröder H, Ushijima H, Dapper J, Bormann J. Eur J Pharmacol (Mol Pharmacol Sect) 1992;226:209–214. doi: 10.1016/0922-4106(92)90063-2. [DOI] [PubMed] [Google Scholar]
- 7.Lannuzel A, Barrier J V, Henry C, Van Tan H, Gilbert B, Gray F, Vincent J D, Tardieu M. Ann Neurol. 1997;42:847–856. doi: 10.1002/ana.410420605. [DOI] [PubMed] [Google Scholar]
- 8.Lipton S A. Neurosci Res Commun. 1994;15:31–37. [Google Scholar]
- 9.Clapham P R. Trends Cell Biol. 1997;7:264–268. doi: 10.1016/S0962-8924(97)01075-1. [DOI] [PubMed] [Google Scholar]
- 10.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, et al. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 11.Reeves J D, McKnight A, Potempa S, Simmons G, Gray P W, Power C A, Wells T, Weiss R A, Talbot S J. Virology. 1997;231:130–134. doi: 10.1006/viro.1997.8508. [DOI] [PubMed] [Google Scholar]
- 12.Hesselgesser J, Halks-Miller M, DelVecchio V, Peiper S C, Hoxie J, Kolson D L, Taub D, Horuk R. Curr Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- 13.Baggiolini M. Nature (London) 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 14.Luster A D. N Engl J Med. 1998;7:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 15.Broder C C, Dimitrov D S. Pathobiology. 1996;64:171–179. doi: 10.1159/000164032. [DOI] [PubMed] [Google Scholar]
- 16.Broder C C, Collman R G. J Leukocyte Biol. 1997;62:20–29. doi: 10.1002/jlb.62.1.20. [DOI] [PubMed] [Google Scholar]
- 17.Bacon K B, Premack B A, Gardner P, Schall T J. Science. 1995;269:1727–1730. doi: 10.1126/science.7569902. [DOI] [PubMed] [Google Scholar]
- 18.Littman D R. Cell. 1998;93:677–680. doi: 10.1016/s0092-8674(00)81429-4. [DOI] [PubMed] [Google Scholar]
- 19.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, et al. Nature (London) 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 20.Lavi E, Kolson D L, Ulrich A M, Fu L, Gonzalez-Scarano F. J Neurovirol. 1998;4:301–311. doi: 10.3109/13550289809114531. [DOI] [PubMed] [Google Scholar]
- 21.Vallat A-V, De Girolami U, He J, Mhashilkar A, Marasco W, Shi B, Gray F, Bell J, Keohane C, Smith T W, et al. Am J Pathol. 1998;152:167–178. [PMC free article] [PubMed] [Google Scholar]
- 22.Heesen M, Berman M A, Benson J D, Gerard E J. Immunology. 1996;157:5455–5460. [PubMed] [Google Scholar]
- 23.Tanabe S, Heesen M, Berman M A, Fischer M B, Yoshizawa I, Luo Y, Dorf M E. J Neurosci. 1997;17:6522–6528. doi: 10.1523/JNEUROSCI.17-17-06522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonini J A, Steiner D F. DNA Cell Biol. 1997;16:1023–1030. doi: 10.1089/dna.1997.16.1023. [DOI] [PubMed] [Google Scholar]
- 25.Zou Y-R, Kottmann A H, Kuroda M, Taniuchi I, Littman D R. Nature (London) 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 26.Abele A E, Scholz K P, Scholz W K, Miller R J. Neuron. 1990;4:413–419. doi: 10.1016/0896-6273(90)90053-i. [DOI] [PubMed] [Google Scholar]
- 27.Imai T, Chantry D, Raport C J, Wood C L, Nishimura M, Godiska R, Yoshie O, Gray P W. J Biol Chem. 1988;273:1764–1768. doi: 10.1074/jbc.273.3.1764. [DOI] [PubMed] [Google Scholar]
- 28.Godinska R, Chantry D, Raport C J, Sozzani S, Allavena P, Leviten D, Mantovani A, Gray P W. J Exp Med. 1997;185:1595–1604. doi: 10.1084/jem.185.9.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholz K P, Miller R J. Neuron. 1992;8:1139–1150. doi: 10.1016/0896-6273(92)90134-y. [DOI] [PubMed] [Google Scholar]
- 30.Masamune A, Igarashi Y, Hakomori S-I. J Biol Chem. 1996;271:9368–9375. doi: 10.1074/jbc.271.16.9368. [DOI] [PubMed] [Google Scholar]
- 31.Imai T, Hieshima K, Haskell C, Baba M, Nagira M, Nishimura M, Kakizaki M, Takagi S, Nomiyama H, Schall T J, et al. Cell. 1997;91:521–530. doi: 10.1016/s0092-8674(00)80438-9. [DOI] [PubMed] [Google Scholar]
- 32.Dreyer E B, Kaiser P K, Offermann J T, Lipton S A. Science. 1990;248:364–367. doi: 10.1126/science.2326646. [DOI] [PubMed] [Google Scholar]
- 33.Flaris N A, Densmore T L, Molleston M C, Hickey W F. Glia. 1993;7:34–40. doi: 10.1002/glia.440070108. [DOI] [PubMed] [Google Scholar]
- 34.Miller R J. Annu Rev Pharmacol Toxicol. 1998;38:201–227. doi: 10.1146/annurev.pharmtox.38.1.201. [DOI] [PubMed] [Google Scholar]
- 35.Bazan J F, Bacon K B, Hardiman G, Wang W, Soo K, Rossi D, Greaves D R, Zlotnik A, Schall T J. Nature (London) 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 36.Weissman D, Rabin R L, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber J M, Fauci A S. Nature (London) 1997;389:981–985. doi: 10.1038/40173. [DOI] [PubMed] [Google Scholar]
- 37.Frazen M F, Choe H, Martin K A, Sun Y, Sidelko M, Mackay C R, Gerard N P, Sodroski J, Gerard C. J Biol Chem. 1997;272:6854–6857. doi: 10.1074/jbc.272.11.6854. [DOI] [PubMed] [Google Scholar]
- 38.Davis C B, Dikic I, Unutmaz D, Hill C M, Arthos J, Siani M A, Thompson D A, Schlessinger J, Littman D R. J Exp Med. 1997;186:1793–1798. doi: 10.1084/jem.186.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutkind J S. J Biol Chem. 1998;273:1839–1842. doi: 10.1074/jbc.273.4.1839. [DOI] [PubMed] [Google Scholar]
- 40.Finkbeiner S, Travazoie S F, Maloratsky A, Jacobs K M, Harris K M, Greenberg M E. Neuron. 1997;19:1031–1047. doi: 10.1016/s0896-6273(00)80395-5. [DOI] [PubMed] [Google Scholar]
- 41.Prehn J H M, Bindokas V P, Marcuccilli C J, Krajewski S, Reed J C, Miller R J. Proc Natl Acad Sci USA. 1994;91:12599–12603. doi: 10.1073/pnas.91.26.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amara A, Le Gall S, Schwartz O, Salamero J, Monte S M, Loetscher P, Baggiolini M, Virelizier J L, Arenzana-Seisdedos F. J Exp Med. 1997;186:139–146. doi: 10.1084/jem.186.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raport C J, Schweickart U L, Eddy R J, Shows T B, Gray P W. Gene. 1995;163:295–299. doi: 10.1016/0378-1119(95)00336-5. [DOI] [PubMed] [Google Scholar]
- 44.Pal R, Garzino-Demo A, Markham P D, Burns J, Brown M, Gallo R C, De Vico A L. Science. 1997;278:695–698. doi: 10.1126/science.278.5338.695. [DOI] [PubMed] [Google Scholar]