Abstract
Urinary biomarkers for diabetes, diabetic nephropathy, and nondiabetic proteinuric renal diseases were sought. For 305 individuals, biomarkers were defined and validated in blinded data sets using high-resolution capillary electrophoresis coupled with electrospray-ionization mass spectrometry. A panel of 40 biomarkers distinguished patients with diabetes from healthy individuals with 89% sensitivity and 91% specificity. Among patients with diabetes, 102 urinary biomarkers differed significantly between patients with normoalbuminuria and nephropathy, and a model that included 65 of these correctly identified diabetic nephropathy with 97% sensitivity and specificity. Furthermore, this panel of biomarkers identified patients who had microalbuminuria and diabetes and progressed toward overt diabetic nephropathy over 3 yr. Differentiation between diabetic nephropathy and other chronic renal diseases reached 81% sensitivity and 91% specificity. Many of the biomarkers were fragments of collagen type I, and quantities were reduced in patients with diabetes or diabetic nephropathy. In conclusion, this study shows that analysis of the urinary proteome may allow early detection of diabetic nephropathy and may provide prognostic information.
Nephropathy is a serious and common complication of diabetes and has become the most prevalent cause of ESRD. Over the years, there has been an ongoing quest to find biomarkers early in the clinical course to identify and treat better individuals at high risk for diabetic nephropathy. Knowledge of the complex molecular and pathophysiologic mechanisms leading to renal disease remains limited, in part because conventional research tools have hampered investigators by restricting their focus to a single or relatively few risk markers at a time. Recent advances in proteomics enable screening of a vast array of proteins simultaneously, aiding assessment of their potential role in the development and progression of disease.1 Urine is well suited for proteomic analysis to identify predictive biomarkers and to unravel the pathogenetic mechanisms of chronic renal disease. The online combination of capillary electrophoresis (CE) and electrospray mass spectrometry (MS) was developed for the rapid (approximately 45 min per sample), sensitive, and automated approach for such an analysis.2
The aim of our study was to examine whether CE-MS can detect differences in the urinary proteome between patients with normo-, micro-, and macroalbuminuric and type 1 diabetes. Furthermore, we sought to evaluate whether CE-MS–defined patterns derived from urinary polypeptides of patients with diabetic nephropathy differ from those of patients with other chronic renal diseases.
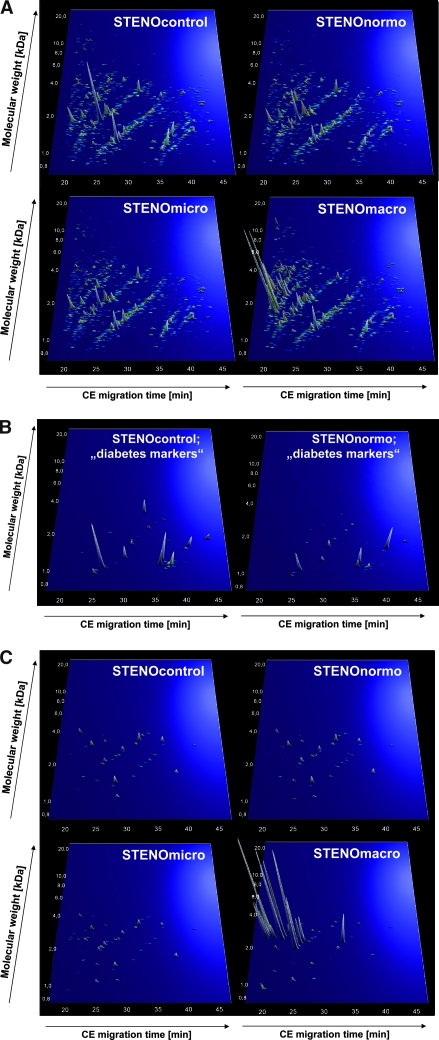
Healthy individuals without diabetes and patients with diabetes were matched with respect to age and gender to establish urinary polypeptide patterns characteristic for diabetes and diabetic nephropathy (Table 1). The three groups of patients with diabetes (persistent normoalbuminuria, microalbuminuria, and diabetic nephropathy) had comparable duration of diabetes, severity of diabetic retinopathy, BP, and serum cholesterol levels. Hemoglobin A1c was lower in normoalbuminuric patients, and serum creatinine was significantly higher in patients with diabetic nephropathy. Urine samples from all participants were analyzed using CE-MS, and the data were normalized using internal standards3 and deposited in adatabase. Only polypeptides present at a frequency >50% in any of the groups were evaluated. The compiled data for the four groups are shown in Figure 1A. Whereas the urinary peptide profiles in samples from the healthy individuals (STENOcontrol), as well as patients with diabetic normalbuminuric (STENOnormo) and microalbuminuric (STENOmicro), seemed similar, the urinary polypeptide patterns of patients with diabetic nephropathy (STENOmacro) were markedly differed.
Table 1.
Characteristics of all participants recruited at the STENO Diabetes Centera
| Characteristic | Healthy Controls(n = 30) | Normoalbuminuria(n = 30) | Microalbuminuria(n = 29) | Diabetic Nephropathy(n = 30) | P (Normo- versus Macroalbuminuria) | P (All Groups) |
|---|---|---|---|---|---|---|
| Age | 51 (1.9) | 54 (2.0) | 56 (1.8) | 50 (1.6) | 0.120 | 0.070 |
| Gender (female/male; mean [SE]) | 7/23 | 7/23 | 7/22 | 7/23 | 0.990 | 0.990 |
| Diabetes duration (yr; mean [SE]) | – | 34 (1.9) | 35 (2.0) | 34 (2.0) | – | – |
| Diabetic retinopathy (nil/simple/proliferative) | – | 2/12/16 | 1/9/19 | 2/7/21 | 0.230 | 0.990 |
| Urinary albumin/creatinine (geometric mean [95% CI] | – | 5 (4 to 6) | 45 (32 to 63) | 765 (564 to 1036) | – | – |
| Systolic BP (mmHg; mean [SE]) | 133 (3) | 136 (3) | 148 (4) | 142 (4) | 0.280 | 0.020 |
| Diastolic BP (mmHg; mean [SE]) | 80 (2) | 77 (2) | 75 (2) | 77 (2) | 0.980 | 0.240 |
| Cholesterol (mmol/L; mean [SE]) | 5.6 (0.1) | 4.6 (0.2) | 4.9 (0.2) | 4.6 (0.2) | 0.370 | <0.001 |
| Creatinine (μmol/L; mean [SE]) | 97 (2) | 94 (2) | 94 (3) | 171 (15) | <0.001 | <0.001 |
| Hemoglobin A1c (%; mean [SE]) | 5.4 (0.1) | 8.0 (0.2) | 8.9 (0.3) | 8.8 (0.2) | 0.0150 | <0.001 |
Figure 1.
(A) Protein patterns of the patients with diabetes and control subjects examined in this study. Shown are compiled patterns consisting of all samples from each of the four groups. The molecular mass (0.7 to 25 kD, on a logarithmic scale) is plotted against normalized migration time (17 to 47 min). Signal intensity is encoded by peak height and color. (B) Distribution of potential differential-diagnostic biomarkers for diabetes in the patients with normoalbuminuria and healthy control subjects. All statistically significant biomarkers from Supplemental Table 1 are shown. (C) Distribution of potential differential-diagnostic biomarkers for diabetic nephropathy in the different groups of patients and healthy control subjects. Shown are all statistically significant biomarkers listed in Supplemental Table 2.
For identification of potential biomarkers for diabetes, urinary polypeptides of healthy individuals were compared with those of patients with diabetes and persistent normoalbuminuria. This analysis identified 40 peptides of statistical significance (P < 0.05 in maxT testing; Supplemental Table 1). A support vector machine-based model (SVM-BM) with these 40 biomarkers discriminated healthy individuals from patients with diabetes and normoalbuminuria with 97% sensitivity and specificity at cross-validation. The distribution of the polypeptides in the two groups is shown in Figure 1B. The validity of the diabetes biomarkers was further evaluated in a test-set cohort of STENOmacro, STENOmicro, and patients with diabetes and normoalbuminuria (HANNOVERcontrol): 53 of 59 patients with diabetes and 32 of 35 healthy control subjects were correctly classified, resulting in 89% sensitivity and 91% specificity.
For definition of biomarkers for diabetic renal disease, urine samples from patients with diabetes and persistent normoalbuminuria (STENOnormo) were compared with those from patients with diabetic nephropathy (STENOmacro). This analysis identified 102 biomarkers of statistical significance (P < 0.05 in maxT testing adjusted for multiple tests, 24 of these biomarkers have been sequenced; Supplemental Table 2). For reduction of the number of variables, a “take-one-out” procedure was used, decreasing the number of biomarkers to 65 (Figure 1C) without losing performance in the classification. An SVM-BM with these 65 polypeptides performed with 93% sensitivity and 97% specificity at cross-validation.
Because SVM-BM do not assign probability, we investigated whether a linear combination of biomarkers may improve disease severity classification. Transformed and calibrated logarithmic amplitudes of all biomarkers were combined for each patient.4 This approach resulted in 100% sensitivity and 93% specificity.
The diabetic renal disease model was validated in an external data set of 35 patients with diabetes and macroalbuminuria (HANNOVERmacro) and 35 healthy individuals (HANNOVERcontrol).5 Using SVM-BM, 34 of 35 controls and all patients were classified correctly. When the linear model was applied, the same control subject and one of the patients were misclassified, resulting in 97% sensitivity and 97% specificity.
When the diabetic renal disease models were applied to the matched 30 patients with diabetes and microalbuminuric in the study (STENOmicro), 21 scored positive for diabetic nephropathy when the linear model was used (six were only marginally positive). On the basis of the same biomarkers in the SVM-BM, 17 of 30 patients scored positive. Eight patients with microalbuminuric later showed an increase in albuminuria of >25% or progressed to macroalbuminuria during a 3-yr follow-up interval. Each scored positive in the linear model, and seven scored positive in the SVM-BM. These data indicate the utility of the biomarkers for not only detection of overt nephropathy but also prediction of its development in patients with diabetes and microalbuminuria with statistical significance (P = 0.0359).
Several peptides were sequenced using MS/MS.6 The sequenced biomarkers that differentiated patients with diabetes from healthy control subjects included fragments of collagen type I (α1 chain) and uromodulin (Supplemental Table 1). For differentiation of patients with and without diabetic nephropathy, the sequences were more diverse, consisting of various collagen types as well as uromodulin fragments (Supplemental Table 2).
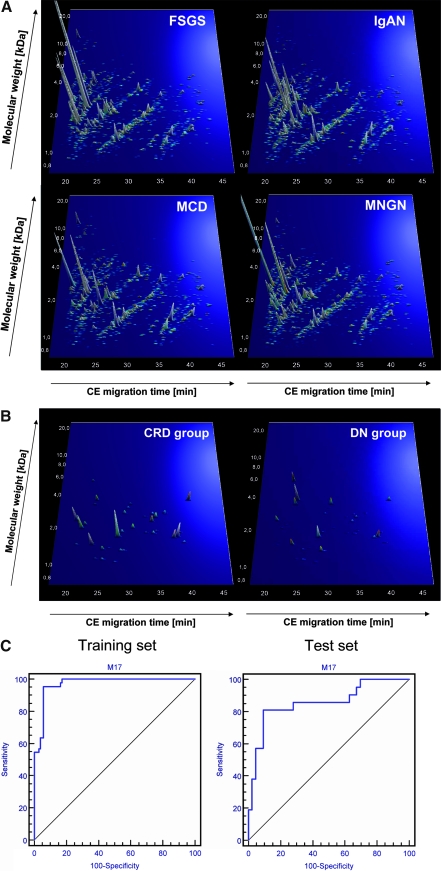
We tested the specificity of the diabetic renal disease pattern using urine samples from patients with other chronic renal diseases: Biopsy-proven IgA nephropathy (n = 57), FSGS (n = 30), membranous glomerulonephritis (n = 35), and minimal-change disease (n = 25). These patients and data sets have been described previously.5,7,8 The compiled polypeptides of the different chronic renal diseases are shown in Figure 2A. As expected, most (n = 104) of these 147 samples scored positive for “diabetic renal disease,” and only 43 scored as normoalbuminuria, indicating that the “diabetic renal disease” pattern reflects chronic renal damage.
Figure 2.
(A) Protein patterns of the patients with chronic renal disease (CRD). Shown are compiled patterns consisting of samples from patients with FSGS (n = 35) IgA nephropathy (IgAN; n = 57), minimal-change disease (MCD; n = 25), and membranous glomerulonephritis (MNGN; n = 29). In comparison with Figure 1A, these compiled data show a much higher degree of similarity to the patients with macroalbuminuria than to any other group with diabetes. The molecular mass (0.7 to 25 kD, on a logarithmic scale) is plotted against normalized migration time (17 to 47 min). Signal intensity is encoded by peak height and color. (B) Distribution of potential differential-diagnostic biomarkers for diabetic nephropathy(DN) in all patients with macroalbuminuria (DN group) and all control subjects with (CRD group) used in the study. All 37 statistically significant biomarkers from Table 2 are shown. (C) Receiver operating characteristic analysis of the performance of the differential diagnostic biomarker pattern for DN. (Left) Data from the training set of 44 case patients and 104 control subjects (sensitivity of 95.5% and specificity of 94.2%; area under the curve was 0.971). (Right) Data from validating the masked test set consisting of 64 samples (81.0% sensitivity and 90.7% specificity; area under the curve was 0.856).
To investigate whether we could define biomarkers to distinguish diabetic nephropathy from the other chronic renal diseases, we randomly selected and compared 104 samples from patients with nondiabetic nephropathy with those from 70% of the patients with diabetic nephropathy (STENOmacro and HANNOVERmacro; n = 44). This process identified 37 biomarkers of statistical significance (P < 0.05 in maxT testing; Table 2, Figure 2B).
Table 2.
The 37 urinary biomarkers that statistically differentiated 104 patients randomly selected from the group of 147 patients with nondiabetic chronic renal diseases from a cohort composed of 70% of patients with clinically confirmed diabetic nephropathya
| Polypeptide
|
P
|
Diabetic Nephropathy
|
Nondiabetic Chronic Renal Disease
|
Sequence | Protein | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Mass (Da) | CE Time (min) | Unadjusted | maxT | Bonferroni | Frequency | Mean Amplitude | Frequency | Mean Amplitude | ||
| 175343b | 5574.25 | 23.20 | 8.77E-06 | 0.0213 | 5.01E-03 | 0.55 | 1.55 | 0.43 | 1.06 | ||
| 140401b | 3669.67 | 24.17 | 1.90E-07 | 0.0017 | 1.10E-04 | 0.82 | 2.51 | 0.48 | 1.26 | ||
| 140112 | 3657.67 | 40.71 | 1.07E-05 | 0.0175 | 6.23E-03 | 0.32 | 0.82 | 0.72 | 1.95 | ||
| 130855b | 3363.54 | 30.22 | 8.03E-08 | 0.0006 | 4.66E-05 | 0.80 | 2.1 | 0.42 | 0.94 | ||
| 129940 | 3333.72 | 23.83 | 3.34E-06 | 0.0456 | 1.91E-03 | 0.55 | 1.53 | 0.29 | 0.67 | ||
| 121444 | 3081.42 | 29.83 | 1.69E-05 | 0.0301 | 9.82E-03 | 0.23 | 0.45 | 0.52 | 1.24 | ||
| 110175b | 2802.82 | 36.34 | 3.94E-14 | 0.0001 | 2.29E-11 | 0.07 | 0.14 | 0.56 | 1.31 | ||
| 107858b | 2751.34 | 29.23 | 1.14E-06 | 0.0043 | 6.62E-04 | 0.20 | 0.52 | 0.61 | 1.52 | ||
| 107763b | 2748.79 | 36.38 | 8.12E-15 | 0.0001 | 4.72E-12 | 0.09 | 0.14 | 0.60 | 1.18 | ||
| 107016 | 2733.78 | 34.16 | 8.38E-12 | 0.0001 | 4.87E-09 | 0.07 | 0.14 | 0.52 | 1.13 | ||
| 99251 | 2574.01 | 32.81 | 3.86E-07 | 0.0027 | 2.24E-04 | 0.16 | 0.29 | 0.53 | 1.04 | ||
| 97736 | 2551.15 | 34.72 | 8.65E-06 | 0.0315 | 4.94E-03 | 0.57 | 1.24 | 0.84 | 2.09 | ||
| 90344 | 2377.10 | 20.80 | 1.29E-05 | 0.0185 | 7.49E-03 | 0.45 | 1.17 | 0.77 | 2.29 | GKNGDDGEAGKhPGRPhGERGPPhGPQ | Collagen α-1 (I) chain (227 to 250; Homo sapiens) |
| 88282 | 2339.00 | 34.01 | 3.74E-05 | 0.0438 | 2.18E-02 | 0.52 | 1.21 | 0.82 | 2.21 | ||
| 80012b | 2191.99 | 22.39 | 6.40E-12 | 0.0001 | 3.72E-09 | 0.14 | 0.31 | 0.63 | 1.62 | NGDDGEAGkPGRpGERGPpGPQ | Collagen α-1 (I) chain |
| 72521b | 2046.81 | 30.72 | 1.89E-05 | 0.0351 | 1.10E-02 | 0.23 | 0.45 | 0.51 | 1.26 | ||
| 72161b | 2039.13 | 21.78 | 1.92E-13 | 0.0001 | 1.11E-10 | 0.14 | 0.31 | 0.61 | 1.83 | SGSVIDQSRVLNLGPITRK | Uromodulin (589 to 607; Homo sapiens) |
| 67382 | 1936.87 | 34.75 | 1.53E-05 | 0.0275 | 8.89E-03 | 0.20 | 0.4 | 0.53 | 1.13 | GEKGPSGEAGTAGPPGTPGPQG | Collagen α-2 (I) chain (844 to 865; Homo sapiens) |
| 64905 | 1893.03 | 28.86 | 3.02E-05 | 0.0429 | 1.75E-02 | 0.27 | 0.65 | 0.63 | 1.57 | ||
| 64431b | 1885.65 | 38.82 | 1.73E-05 | 0.0276 | 1.01E-02 | 0.23 | 0.47 | 0.61 | 1.29 | ||
| 63427b | 1865.81 | 32.98 | 2.73E-09 | 0.0003 | 1.59E-06 | 0.18 | 0.34 | 0.62 | 1.31 | DAGPAGPKGEPhGSPhGENGAPhG | Collagen α-1 (I) chain (279 to 299; Homo sapiens) |
| 62387 | 1844.48 | 34.27 | 3.82E-07 | 0.0025 | 2.22E-04 | 0.18 | 0.43 | 0.58 | 1.39 | ||
| 57265b | 1732.77 | 28.18 | 1.08E-06 | 0.0023 | 6.30E-04 | 0.55 | 1.69 | 0.93 | 3.04 | ||
| 56884b | 1725.59 | 38.32 | 5.12E-05 | 0.0494 | 2.97E-02 | 0.52 | 1.52 | 0.88 | 2.62 | ||
| 56514 | 1716.77 | 28.00 | 3.09E-06 | 0.0060 | 1.79E-03 | 0.45 | 1.04 | 0.79 | 2.07 | ||
| 55637b | 1698.57 | 37.73 | 3.60E-06 | 0.0082 | 2.09E-03 | 0.27 | 0.68 | 0.63 | 1.79 | ||
| 45863 | 1549.70 | 39.49 | 2.28E-05 | 0.0366 | 1.32E-02 | 0.18 | 0.43 | 0.53 | 1.21 | ||
| 44802 | 1526.69 | 23.92 | 2.62E-06 | 0.0065 | 1.52E-03 | 0.23 | 0.48 | 0.62 | 1.31 | ||
| 42378 | 1486.68 | 21.15 | 1.39E-09 | 0.0002 | 8.09E-07 | 0.11 | 0.28 | 0.59 | 1.35 | ||
| 42304b | 1485.67 | 23.77 | 7.18E-06 | 0.0072 | 4.17E-03 | 0.64 | 1.76 | 0.94 | 2.81 | DGQPhGAKGEPhGDAGAK | Collagen α-1 (I) chain (820 to 835; Homo sapiens) |
| 36350 | 1396.62 | 26.67 | 1.04E-05 | 0.0194 | 6.01E-03 | 0.18 | 0.38 | 0.53 | 1.07 | ||
| 33973 | 1353.66 | 25.63 | 2.10E-05 | 0.0241 | 1.22E-02 | 0.61 | 1.31 | 0.87 | 2.16 | ||
| 32470 | 1326.55 | 29.20 | 1.18E-06 | 0.0044 | 6.86E-04 | 0.23 | 0.48 | 0.62 | 1.36 | SpGGpGSDGKpGPpG | Collagen type 3 α1 |
| 32343 | 1324.59 | 28.70 | 7.45E-07 | 0.0032 | 4.33E-04 | 0.77 | 2.04 | 0.40 | 0.95 | TGPGGDKGDTGPpGP | Collagen α-1 (III) chain |
| 20750 | 1141.52 | 24.51 | 1.89E-07 | 0.0017 | 1.10E-04 | 0.20 | 0.39 | 0.59 | 1.29 | ||
| 19791b | 1128.49 | 25.65 | 1.48E-08 | 0.0003 | 8.59E-06 | 0.34 | 0.76 | 0.81 | 1.93 | ||
| 16497b | 1078.47 | 27.76 | 1.01E-05 | 0.0195 | 5.89E-03 | 0.25 | 0.46 | 0.55 | 1.20 | ||
Shown are the protein/peptide identification number in the data set (ID), mass (in Da) and normalized migration time (CE Time), the P values (unadjusted and adjusted using maxT and Bonferroni), and the frequency and mean amplitude in the two groups, diabetic nephropathy (DN) and other chronic renal disease (CRD). Where available, sequence and original protein are shown.
Subsequently used in the classification model.
Using a take-one-out procedure, the number of biomarkers decreased from 37 to 17. A SVM-BM based on these 17 biomarkers distinguished diabetic renal disease from the four other chronic kidney diseases. This model correctly classified 42 of 44 patients with diabetic nephropathy and 98 of 104 patients with other chronic renal diseases. Cross-validation of this training set showed 91% sensitivity and 89% specificity. To validate these biomarkers further, we analyzed the 30% of initially recruited patients and control subjects who had been omitted. Classification of this masked data set showed 81% sensitivity and 91% specificity (Figure 2C).
In this study we demonstrate urinary proteomic biomarkers distinct for diabetes, diabetic nephropathy, and nondiabetic proteinuric renal diseases. We sequenced only some biomarkers, because the procedure is challenging.1,9 The most striking observation was the decreased excretion of specific collagen fragments in patients with diabetes compared with healthy control subjects; several additional collagen fragments were less common in patients with diabetic nephropathy compared with normoalbuminuric diabetic patients. This observation supports previous reports10 and extends earlier findings by using validation in external data sets; CE-MS with ultrafiltration to remove larger proteins; and a new generation of mass spectrometers with better sensitivity, mass accuracy, and resolution.
Collagen increases in renal tissue of patients with diabetic nephropathy,11,12 and the decreased proteolysis as a result of increased synthesis of protease inhibitors diminishes excretion of collagen fragments.13,14 Furthermore, advanced glycation end products cross-link collagen and thus increase its resistance against proteases.15,16 A similar principle was shown for tubulointerstitial elastin deposition in experimental diabetic nephropathy: Decreased activity of elastase IIIB and increased production of elastase inhibitor diminished elastin turnover and increased its quantity.17
Urine contains a plethora of collagen fragments. In a study of >3000 individuals, 231 of 353 sequences were from collagen.18 Several fragments were found in >90% of all samples, with similar amplitudes. Other collagen fragments showed significant differences between control subjects and patients. These fragments are likely products of specific proteases and may serve as indicators of the activity of these enzymes.
Patients with diabetic nephropathy showed decreased levels of collagen type I and uromodulin fragments, whereas albumin fragments were highly upregulated. Assessment of these peptides by immunologic technologies, such as Western blot, is difficult because of their small size. Alternative approaches, such as ELISA, seem inappropriate, because several peptides sharing an antigenic epitope cannot be easily distinguished immunologically.19
In conclusion, we demonstrate that the urinary proteome differentiates healthy individuals from diabetic patients with persistent normoalbuminuria, low-grade albuminuria, or nephropathy. Furthermore, it distinguishes patients with diabetic nephropathy from patients with other chronic renal diseases. This finding could have great clinical value among patients with type 2 diabetes and nephropathy, because they are known to have increased albuminuria on a very heterogenic background when proteinuria occurs in the absence of diabetic retinopathy.20 The data from the blinded prospective evaluation of the patients with microalbuminuria indicated that urinary proteome analysis identified patients who had microalbuminuria and were at greater risk for diabetic nephropathy. On the basis of sequencing of some biomarkers, we speculate that changes in the collagen metabolism are closely linked with renal damage in patients with diabetes.
CONCISE METHODS
Patients, Procedures, and Demographics
This study included an internal data set used to identify biomarkers of diabetes and diabetic nephropathy by proteomic analysis. The internal data set consisted of patients examined at the Steno Diabetes Center in 2004 and was composed of 30 white, healthy individuals (STENOcontrol) and three groups of white patients who had type 1 diabetes and attended the Steno Diabetes Center: (1) 30 patients with persistent normoalbuminuria (STENOnormo; <30 mg/24 h and long-standing diabetes [>15 yr]), (2) 29 patients with persistent microalbuminuria (STENOmicro; ≥30 but <300 mg/24 h in at least two of three consecutive samples), and (3) 30 patients with diabetic nephropathy (persistent macroalbuminuria [STENOmacro; ≥300 mg/24 h] and coexistence of diabetic retinopathy or a renal biopsy showing diabetic glomerulosclerosis). The groups were matched for age, gender, and duration of diabetes. The external data sets consisted of 35 healthy individuals without diabetes (HANNOVERcontrol) and 35 patients with diabetic nephropathy, most with type 2 diabetes (HANNOVERmacro). Furthermore, biomarkers of diabetic nephropathy were evaluated in patients with biopsy-verified IgA nephropathy (n = 57), FSGS (n = 30), membranous glomerulonephritis (n = 35), and minimal-change disease (n = 25). Patients in the external data sets had been evaluated in other hospitals.5,8,21,22 The local ethics committees approved the study, and all participants gave informed consent. The study was performed in accordance with the Declaration of Helsinki.
Sample Preparation and CE-MS Analysis
All analyses and data processing were performed in accordance with the recently published minimum information about a proteomics experiment guidelines23 and in agreement with the suggested guidelines for clinical proteome analysis.24 All urine samples for CE-MS analyses were from spontaneously voided urine and stored at −80°C until analysis. Sample preparation and CE-MS analysis were performed.3 Briefly, a 0.7-ml sample was prepared and resuspended in 10 to 100 μl of H2O (depending on the peptide concentration) to a concentration of 1 to 5 μg/μl. Between 100 and 500 nl of sample was injected hydrodynamically, aiming for a total amount of 0.5 μg peptide per analysis, corresponding to 0.7 to 14.0 μl of the original sample. The data on all CE-MS analyses of all patients in the study are available in Supplemental Table 3 (downloadable at http://www.mosaiques-diagnostics.de/GenexPivot_12092007.exe password: 99Mischak123). A total of 5616 different polypeptides were tentatively identified (annotated), and, on average, 1632 polypeptides were found in each urine sample; on average, 1185 of these were annotated.
Data Processing and Analysis
Mass spectral ion peaks representing identical molecules at different charge states were deconvoluted into a single mass using MosaiquesVisu software.3 In addition, the migration time and ion signal intensity (amplitude) were normalized using internal polypeptide standards as described in detail previously.25 This approach results in superior comparability of the data sets as compared with normalization of the signal intensity to urinary creatinine or total protein content, because especially the latter will give poor results in patients with proteinuria. Each polypeptide is characterized by its molecular mass (kD), normalized migration time (min), and normalized signal intensity as the measure for relative abundance. All detected polypeptides were deposited, matched, and annotated in a Microsoft SQL database, allowing further analysis and comparison of multiple samples (patient groups). Polypeptides within different samples were considered identical when the mass deviation was <50 ppm and the migration time deviation was <1 min. CE-MS data of all individual samples can be accessed in Supplemental Table 3 (downloadable at http://www.mosaiques-diagnostics.de/GenexPivot_12092007.exe password: 99Mischak123).
Definition of Biomarkers and Sample Classification
The unadjusted P values were calculated using the arcsinh-transformed intensities and the Gaussian approximation to the t-distribution. Bonferroni adjustments were made by applying the standard Bonferroni criterion to markers that passed the frequency threshold of 70%. MaxT P values were calculated using the Westfall and Young maxT procedure.26
Estimates of sensitivity and specificity were calculated by tabulating the number of correctly classified samples. Confidence intervals were based on exact binomial calculations using MedCalc 8.1.1.0 (MedCalc Software; Mariakerke, Belgium; http://www.medcalc.be). The receiver operating characteristic plot was generated by plotting all sensitivity values (true-positive fraction) on the y axis against their equivalent (1 − specificity) values (false-positive fraction) for all available thresholds on the x axis (MedCalc Software).27 Disease-specific polypeptide patterns based on predefined polypeptides were generated using the SVM-BM MosaCluster software.28
For linear combination, all normalized signal intensity values of biomarkers were log-transformed. Values <1 were substituted with a value of 1 to avoid negative values. The average signal intensity for a specific biomarker over all cases was compared with the average intensity for the same biomarker over all controls. To avoid artificial weighting of specific biomarkers in the set as a result of the difference in observed signal intensities for case and control, the distance between the two averages (case and control) was set relative to a value of 2. The relative distance of signal intensities between the disease and control samples was provided using the following formula:
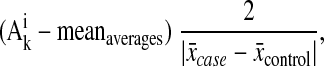 |
where Aki is the log-transformed signal intensity of the ith biomarker in the kth sample in either the test set or the blinded set, meanaverages is the average of the mean intensity of all possible markers for test set samples, x̄case represents the mean observed signal intensity of the possible biomarker from all case samples, and x̄control represents the mean signal intensity of the possible biomarker from the control samples.
Cross-validation was performed and defined by the take-one-out procedure.29 To reduce the number of biomarkers in a model, a take-one-out procedure was used.3 Briefly, models were generated that were each based on n − 1 biomarkers. These were evaluated using complete cross-validation and compared with the classification results of the model based on n biomarkers. A single biomarker that apparently did not improve classification was removed.
Sequencing of Polypeptides
Candidate biomarkers were sequenced using liquid chromatography–MS/MS analysis (on a quadrupole time-of-flight mass spectrometer instrument).6 Further analysis was performed using instruments with electron transfer dissociation capability.4,30,31
DISCLOSURES
H. Mischak is founder and co-owner of Mosaiques Diagnostics, which developed the CE-MS technology and the MosaiquesVisu software. M. Dakna and P. Zürbig are employees of Mosaiques Diagnostics.
Acknowledgments
This work was supported in part by a grant LSHM-CT-2005-018733 from the European Union to the PREDICTIONS (PREvention of DIabetic ComplicaTIONS) network to H.M. and L.T. and by a EUROTRANS-BIO grant ETB-2006-016 to H.M. via the Urosysteomics consortium. D.M.G. gratefully acknowledges support from a National Institutes of Health predoctoral fellowship: The Biotechnology Training Program (NIH 5T32GM08349).
We are grateful to Danilo Fliser, Marion Haubitz, Jürgen Floege, and Harald Ruprecht for supplying samples from patients with chronic renal disease.
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental information for this article is available online at http://www.jasn.org/.
REFERENCES
- 1.Fliser D, Novak J, Thongboonkerd V, Argiles A, Jankowski V, Girolami M, Jankowski J, Mischak H: Advances in urinary proteome analysis and biomarker discovery. J Am Soc Nephrol 18: 1057–1071, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Kolch W, Neususs C, Pelzing M, Mischak H: Capillary electrophoresis-mass spectrometry as a powerful tool in clinical diagnosis and biomarker discovery. Mass Spectrom Rev 24: 959–977, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Theodorescu D, Wittke S, Ross MM, Walden M, Conaway M, Just I, Mischak H, Frierson HF: Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol 7: 230–240, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Zimmerli LU, Schiffer E, Zurbig P, Kellmann M, Mouls L, Pitt A, Coon JJ, Schmiederer RE, Mischak H, Peter K, Kolch W, Delles C, Dominiczak AF: Urinary proteomic biomarkers in coronary artery disease. Mol Cell Proteomics 7: 290–298, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Mischak H, Kaiser T, Walden M, Hillmann M, Wittke S, Herrmann A, Knueppel S, Haller H, Fliser D: Proteomic analysis for the assessment of diabetic renal damage in humans. Clin Sci (Lond) 107: 485–495, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Zurbig P, Renfrow MB, Schiffer E, Novak J, Walden M, Wittke S, Just I, Pelzing M, Neususs C, Theodorescu D, Root C, Ross M, Mischak H: Biomarker discovery by CE-MS enables sequence analysis via MS/MS with platform-independent separation. Electrophoresis 27: 2111–2125, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Haubitz M, Fliser D, Rupprecht H, Floege J, Haller H, Rossing K, Walden M, Wittke S, Mischak H: Defining renal diseases based on proteome analysis. Nephrol Dial Transplant 20: V20, 2005 [Google Scholar]
- 8.Julian BA, Wittke S, Novak J, Coon JJ, Zurbig P, Schiffer E, Haubitz M, Moldoveanu Z, Calcatera SM, Wyatt RJ, Sykora J, Sladkova E, Hes O, Mischak H, McGuire BM: Electrophoretic methods for analysis of urinary polypeptides in IgA-associated renal diseases. Electrophoresis 28: 4469–4483, 2007 [DOI] [PubMed] [Google Scholar]
- 9.Mischak H, Julian BA, Novak J: High-resolution proteome/peptidome analysis of peptides and low-molecular-weight proteins in urine. Proteomics Clin Appl 1: 792–804, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossing K, Mischak H, Parving HH, Christensen PK, Walden M, Hillmann M, Kaiser T: Impact of diabetic nephropathy and angiotensin II receptor blockade on urinary polypeptide patterns. Kidney Int 68: 193–205, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Mohan PS, Carter WG, Spiro RG: Occurrence of type VI collagen in extracellular matrix of renal glomeruli and its increase in diabetes. Diabetes 39: 31–37, 1990 [DOI] [PubMed] [Google Scholar]
- 12.Umezono T, Toyoda M, Kato M, Miyauchi M, Kimura M, Maruyama M, Honma M, Yagame M, Suzuki D: Glomerular expression of CTGF, TGF-β1 and type IV collagen in diabetic nephropathy. J Nephrol 19: 751–757, 2006 [PubMed] [Google Scholar]
- 13.Krag S, Nyengaard JR, Wogensen L: Combined effects of moderately elevated blood glucose and locally produced TGF-β1 on glomerular morphology and renal collagen production. Nephrol Dial Transplant 22: 2485–2496, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Han SY, Jee YH, Han KH, Kang YS, Kim HK, Han JY, Kim YS, Cha DR: An imbalance between matrix metalloproteinase-2 and tissue inhibitor of matrix metalloproteinase-2 contributes to the development of early diabetic nephropathy. Nephrol Dial Transplant 21: 2406–2416, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Bakris GL, Bank AJ, Kass DA, Neutel JM, Preston RA, Oparil S: Advanced glycation end-product cross-link breakers: A novel approach to cardiovascular pathologies related to the aging process. Am J Hypertens 17: 23S–30S, 2004 [DOI] [PubMed] [Google Scholar]
- 16.Susic D, Varagic J, Ahn J, Frohlich ED: Crosslink breakers: A new approach to cardiovascular therapy. Curr Opin Cardiol 19: 336–340, 2004 [DOI] [PubMed] [Google Scholar]
- 17.Thongboonkerd V, Barati MT, McLeish KR, Benarafa C, Remold-O'Donnell E, Zheng S, Rovin BH, Pierce WM, Epstein PN, Klein JB: Alterations in the renal elastin-elastase system in type 1 diabetic nephropathy identified by proteomic analysis. J Am Soc Nephrol 15: 650–662, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Coon JJ, Zürbig P, Dakna M, Dominiczak AF, Decramer S, Fliser D, Frommberger M, Golovko I, Good DM, Herget-Rosenthal S, Jankowski J, Julian BA, Kellmann M, Kolch W, Massy Z, Novak J, Rossing J, Schanstra JP, Schiffer E, Theodorescu D, Vanholder R, Weissinger EM, Mischak H, Schmitt-Kopplin P: CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteomics 2008, in press [DOI] [PMC free article] [PubMed]
- 19.Good DM, Thongboonkerd V, Novak J, Bascands JL, Schanstra JP, Coon JJ, Dominiczak A, Mischak H: Body fluid proteomics for biomarker discovery: Lessons from the past hold the key to success in the future. J Proteome Res 6: 4549–4555, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Christensen PK, Larsen S, Horn T, Olsen S, Parving H-H: The causes of albuminuria in patients with type 2 diabetes without diabetic retinopathy. Kidney Int 58: 1719–1731, 2000 [DOI] [PubMed] [Google Scholar]
- 21.Haubitz M, Wittke S, Weissinger EM, Walden M, Rupprecht HD, Floege J, Haller H, Mischak H: Urine protein patterns can serve as diagnostic tools in patients with IgA nephropathy. Kidney Int 67: 2313–2320, 2005 [DOI] [PubMed] [Google Scholar]
- 22.Weissinger EM, Wittke S, Kaiser T, Haller H, Bartel S, Krebs R, Golovko I, Rupprecht HD, Haubitz M, Hecker H, Mischak H, Fliser D: Proteomic patterns established with capillary electrophoresis and mass spectrometry for diagnostic purposes. Kidney Int 65: 2426–2434, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Taylor CF, Paton NW, Lilley KS, Binz PA, Julian RK Jr, Jones AR, Zhu W, Apweiler R, Aebersold R, Deutsch EW, Dunn MJ, Heck AJ, Leitner A, Macht M, Mann M, Martens L, Neubert TA, Patterson SD, Ping P, Seymour SL, Souda P, Tsugita A, Vandekerckhove J, Vondriska TM, Whitelegge JP, Wilkins MR, Xenarios I, Yates JR III, Hermjakob H: The minimum information about a proteomics experiment (MIAPE). Nat Biotechnol 25: 887–893, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Mischak H, Apweiler R, Banks RE, Conaway M, Coon JJ, Dominizak A, Ehrich JH, Fliser D, Girolami M, Hermjakob H, Hochstrasser DF, Jankowski V, Julian BA, Kolch W, Massy Z, Neususs C, Novak J, Peter K, Rossing K, Schanstra JP, Semmes OJ, Theodorescu D, Thongboonkerd V, Weissinger EM, Van Eyk JE, Yamamoto T: Clinical Proteomics: A need to define the field and to begin to set adequate standards. Proteomics Clin Appl 1: 148–156, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Theodorescu D, Fliser D, Wittke S, Mischak H, Krebs R, Walden M, Ross M, Eltze E, Bettendorf O, Wulfing C, Semjonow A: Pilot study of capillary electrophoresis coupled to mass spectrometry as a tool to define potential prostate cancer biomarkers in urine. Electrophoresis 26: 2797–2808, 2005 [DOI] [PubMed] [Google Scholar]
- 26.Westfall PH, Young SS: Resampling-Based Multiple Testing: Examples and Methods for P-Value Adjustment, New York, Wiley, 1993
- 27.Zou KH, O'Malley AJ, Mauri L: Receiver-operating characteristic analysis for evaluating diagnostic tests and predictive models. Circulation 115: 654–657, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Decramer S, Wittke S, Mischak H, Zurbig P, Walden M, Bouissou F, Bascands JL, Schanstra JP: Predicting the clinical outcome of congenital unilateral ureteropelvic junction obstruction in newborn by urinary proteome analysis. Nat Med 12: 398–400, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Levner I: Feature selection and nearest centroid classification for protein mass spectrometry. BMC Bioinformatics 6: 68, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coon JJ, Shabanowitz J, Hunt DF, Syka JE: Electron transfer dissociation of peptide anions. J Am Soc Mass Spectrom 16: 880–882, 2005 [DOI] [PubMed] [Google Scholar]
- 31.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF: Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A 101: 9528–9533, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]