Abstract
Background
One genetic mechanism known to be associated with autism spectrum disorders (ASD) is chromosomal abnormalities. The identification of copy number variants (CNV) i.e. microdeletions and microduplications that are undetectable at the level of traditional cytogenetic analysis allows the potential association of submicroscopic chromosomal imbalances and human disease.
Methods
We performed array comparative genomic hybridization (aCGH) utilizing a 19K whole genome tiling path bacterial artificial chromosome (BAC) microarray on 397 unrelated subjects with autism spectrum disorder (ASD). Common CNV were excluded using a control group comprised of 372 individuals from the NIMH Genetics Initiative Control samples. Confirmation studies were performed on all remaining CNV using FISH (Fluorescence In Situ Hybridization), microsatellite analysis and/or quantitative PCR analysis.
Results
A total of 51 CNV were confirmed in 46 ASD subjects. Three maternal interstitial duplications of 15q11-q13 known to be associated with ASD were identified. The other 48 CNV ranged in size from 189 kb to 5.5 Mb and contained from 0 to ~40 RefSeq genes. Seven CNV were de novo and 44 were inherited.
Conclusions
51 autism-specific CNV were identified in 46/397 ASD patients using a 19K BAC microarray for an overall rate of 11.6%. These microdeletions and microduplications cause gene dosage imbalance in 272 genes many of which could be considered as candidate genes for autism.
Keywords: autism, array comparative genomic hybridization, microdeletions, microduplications
Introduction
Autism was first described by Leo Kanner in 1943 as a childhood developmental disorder characterized by the “inability to relate themselves in the ordinary way to other people” and by “insistence on sameness” (1). The diagnostic criteria include qualitative impairment of reciprocal social interaction and communicative development, and restricted interests and repetitive behaviors. The prevalence of autism is 1:500 with a 4:1 male to female ratio (2,3). In addition to autism, autism spectrum disorders (ASDs) include Asperger syndrome, pervasive developmental disorder not otherwise specified, and a few specific conditions such as Rett syndrome. Together the prevalence of autism and ASD is ~1 in 160 (2,3).
The heritability of autism is well-documented. Overall, five twin studies have shown that an average of 60% of monozygotic twins (MZ) are concordant for autism while more than 90% of the monozygotic co-twins of probands with autism had significant social impairment. In contrast, dizygotic twins (DZ) have only a 10% concordance rate for autism or autism related social deficits (4).
The development of microarray based technologies for CGH (comparative genomic hybridization) analysis has enabled the detection of submicroscopic microdeletions and microduplications (also referred to as copy number variants (CNV)). Three independent array CGH platforms have been reported recently that identified small CNV in autism: 1) ~1 Mb resolution BAC microarrays (5–9); 2) the Affymetrix 10K SNP array (10); and 3) 85K representational oligonucleotide microarray analysis (ROMA) (11). One limitation of these studies is incomplete coverage of the human genome.
In this study we performed array comparative genomic hybridization (aCGH) on 397 unrelated probands with ASD using a 19K BAC microarray developed at the Roswell Park Cancer Institute. Common CNV specific to this 19K BAC microarray were identified in 372 individuals from the NIMH Genetics Initiative Control sample set and excluded from the autism sample data. We have identified 51 CNV, not present in the controls, in 46 ASD subjects.
Subjects and Methods
ASD subjects
The ASD probands selected for this study are a subset of the Autism Genetic Resource Exchange (AGRE) subjects that were collected with informed consent and institutional review board (IRB) approval (12). We are reporting results on 397 of these subjects including 232 males and 165 females. Females were chosen preferentially to partially balance the 4:1 male to female ratio observed in ASD. The more severely affected sibling was chosen based on the strictness of their AGRE ADI-R classification (i.e. Autism, then Not Quite Autism, then Broad Spectrum); if both siblings had the same ADI-R classification then severity of their ADOS classification (i.e. Autism over Broad Spectrum) was used when available. The race/ethnicities of the autism subjects included 235 White/Not Hispanic, 28 White/Hispanic, 9 Asians, 4 Blacks, 14 with more than 1 race/Not Hispanic, 7 with more than 1 race/Hispanic and 98 unknown. Follow-up studies of subjects with one or more CNV included parents and available affected and unaffected siblings. Thirty-five probands were from simplex families and 362 from multiplex families.
The NIMH Genetics Initiative Control sample set
372 control subjects were selected for this study including 262 Caucasians and 100 African-Americans. All subjects were characterized for Axis I disorders; although these individuals were not excluded from the sample set. Autistic traits were not assessed. Although the patient sample set contains only 4 African-Americans, 100 African-American control subjects were analyzed to give a more accurate representation of the CNV distinctive for this population. No Hispanic or Asian control samples are currently available in this collection.
Construction of the Human BAC CGH array
The minimal tiling RPCI BAC array contains ~19,000 BAC clones that were chosen by virtue of their STS content, paired BAC end-sequence and association with heritable disorders and cancer. The 19K BAC array represents an evolution of an earlier design that originally contained 6,102 clones (13). The additional ~13,000 clones were selected using bioinformatics to minimize gap sizes between the ends of clones and the total number of clones required. A detailed description of the methodology for this assay is presented in supplementary methods.
Fluorescence in situ hybridization (FISH)
Lymphoblastoid cell lines for each proband and selected family members were acquired from the Rutgers University Cell and DNA Repository and cultured using standard techniques. RPCI-11 BACs that defined the boundaries of the selected CNV were acquired from several sources including the Wellcome Trust Sanger Institute and the Roswell Park Cancer Institute. BAC probes were labeled by nick translation using either Spectrum Green or Spectrum Orange fluorescent dyes (Abbott Labs). FISH was performed using standard techniques. Slides were analyzed with a Zeiss Axioplan 2 fluorescent microscope with a cooled CCD camera and Applied Imaging CytoVision v3.7 software.
Microsatellite Analysis
Microsatellites were selected from the UCSC Genome Browser microsatellite or simple repeat tracks and primers were designed using the MIT Primer3 program. For a single reaction, a master mix of 1ul 10x PCR buffer with 15 mM MgCl2, 1 μl 10mM dNTP, 0.1 μl Ampli Taq Gold enzyme, 0.8 μl 10 mM primers (forward & reverse), and 7.1 μl sterile H2O was prepared. 1 μl DNA (10 ng/μl) was added to each reaction. The PCR reactions were run in ABI 9700 thermocyclers using the following conditions: hot start at 94°C for 10 min, 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec for 35 cycles followed by a final extension step at 72°C for 10 min. Samples were analyzed on an ABI 3730 XL DNA sequencing analyzer and processed using GeneMapper 3.7 software (Applied Biosystems).
Quantitative PCR
Quantitative PCR was performed with the Power SYBR Green PCR master mix kit (Applied Biosystems) according to the manufacturer’s recommendations. Assays were analyzed using an ABI 7900 HT Real-time PCR instrument with SDS 2.2.1 software (Applied Biosystems). Each sample was run in triplicate and the RQ data averaged.
Results
aCGH control samples
A total of 272 Caucasian and 100 African-American control samples were analyzed using the same 19K BAC microarray as the autism subjects. To insure a high level of specificity in the controls, a threshold of 5 SD above and below the mean log2 ratios were used to distinguish “normal” versus “abnormal” clones. The Caucasian and African-American data were evaluated independently to identify CNV specific for each race. A summary of the control data is presented in Table 1 with a complete listing of the data in Table S1.
Table 1.
Summary of NIMH Control Data using the 19K whole genome tiling path BAC microarray.
| Caucasian only | African-American only | Both African-American and Caucasian | |
|---|---|---|---|
| (N=272) | (N=100) | ||
| Number of BACs > 5 SD | 1276 | 720 | 384 |
| BAC clones in only 1 subject | 882 | 547 | - |
| BAC clones in >1 subject | 394 | 173 | 384 |
The data for BACs representing CNV in a single subject and BACs representing CNV in >1 subject is presented separately. The total number of BACs identified was 2,380 of which 1276 were specific for Caucasians and 720 specific for African-Americans. Only 384 BACs were found in both populations, indicating the presence of a distinctive set of common CNV for each population (Table 1). We then compared our control CNV data with the published control data as compiled in the Database of Genomic Variants (DGV) (14). Only 506/1154 (30.5%) of the Caucasian CNV and 320/1103 (29.0%) of the African-American CNV were present in the DGV. In addition, pools of 20 males or 20 females served as internal standard reference DNA for aCGH analyses from which the test sample was compared. Therefore, the list of CNV in the controls that are presented in Table S1 are specific for this 19K BAC microarray.
aCGH autism samples
A total of 397 ASD probands were analyzed by aCGH using the RPCI 19K BAC microarray. A threshold of 5 SD and manual curation were used to distinguish BACs with a normal copy number from those containing microdeletions and microduplications. Additionally, only CNV with 2 contiguous BACs were studied further to reduce false positives. Note that the rigorous parameters we used for selection of CNV may have missed small CNV that are present in a single clone. Two levels of confirmation were performed on the autism-specific CNV. FISH was performed on all probands to confirm both the location of the abnormal BAC and the boundaries of the CNV. Microsatellite and/or qPCR were performed on the proband and all family members to determine the parent of origin and inheritance pattern, and to confirm paternity. Analyses included both affected and unaffected siblings. Table 2 summarizes the CNV identified in these ASD subjects and Table S2 provides additional detail. A complete list of the ASD patients with normal aCGH results is presented in Table S3. Figures 1 & 2 show the aCGH and FISH confirmation of a patient with a duplication of 2p15. Figures 3 & 4 show the aCGH and FISH confirmation of a patient with a deletion of 17p12.
Table 2.
AGRE confirmed abnormal aCGH results
| Family ID | Individual ID | BloodID | Sex | Family Structure | Ethnicity | Race | 19K aCGH result | Size | # genes | transmission | Confirmed |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AU0385 | AU038503 | HI0309 | F | Multiplex | Unknown | Unknown | del(2)(p25.3) | 1.2 Mb | 5 | inherited | F |
| AU0230 | AU023012 | HI1001 | M | Multiplex | Unknown | Unknown | dup(2)(p24.2) | 970 kb | 4 | inherited | F,S |
| AU0039 | AU0039301* | HI1481 | M | Multiplex | NOT Hispanic | White | dup(2)(p22.3) | 700 kb | 3 | inherited | F,S |
| AU0640 | AU064004 | HI2215 | F | Multiplex | NOT Hispanic | White | dup(2)(p21) | 530 kb | 3 | inherited | F,S |
| AU0355 | AU035505 | HI0234 | F | Simplex | NOT Hispanic | White | dup(2)(p15) | 770 kb | 4 | inherited | F,S |
| AU0483 | AU048304* | HI0911 | F | Multiplex | Hispanic | White | del(2)(q32.3) | 450 kb | 1 | inherited | F,S |
| AU0660 | AU066005* | HI1928 | F | Multiplex | NOT Hispanic | White | del(3(p26.3) | 700 kb | 1 | inherited | F,S |
| AU0053 | AU005303* | HI1102 | M | Multiplex | NOT Hispanic | White | dup(3)(p26.2) | 285 kb | 2 | inherited | F,S |
| AU0034 | AU003404* | HI1289 | F | Multiplex | NOT Hispanic | White | del(3)(p14.1) | 1.3 Mb | 4 | inherited | F,S |
| AU0215 | AU021503* | HI0020 | M | Multiplex | Hispanic | White | dup(3)(q12.1) | 300 kb | 4 | inherited | F,S,Q |
| AU0293 | AU029303 | HI0525 | M | Multiplex | NOT Hispanic | White | dup(5)(q12.1) | 575 kb | 3 | inherited | F,S,Q |
| AU0994 | AU0994301 | HI2775 | F | Multiplex | NOT Hispanic | White | del(5)(q12.1) | 810 kb | 1 | inherited | F,S,Q |
| AU0385 | AU038503 | HI0309 | F | Multiplex | Unknown | Unknown | dup(7)(p22.3) | 1.1 Mb | 12 | inherited | F |
| AU0386 | AU038603 | HI0289 | M | Simplex | Unknown | Unknown | dup(7)(p21.1) | 600 kb | 2 | inherited | F,S |
| AU0672 | AU067209 | HI1686 | F | Multiplex | Unknown | Unknown | del(7)(q21.11) | 2.9 Mb | 3 | inherited | F,S,Q |
| AU0340 | AU034004 | HI0125 | F | Multiplex | NOT Hispanic | White | dup(7)(q21.13) | 615 kb | 2 | inherited | F,S |
| AU0029 | AU002905 | HI0128 | F | Multiplex | NOT Hispanic | White | del(7)(q31.32) | 900 kb | 4 | inherited | F,S |
| AU0785 | AU078503 | HI1954 | F | Multiplex | NOT Hispanic | White | dup(8)(q11.23) | 310 kb | 3 | inherited | F,S |
| AU0305 | AU030504* | HI0182 | M | Multiplex | NOT Hispanic | White | del(10)(p13) | 318 kb | 6 | inherited | F,S |
| AU0845 | AU084505 | HI2168 | F | Multiplex | NOT Hispanic | White | dup(10)(q11.22-q11.23) | 5.5 Mb | 28 | inherited | F,S |
| AU0562 | AU056204 | HI0799 | F | Multiplex | NOT Hispanic | White | dup(11)(p11.12) | 600 kb | 2 | inherited | F,S |
| AU0487 | AU048703* | HI1000 | F | Multiplex | NOT Hispanic | White | dup(12)(p11.1) | 1.2 Mb | 3 | inherited | F,S |
| AU0383 | AU038304 | HI0299 | M | Multiplex | NOT Hispanic | White | dup(13)(q14.2) | 393 kb | 1 | inherited | F,S |
| AU0173 | AU017305 | HI0933 | M | Multiplex | NOT Hispanic | White | dup(14)(q23.1) | 189 kb | 3 | inherited | F,S,Q |
| AU0065 | AU006504* | HI0473 | M | Multiplex | NOT Hispanic | White | dup(15)(q11-q13) | 6.1 Mb | 18 | inherited | F,S,Q |
| AU0106 | AU010603* | HI0121 | M | Multiplex | NOT Hispanic | White | dup(15)(q11-q13) | 4.7 Mb | 18 | de novo | F,S |
| AU0233 | AU023304 | HI0304 | M | Multiplex | NOT Hispanic | White | dup(15)(q11-q13) | 5.9 Mb | 18 | de novo | F,S |
| AU0132 | AU013203 | HI0385 | M | Simplex | Unknown | Unknown | dup(15)(q12.1) | 195 kb | 3 | de novo | F,S |
| AU0520 | AU052003* | HI2139 | M | Multiplex | NOT Hispanic | White | dup(15)(q13.1) | 289 kb | 3 | inherited | F,S |
| AU0520 | AU052003* | HI2139 | M | Multiplex | NOT Hispanic | White | dup(15)(q13.3) | 372 kb | 2 | inherited | F,S |
| AU0467 | AU046703 | HI1030 | M | Multiplex | Unknown | Unknown | dup(15)(q21.3) | 1.2 Mb | 12 | inherited | F,S |
| AU0561 | AU056103* | HI0972 | M | Multiplex | Unknown | Unknown | del(15)(q25.3) | 600 kb | 9 | inherited | F,S |
| AU0154 | AU0154303* | HI2467 | F | Multiplex | NOT Hispanic | Pacific Islander | del(16)(p11.2) | 980 kb | 24 | de novo | F,S,Q |
| AU0298 | AU029803 | HI0624 | F | Multiplex | NOT Hispanic | White | del(16)(p11.2) | 590 kb | 24 | de novo | F,S |
| AU0419 | AU041905 | HI0646 | F | Multiplex | NOT Hispanic | White | del(16)(p11.2) | 700 kb | 24 | de novo | F,S |
| AU0938 | AU0938301 | HI2997 | M | Multiplex | NOT Hispanic | White | del(16)(p11.2) | 980 kb | 24 | de novo | F,S |
| AU0029 | AU002905 | HI0128 | F | Multiplex | NOT Hispanic | White | dup(16)(q22.1) | 890 kb | 15 | de novo | F,S |
| AU0796 | AU079605 | HI1937 | M | Multiplex | Unknown | Unknown | dup(16)(q23.3) | 527 kb | 2 | inherited | F,S |
| AU0796 | AU079605 | HI1937 | M | Multiplex | Unknown | Unknown | dup(16)(q24.1) | 510 kb | 6 | inherited | F,S |
| AU0779 | AU077903 | HI2043 | F | Multiplex | NOT Hispanic | White | dup(17)(p13.1) | 400 kb | 4 | inherited | F,S |
| AU0149 | AU014905* | HI0563 | M | Multiplex | NOT Hispanic | White | del(17)(p12) | 1.1 Mb | 5 | inherited | F,S |
| AU0611 | AU061103 | HI1252 | M | Multiplex | NOT Hispanic | White | dup(18)(q22.3) | 360 kb | 1 | inherited | F,S,Q |
| AU0276 | AU027604 | HI0937 | F | Multiplex | Unknown | Unknown | dup(19)(q12) | 900 kb | 5 | inherited | F,S |
| AU1368 | AU1368302* | HI3629 | F | Multiplex | Unknown | Unknown | del(20)(p12.3) | 480 kb | 1 | inherited | F,S |
| AU0187 | AU018704 | HI0340 | F | Multiplex | Unknown | Unknown | dup(20)(q12) | 264 kb | 1 | inherited | F,S,Q |
| AU0477 | AU047704 | HI1876 | F | Multiplex | Hispanic | More Than One Race | dup(21)(q22.12) | 210 kb | 5 | inherited | F,S |
| AU0049 | AU004903 | HI0322 | F | Multiplex | Unknown | Unknown | dup(22)(q11.21) | 2.2 Mb | 40 | de novo | F,S |
| AU0018 | AU001804 | HI1183 | M | Multiplex | NOT Hispanic | White | dup(22)(q11.21) | 2.2 Mb | 40 | inherited | F,S |
| AU0034 | AU003404* | HI1289 | F | Multiplex | NOT Hispanic | White | dup(22)(q13.33) | 570 kb | 2 | inherited | F,S |
| AU0930 | AU0930202 | HI2506 | F | Multiplex | NOT Hispanic | White | dup(X)(p22.32) | 360 kb | 0 | inherited | F,S |
| AU0709 | AU070908 | HI1920 | F | Multiplex | Unknown | Unknown | dup(X)(p22.22) | 510 kb | 4 | inherited | F,S |
= concordant with affected sibs and discordant with unaffected sibs.
del = deletion; dup = duplication; F = Fluorescence in situ hybridization; S = short tandem repeat polymorphisms/microsatellites; Q = quantitative PCR
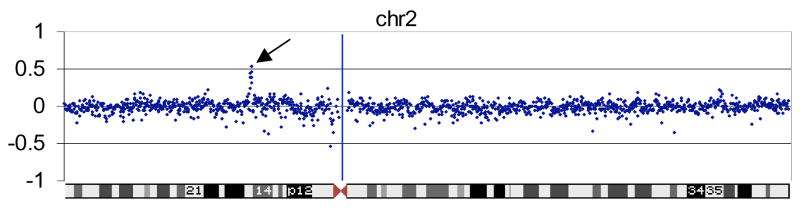
Figure 1. Duplication 2p15 aCGH results.
The aCGH results show the log2 ratio of the patient vs. reference DNA on the vertical axis. Each individual BAC is represented as a single blue dot and the horizontal axis shows the position of each BAC along chromosome 2. The duplication of 2p15 is indicated by an arrow pointing to a vertical line of dots.
Figure 2. Duplication 2p15 FISH results.
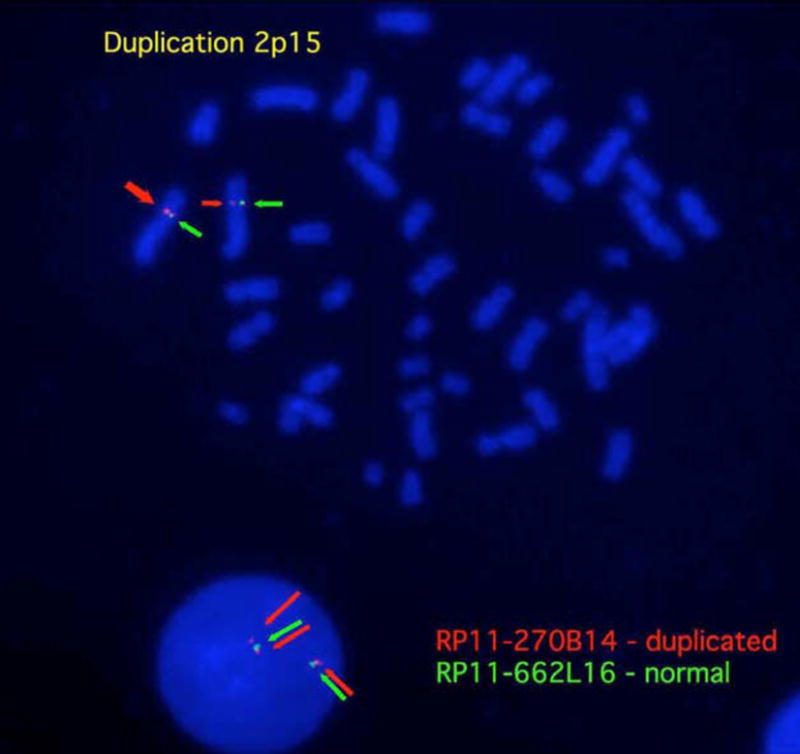
The proximal breakpoint boundary is demonstrated where RP11-662L16 (green) shows 2 normal signals while RP11-270B14 (red) shows a larger signal on one metaphase chromosome and an extra signal in the interphase nucleus.
Figure 3. Deletion 17p12 aCGH results.
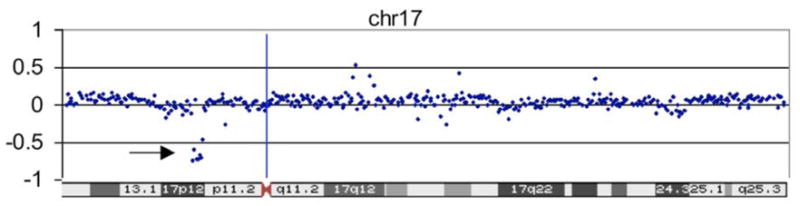
In this case the arrow indicates a cluster of dots, located between −0.5 and −1.0, that identifies the deletion.
Figure 4. Deletion 17p12 FISH results.
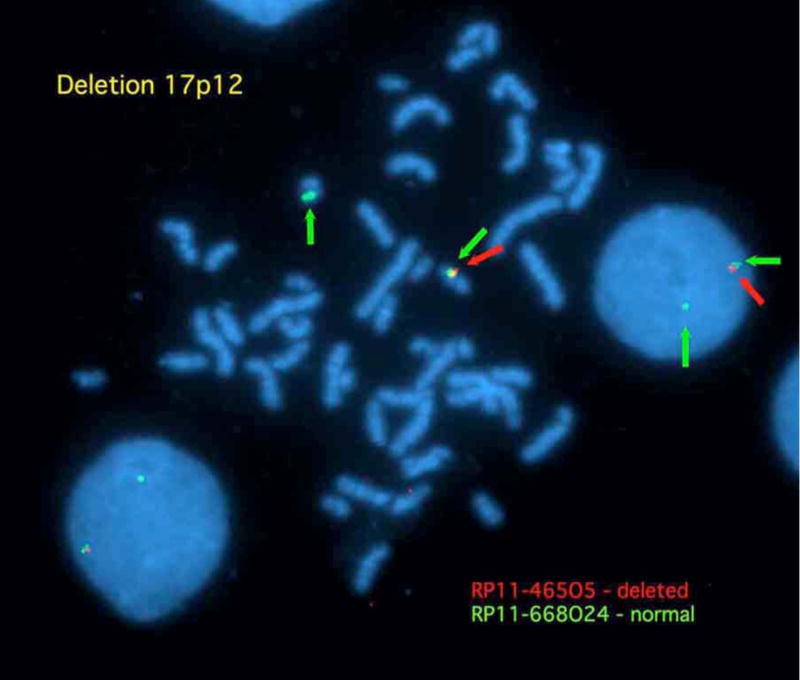
The distal breakpoint boundary is demonstrated where RP11-668O24 (green) shows 2 normal signals while RP11-465O5 (red) shows a single signal in both the metaphase chromosomes and the interphase nucleus.
A total of 51 CNV were identified in 46 ASD probands including 21 males and 25 females (Table S2). Forty-one patients showed a single CNV and 5 patients had 2 CNV (Table 2). Forty-two CNV were familial and 9 were de novo, consistent with the multiplex family structure of the AGRE autism patient collection that focused on multiplex families for linkage studies. Hence an overrepresentation of multiplex versus simplex families is present in this sample set resulting in the identification of mostly inherited CNV. One limitation of this study was the use of genomic DNA extracted from lymphoblastoid cell lines leading to the remote possibility of cultural artifacts in the 9 de novo CNV detected.
Recurrent CNV were identified for 3 loci. Three interstitial maternal duplications of 15q11-q13 were identified in this study. Subsequently, two of these patients were found to have had abnormal methylation analyses performed by another laboratory that were consistent with the aCGH data. In two cases 22q11 duplications were identified, one de novo and one inherited both of paternal origin. In 4 cases de novo16p11.2 microdeletions were identified 3 of maternal origin and 1 paternal. The other 42 CNV ranged in size from 189 kb to 5.5 Mb, well below the level of resolution of traditional cytogenetics. Overall, in this study autism-specific CNV were identified in 46/397 (11.6%) of the ASD patients analyzed.
Within the AGRE data set, non-verbal IQ (estimated from the Raven Colored Progressive Matrices) (15) was available for 9 males and 9 females with CNV. Males’ scores (M = 102.78, SD = 18.25) did not significantly differ from females’ scores (M = 104.00, SD = 18.00), t(16) = −0.14, p = 0.89.
Discussion
Although ASDs are well known to be strongly genetic, no single genetic cause has been identified that accounts for (or contributes to) autism in all or even a significant proportion of patients. Due to this genetic heterogeneity, many unique genetic variants are likely to result in the ASD phenotype including single genes affecting multiple cellular processes or function in the same neurodevelopmental pathway and cytogenetic abnormalities affecting multiple contiguous genes. The identification of CNV below the detection threshold of traditional cytogenetics has opened the door for discovery of submicroscopic deletions and duplications that may either cause or contribute to the ASD phenotype. In this study we used a 19K BAC array at a level of resolution of ~200 kb to detect CNV specific for autism.
The higher level of resolution provided by a BAC microarray relative to traditional cytogenetics increases the complexity for interpreting the significance of these autism-specific CNV. In the past the assumption has been that a large de novo chromosomal abnormality would be associated with disease, although the very low penetrance of paternally inherited 15q11-q13 duplications demonstrates that size is not entirely predictive of phenotypic effect. Submicroscopic CNV may have even more subtle effects. Multiple follow-up analyses are needed to assess each individual CNV. Some examples of the types of analyses that can be performed include analysis of very large samples, concordance with affected siblings, genotype-phenotype correlation between the affected child in de novo cases and both the affected child and carrier parent in inherited cases and an assessment of the gene content of the CNV.
Concordance
One line of evidence that would increase support for these CNV as ASD risk loci are concordance between affected siblings. In the 41 families with inherited CNV, 21 sibs affected with autism were concordant and 12 affected sibs were discordant for the presence of the specific CNV. In addition 14 sibs carried a diagnosis of broad spectrum of which 5 were concordant and 9 were discordant. However, the interpretation of concordance data in affected siblings with discordance in the severity of ASD is problematic. These CNV could be responsible for the more severe phenotype in the autistic proband compared to the broad spectrum sibling and affect the overall concordance rate observed in this study. Discordance for a CNV between sibs classified with the same severity of autism does not rule out the role of the CNV in susceptibility to autism where other non-shared and shared loci between the CNV discordant sibs contribute to the similarity between the phenotypes. While ascertainment through multiplex families and simplex families may detect different sets of CNV, we believe that discovery of both sets allows a fuller picture of the role of CNV in autism susceptibillty.
Genotype-phenotype correlations
Although phenotypic data are available for most of the probands with CNV, 42/51 CNV were only observed in a single subject and no reliable assessment can be made. In the CNV with multiple affected patients, dup 15q11-q13 has been well-characterized. However, dup 22q11 and del 16p11.2 have a small number of patients for whom preliminary phenotypic characterization can begin. Currently, one 22q11 duplication has been reported in a single patient with autism (16), while additional subjects have been reported with features characteristic of autism without a formal diagnosis (17, 18). The most consistent features of 22q11 duplication subjects were intellectual disability, neuropsychological problems and speech disorder (17). The phenotypic data on the 4 patients with 16p11.2 microdeletions did not show any consistent features that would distinguish this group of patients although behavioral difficulties involving aggression and overactivity were noted. A detailed study of this recurrent microdeletion has been performed and is reported elsewhere (19).
Gene content
Another important analysis to perform to interpret CNV is the gene content within the abnormalities. The 51 CNV reported in this study contain a total of 269 RefSeq genes, some of which could be considered as candidate genes for autism (Table S3). Although these 51 CNV are located in multiple loci within the human genome, groups of these genes could be integrated within the same neurodevelopmental pathways including axon guidance, nervous system development and synaptic transmission. Ongoing efforts from several sources pathways including the Gene Ontology database (20), GeneWays (21) and KEGG (22) are compiling published data on all human genes to create interacting networks.
Summary
This autism CNV study on autism subjects used a 19K whole genome tiling path BAC microarray to identify 51 CNV in 46/397 (11.6%) which is significantly higher than the ~5% identified using traditional cytogenetics. These smaller microdeletions and microduplications will allow a more focused effort to study the individual genes within these CNV. However, we are unable to differentiate for each specific CNV whether they 1) cause ASD, 2) contribute to ASD or 3) have no phenotypic effect without analysis of much larger sample sizes or other methods of investigation.
Supplementary Material
Acknowledgments
This work was partially funded by the National Alliance for Autism Research (S.L.C.) and the National Institute of Neurological Diseases and Stroke (R01 NS51812) (S.L.C.). This work was also supported by the National Institutes of Health/National Cancer Institute, (P30 CA016056) (RPCI Cancer Center Support Grant).
We gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange (AGRE) Consortium and the participating AGRE families. The Autism Genetic Resource Exchange is a program of Cure Autism Now and is supported, in part, by a grant to Daniel H. Geschwind (PI) from the National Institute of Mental Health (MH64547).
Footnotes
Financial disclosures and conflicts of interest
Susan L. Christian reported no biomedical interests or potential conflicts of interest. Camille W. Brune reported no biomedical interests or potential conflicts of interest. Jyotsna Sudi reported no biomedical interests or potential conflicts of interest. Ravinesh A. Kumar reported no biomedical interests or potential conflicts of interest. Shaung Liu reported no biomedical interests or potential conflicts of interest. Samer KaraMohamed reported no biomedical interests or potential conflicts of interest. Judith A. Badner reported no biomedical interests or potential conflicts of interest. Seiichi Matsui reported no biomedical interests or potential conflicts of interest. Jeffrey Conroy reported no biomedical interests or potential conflicts of interest. Devin McQuaid reported no biomedical interests or potential conflicts of interest. James Gergel reported no biomedical interests or potential conflicts of interest. Eli Hatchwell reported no biomedical interests or potential conflicts of interest. T. Conrad Gilliam reported no biomedical interests or potential conflicts of interest. Elliot S. Gershon reports that he is a consultant to Epix Pharmaceuticals, which has no interest in these findings. Norma J. Nowak reported no biomedical interests or potential conflicts of interest. William B. Dobyns reported no biomedical interests or potential conflicts of interest. Edwin H. Cook, Jr reported no biomedical interests or potential conflicts of interest.
Web Resources
AGRE website: http://www.agre.org/
NIMH Genetics Initiative: http://nimhgenetics.org/
Roswell Park Cancer Institute Microarray Facility: http://microarrays.roswellpark.org
UCSC Genome Bioinformatics: http://genome.ucsc.edu
MIT Primer3 program for PCR primer design: http://frodo.wi.mit.edu/
Rutgers University Cell and DNA Repository: http://www.rucdr.org/
Wellcome Trust Sanger Institute: http://www.sanger.ac.uk
Database of Genomic Variants (DGV): http://projects.tcag.ca/variation/
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kanner L. Autistic Disturbances of affective contact. Nervous Child. 1943;2:217–250. [PubMed] [Google Scholar]
- 2.Chakrabarti S, Fombonne E. Pervasive developmental disorders in preschool children: confirmation of high prevalence. Am J Psychiatry. 2005;162:1133–1141. doi: 10.1176/appi.ajp.162.6.1133. [DOI] [PubMed] [Google Scholar]
- 3.Fombonne E. Epidemiology of autistic disorder and other pervasive developmental disorders. J Clin Psychiatry. 2005;66(Supp 10):3–8. [PubMed] [Google Scholar]
- 4.Folstein SE, Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet. 2001;2:943–955. doi: 10.1038/35103559. [DOI] [PubMed] [Google Scholar]
- 5.Harvard C, Malenfant P, Koochek M, Creighton S, Mickelson ECR, Holden JJA, et al. A variant of Cri du Chat phenotype and autism spectrum disorder in a subject with de novo cryptic microdeletions involving 5p15.2 and 3p24–25 detected using whole genomic array CGH. Clin Genet. 2005;67:341–351. doi: 10.1111/j.1399-0004.2005.00406.x. [DOI] [PubMed] [Google Scholar]
- 6.Koochek M, Harvard C, Hildebrand MJ, Van Allen M, Wingert H, Mickelson E, et al. 15q duplication associated with autism in a multiplex family with a familial cryptic translocation t(14;15)(q11.2;q13.3) detected using array-CGH. Clin Genet. 2006;69:124–134. doi: 10.1111/j.1399-0004.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- 7.Bonnet C, Gregoire MJ, Brochet K, Raffo E, Leheup B, Jonveaux P. Pure de-novo 5 Mb duplication at Xp11.22-p11.23 in a male: phenotypic and molecular characterization. J Hum Genet. 2006;51:815–823. doi: 10.1007/s10038-006-0023-3. [DOI] [PubMed] [Google Scholar]
- 8.Jacquemont M-L, Sanlaville D, Redon R, Raoul O, Cormier-Daire V, Lyonnet S, et al. Array-based comparative genomic hybridization identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43:842–848. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajcan-Separovic E, Harvard C, McGillvray B, Jall J, Qiao Y, Hurlburt J, et al. Clinical and molecular cytogenetic characterization of a newly recognized microdeletion syndrome involving 2p15–16.1. J Med Genet. 2007;44(4):269–276. doi: 10.1136/jmg.2006.045013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Autism Genome Project Consortium. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nature Genetics. 2007;39(3):319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, et al. The Autism Genetic Resource Exchange: A resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet. 2001;69:463–466. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowak NJ, Snijders A, Conroy JM, Albertson D. Current Protocols in Human Genetics. 2005a. The BAC Resource: Tools for Array CGH and FISH; pp. 1–34. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet Genome Res. 2006;115(3–4):205–14. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]
- 15.Raven J. Coloured Progressive Matrices-Sets A, AB, B. Oxford Psychologists Press; Oxford, UK: 1993. [Google Scholar]
- 16.Mukaddes NM, Herguner S. Autistic disorder and 22q11 duplication. World J Biol Psychiatry. 2007;8(2):127–130. doi: 10.1080/15622970601026701. [DOI] [PubMed] [Google Scholar]
- 17.Portnoï M-F, Lebas F, Gruchy N, Ardalan A, Biran-Mucignat V, Malan V, et al. 22q11 duplication syndrome: Two new familial cases with some overlapping features with DiGeorge/Velocardiofacial syndromes. Am J Med Genet. 2005;137A:47–51. doi: 10.1002/ajmg.a.30847. [DOI] [PubMed] [Google Scholar]
- 18.Alberti A, Romano C, Falco M, Cali F, Schinocca P, Galesi O, et al. 1.5 Mb de novo 22q11 microduplication in a patient with cognitive deficits and dysmorphic facial features. Clin Genet. 2007;71:177–182. doi: 10.1111/j.1399-0004.2007.00750.x. [DOI] [PubMed] [Google Scholar]
- 19.Kumar RA, KaraMohamed S, Sudi J, Conrad D, Brune C, Badner JA, et al. Recurrent 16p11.2 microdeletions in Autism. Hum Molec Genet. 2008 doi: 10.1093/hmg/ddm376. in press. [DOI] [PubMed] [Google Scholar]
- 20.Lomax J. Get ready to GO! A biologist’s guide to the Gene Ontology. Brief Bioinform. 2005;6:298–304. doi: 10.1093/bib/6.3.298. [DOI] [PubMed] [Google Scholar]
- 21.Rzhetsky A, Iossifov I, Koike T, Krauthammer M, Kra P, Morris M, Yu H, et al. GeneWays: a system for extracting, analyzing, visualizing, and integrating molecular pathway data. J Biomed Inform. 2004;37:43–53. doi: 10.1016/j.jbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Aoki-Kinoshita KF, Kanehisa M. Gene annotation and pathway mapping in KEGG. Methods Mol Biol. 2007;396:71–92. doi: 10.1007/978-1-59745-515-2_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.