Abstract
The sophistication of the insect olfactory system is elegantly demonstrated by the reception of sex pheromone by the Japanese beetle. In this insect, two olfactory receptor neurons housed in antennal sensilla placodea are highly sensitive. One neuron specifically detects the sex pheromone produced by conspecific females (R,Z)-5-(−)-(1-decenyl)oxacyclopentan-2-one [(R)-japonilure]. The other neuron is tuned to (S)-japonilure, a sex pheromone from a closely related species and a behavioral antagonist for the Japanese beetle. These chemical signals are enzymatically terminated by antennal esterases that open the lactone rings to form physiologically inactive hydroxyacids. We have isolated a pheromone-degrading enzyme, PjapPDE, from >100,000 antennae of the Japanese beetle. PjapPDE was demonstrated to be expressed only in the antennal tissues housing the pheromone-detecting sensilla placodea. Baculovirus expression generated recombinant PjapPDE with likely the same posttranslational modifications as the native enzyme. Kinetic studies with pure native and recombinant PjapPDE showed a clear substrate preference, with an estimated half-life in vivo for the sex pheromone and a behavioral antagonist of ≈30 and ≈90 ms, respectively.
Keywords: olfaction, (R)-japonilure, esterase, LC-ESI/MS, signal inactivation
The ultimate refinement of the insect olfactory system is the ability to discriminate stereoisomers. The importance of stereochemistry discrimination with respect to (E)- and (Z)-isomerism was recognized with the identification of the first insect sex pheromone, bombykol (1). The existence of chiral pheromones has been recognized in the pioneering work of Silverstein, one of the founding fathers of chemical ecology (2), and a few studies of enantiomeric effects on insect behavior (for review, see ref. 3). However, the most compelling evidence for the significance of (R)/(S) configurations in insect semiochemicals was demonstrated with the sex pheromone for the Japanese beetle, Popillia japonica (4). Although female beetles release predominantly (R,Z)-5-(−)-(1-decenyl)oxacyclopentan-2-one [(R)-japonilure, in short (4)], the presence of (S)-japonilure in synthetic formulations dramatically reduced male capture (4). This behavioral antagonist, (S)-japonilure, was later identified as the sex pheromone for the Osaka beetle, Anomala osakana, a species sharing the same native habitat with the Japanese beetle (5). Interestingly, the sex pheromone of the Japanese beetle, (R)-japonilure, functions as a behavioral antagonist for the Osaka beetle. These earlier studies thus unveiled an interesting mechanism of isolation of chemical communication channels with enantiomers of a semiochemical functioning mutually as sex pheromone for the emitting species and a behavioral antagonist for the allospecific receiver.
Males of the Japanese beetle detect these semiochemicals with antennal olfactory receptor neurons (ORNs) extremely specific to either the sex pheromone or a behavioral antagonist (6). These ORNs are cocompartmentalized in sensilla placodea that are distributed over the internal surfaces of the lamellate antennae (7). Although morphologically indistinguishable, the ORNs in a subset of these sensilla differ in their physiological properties (8). One ORN is tuned to the sex pheromone and the other to the behavioral antagonist. Both of these neurons are mutually inhibited when challenged with a racemic mixture (8). In addition, the male antennae possess sensilla housing ORNs that are silent to either stereoisomer but are specialized for the detection of racemic mixtures (8). These studies also indicated that the recovery of the ORNs tuned to a behavioral antagonist is significantly slower than that of the sex pheromone ORNs. For example, the time for repolarization of the receptor membrane was significantly longer after stimulation with 100 ng of (S)- than (R)-japonilure, 305 ± 27 and 217 ± 21 ms, respectively (8). Thus, we hypothesized that the differential kinetics of inactivation of the sex pheromone and a behavioral antagonist might contribute at least in part to the observed inhibition and, consequently reduced capture. We have isolated, identified, cloned, and expressed an antennae-specific pheromone-degrading enzyme from the Japanese beetle (PjapPDE). Kinetic studies indicated that PjapPDE is involved in the rapid inactivation of the sex pheromone (estimated half-time in vivo, t½, ≈30 ms) and slower degradation of a behavioral antagonist (t½, ≈90 ms).
Results
Monitoring Japonilure Degradation.
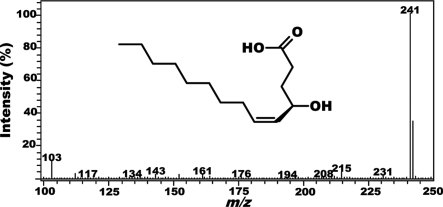
We have developed an analytical method to monitor the degradation of japonilure with nonradioactive substrates. Because the hydroxyacid generated by the opening of the lactone ring (9) is thermolabile, it was not possible to monitor the reaction by gas chromatography (GC) and/or gas chromatography–mass spectrometry. The hydroxyacid undergoes cyclization in the injection port even if a cool-on-column injector is used. Thus, GC-based analysis would require protection of the free acid and/or separation of substrate and product because the hydroxyacid that is enzymatically produced would be reconverted into japonilure by an analytical artifact. We, therefore, explored alternative analytical tools for fast monitoring of pheromone degradation. The hydroxyacid can be detected at low concentrations by liquid chromatography–mass spectrometry with an electrospray ionization probe (LC-ESI/MS) operating in negative mode. Samples of authentic hydroxyacid of japonilure gave a mass spectrum with predominantly the base peak at m/z 241 [M-H]−1 (Fig. 1) caused by deprotonation of the carboxylic acid. Because the contribution of all of the other peaks combined was small, we operated the mass spectrum in the selected ion mode and observed a linear detector response even with very low amounts of the hydroxyacid [supporting information (SI) Fig. S1]. The kinetics of japonilure degradation by antennal esterases was monitored by determining the formation of the derived hydroxyacid over time and by using freshly generated calibration curves for quantifications. A background signal derived from the reaction buffer (Fig. S2a) did not interfere with the measurements because the intensity was very small [<500 relative units of area (u.a.); 0.1 ng of hydroaxyacid, 130,000 u.a.]. We also confirmed that japonilure was not degraded merely by incubation in the reaction buffer (see Fig. S2b).
Fig. 1.
Mass spectral data for 4-(R)-hydroxy-5-tetradecenoic acid obtained by flow injection analysis with an ESI mass spectrometer operated in the negative ion mode and full scan. The only significant peak detected was the base peak at m/z 241.
Identification and Isolation of PjapPDE.
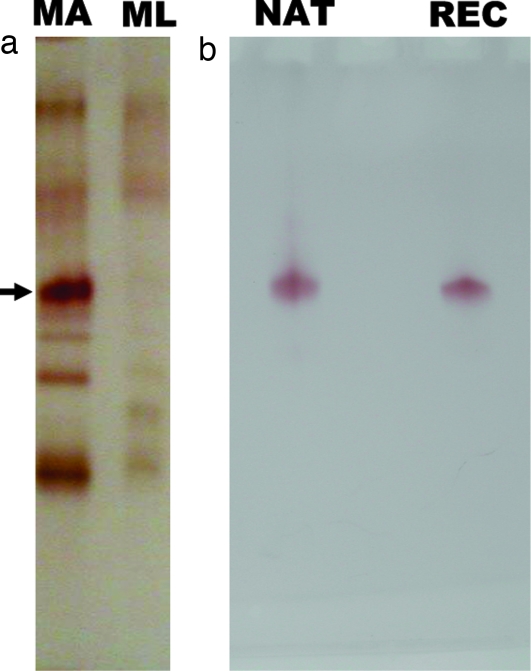
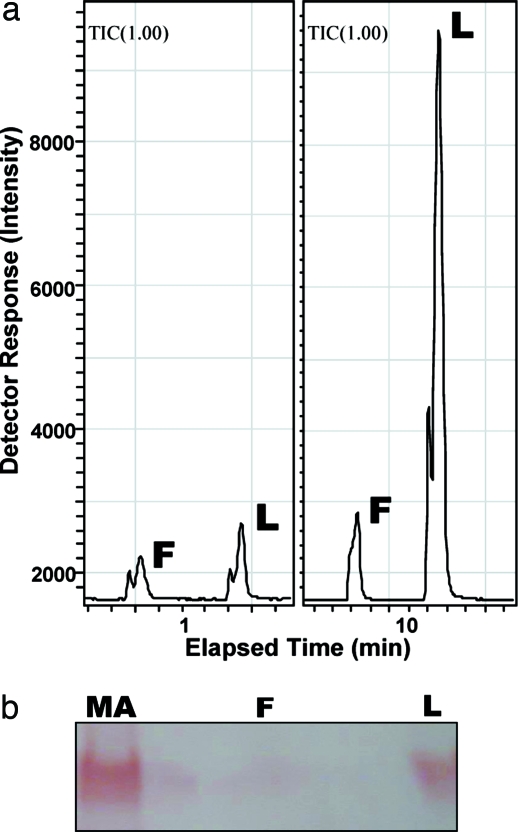
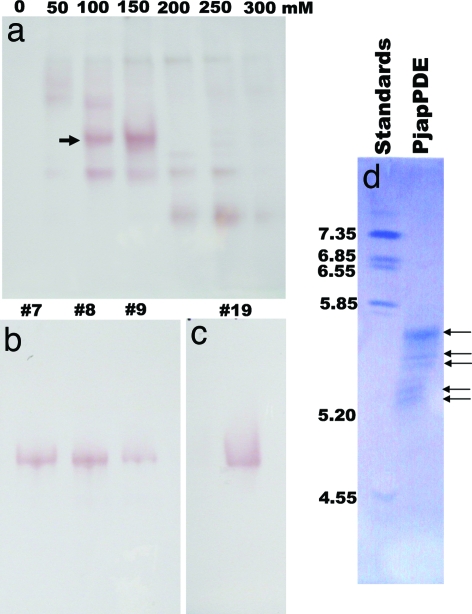
Analysis of the protein extracts from antennae and nonolfactory tissues (legs) of the Japanese beetle, separated by native PAGE and visualized by naphthyl acetate assay, showed a number of esterase bands, with one band specific to the antennae (Fig. 2a). Despite the larger amount of tissue in the control extracts, the overall amount of proteins was smaller in legs than in antennal extracts. Apparently there was a second, faster migrating, antennae-specific band, but further analysis by comparing antennal extracts with larger amounts of control tissues confirmed that indeed only one esterase is antennae-specific (SI Fig. S3). Although esterase from olfactory and nonolfactory tissues degraded both radiolabeled isomers of japonilure into their corresponding hydroxyacids, the antennae-specific band, isolated directly from a gel, demonstrated a clear preference for the pheromone over a behavioral antagonist and rapid degradation of these semiochemicals (9). To address this substrate preference and study the kinetics of pheromone degradation, we isolated the antennae-specific pheromone-degrading enzyme(s). With an analytical tool to monitor degradation of japonilure, we examined first whether the antennae-specific esterase was expressed in olfactory tissues. We prepared separated samples of the whole antennae, antennal flagella, and lamellae, the antennal tissue housing pheromone-detecting sensilla (7). Faster pheromone degradation by esterase(s) in lamellae extract showed that indeed japonilure-degrading anzyme(s) is (are) expressed in olfactory tissues (Fig. 3a). Native PAGE analysis (Fig. 3b) indicated that the antennae-specific band is expressed in lamellae, but not in flagella. We thus named the antennae-specific esterase(s) PjapPDE. To purify PjapPDE, we extracted proteins from >100,000 antennae. PjapPDE activity was detected in the fractions eluted from an anion exchange column (DEAE) with 100 and 150 mM NaCl (Fig. 4a). With these fractions separated on a gel filtration column, the molecular mass of PjapPDE was estimated to be 59 kDa (data not shown). The eluted fractions (Fig. 4b) were pooled and loaded onto high-resolution ion exchange column (Mono Q) to generate one fraction containing a single band with esterase activity (Fig. 4c). This purest fraction was further separated by isoelectric focusing (IEF) gel electrophoresis into five bands with pI ranging from 5.2 to 5.8 (Fig. 4d). As determined by Edman degradation, all of these bands had an identical N-terminal sequence, NNPFPTLEISTGVLQ. Thus, these five bands could be derived from proteins with the same primary sequence and different posttranslational modifications, or proteins having the same N-terminal and different internal sequences.
Fig. 2.
Analyses of esterases separated by 10% native PAGE and visualized by α/β-naphthyl acetate assay. (a) Comparison of esterases expressed in olfactory and nonolfactory tissues suggests that PjapPDE (arrow) is specifically expressed in antennae. MA, 50 male antennae extract; ML, 5 legs extract. (b) Migration of purified native (NAT) and recombinant (REC) PjapPDE (50 antennae-equivalent of each) suggests similar posttranslational modifications.
Fig. 3.
Analysis of enzymatic activity of extracts from olfactory and nonolfactory antennal tissues (L, lamellae; F, flagella) by LC-ESI/MS (a) and gel electrophoresis (b). (a) The two points of the time course monitoring shown demonstrated a faster degradation of (R)-japonilure by lamellar extract and slow or almost no degradation by the extract from flagella, thus indicating that pheromone-degrading enzyme(s) is (are) expressed in lamellae. (b) Gel electrophoresis analysis of the same extracts compared with male antennae extracts (MA) showed that lamellae but not flagella express PjapPDE.
Fig. 4.
Purification of PjapPDE by anion exchange, gel filtration, and IEF chromatography. (a) PjapPDE (arrow) was separated from nonolfactory esterases by DEAE, recovered into three gel filtration fractions (b), and purified into a single protein by Mono Q (c). The distortion of the band (c) is caused by high salt concentration. Pure PjapPDE was separated by IEF (d) into five bands with the same N-terminal amino acid sequences. pI markers are indicated on the left of the IEF gel.
cDNA Cloning and PjapPDE Expression.
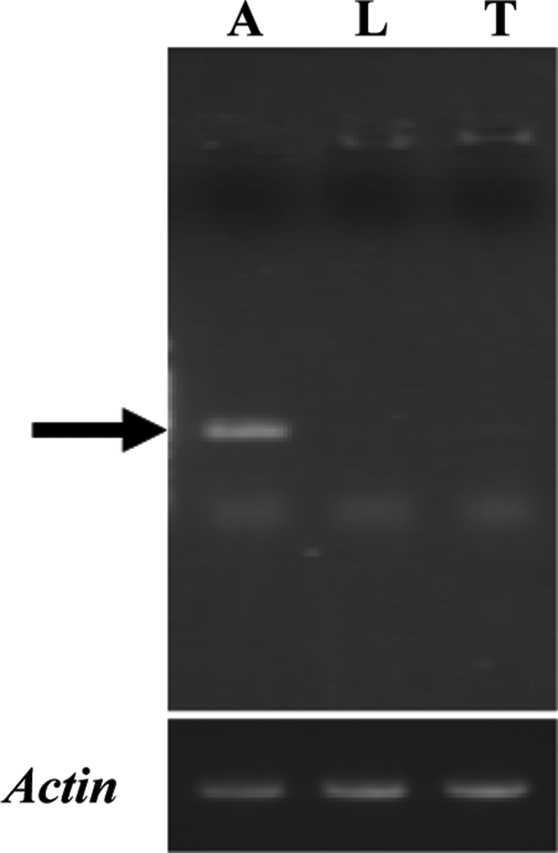
For cloning the cDNA(s) encoding PjapPDE, we used degenerate primers designed from the N-terminal sequence (NNPFPTLEI) and a conserved region in insect esterases (10). On the basis of the 500-bp partially amplified cDNA fragment we designed gene-specific primers and cloned the entire cDNA. The PjapPDE cDNA included 1,871 bp and encoded 554 aa residues (GenBank accession no. AY866482). Sequencing 16 independent clones verified that PjapPDE cDNA does not have any polymorphisms. Fifteen amino acid residues at the N-terminal region of the predicted mature protein were identical to those obtained from Edman degradation of the purified protein. The PjapPDE gene expression was detected by RT-PCR in the antennae but not in nonolfactory tissues, hindleg and thorax (Fig. 5). This antennal tissue-specificity determined by gene expression is in agreement with protein expression pattern (Figs. 2a and 3 and SI Fig. S3). We attempted to express PjapPDE by using bacterial expression to determine whether posttranslational modifications are important for enzymatic activity. Several attempts using two expression systems, different host cells, and innumerous experimental conditions were unsuccessful. We succeeded, however, in expressing PjapPDE by using the baculovirus expression system. Infection of the recombinant virus AcMNPV-PjapPDE induced a de novo esterase expression in the medium (Fig. 2b) but not in mock infection (data not shown). This baculovirus expression system generated enough samples for purification and kinetic studies, but small samples prevented accurate determination of protein amounts, probably because of difficulties related to expression of coleopteran esterase with lepidopteran cells. Thus, kinetic studies were carried out by using antenna-equivalent amounts of recombinant PjapPDE.
Fig. 5.
RT-PCR analyses of the expression of PjapPDE. Each cDNA pool was synthesized from male antennae (A), hindlegs (L), and thorax (T). The PjapPDE cDNA fragment (arrow) was amplified by using gene-specific primers. Actin gene expression was detected as an internal control.
Kinetics of Japonilure Degradation.
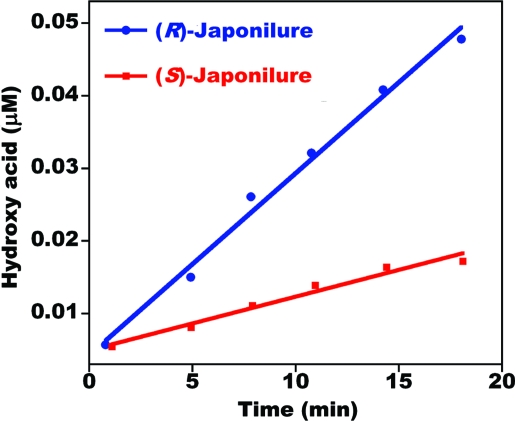
We studied the kinetics of pheromone inactivation with crude antennal extracts and recombinant PjapPDE by monitoring the production of the corresponding hydroxyacids. As indicated by the velocity of the reactions under the same conditions, PjapPDE showed substrate preference for the sex pheromone over a behavioral antagonist (Fig. 6). The kinetic parameters (Km ± SEM) and maximal velocities (Vmax ± SEM) obtained with isolated native PjapPDE for (R)-japonilure (Km, 754 ± 108 μM; Vmax, 3.1 ± 0.022 μM·min−1) and (S)-japonilure (Km, 812 ± 95 μM; Vmax, 1.1 ± 0.06 μM·min−1) confirmed a clear chiral discrimination with preference for the pheromone over the behavioral antagonist. Likewise, purified recombinant PjapPDE showed a preference for (R)-japonilure (Km, 660 ± 84 μM; Vmax, 2.7 ± 0.3 μM·min−1) and slower degradation of (S)-japonilure (Km, 858 ± 116 μM; Vmax, 1.3 ± 0.11 μM·min−1).
Fig. 6.
Degradation of enantiomers of japonilure by native PjapPDE. Both reactions were conducted under the same conditions by incubating 2.2 μM substrate with 50 antennae-equivalent of purified PjapPDE.
Discussion
The calculated molecular mass of PjapPDE (60,410 Da) is similar to that of purified PjapPDE (59 kDa) measured by gel filtration. The deduced amino acid sequence of PjapPDE included the conserved catalytic triad, Ser209, Glu338, His453, and salt bridges, Arg63, Glu101, Arg159, and Asp181 (11). The isoelectric point, pI 5.6, calculated from the deduced amino acid sequence was similar to that of five of the purified bands (Fig. 4d). The recombinant enzyme showed a migration similar to that of native PjapPDE, thus suggesting that expressing with insect cells led to similar posttranslational modification(s) (Fig. 2b). The amino acid sequence of PjapPDE contained three potential N-linked glycosylation sites. FASTA analyses indicated that PjapPDE showed 42% identities to a previously identified pheromone-degrading enzyme from the wild silk moth, Antheraea polyphemus, ApolPDE1 (GenBank accession no. AY866480; ref. 12).
Kinetic studies with both recombinant and native enzymes indicated that PjapPDE degrade both enantiomers of japonilure. Direct applications of the hydroxyacids into the sensillar lymph of pheromone-detecting sensilla on the Japanese beetle antennae showed that the degradation products of japonilure are inactive. Specifically, these hydroxyacids applied through recording electrodes did not stimulate ORNs that are tuned to the sex pheromone or a behavioral antagonist (A. A. Nikonov and W.S.L., unpublished data). Thus, degradation of japonilure by a sensillar esterase inactivates/terminates the chemical signals carried by both sex pheromone and behavioral antagonist. However, if enzymatic inactivation plays a significant role in olfaction, PjapPDE must degrade japonilure within milliseconds (13). In odorant-mediated flights, insects must reset their olfactory systems quickly to detect intermittent signals encountered while flying (14, 15). With the kinetic parameters adjusted for in vivo conditions, it has been estimated that the sex pheromone of the wild silkmoth has a half-life of 15 ms within the sensillar lymph (16). Kinetic studies with native and recombinant pheromone-degrading enzymes from the wild silkmoth confirmed the role of ApolPDE in rapid inactivation of the sex pheromone (12). To adjust the kinetic parameters (Vmax) obtained in vitro with native and recombinant PjapPDE to physiologically relevant values based on the in vivo enzyme concentration, we followed Vogt's assumption (16). Note that enzymatic activity in vitro is dramatically slower as nanoliters of sensillar lymph are diluted into milliliters in a reaction vial. Because of the limitations to estimate the volume of the irregular sensillar cavity in the Japanese beetle antennae (7), we also considered that the volume of sensillar lymph in the Japanese beetle to be equivalent to that of the wild silk moth [≈60 nl (16–19)]. By using Vmax adjusted for native PjapPDE, we estimate that the half-life of the sex pheromone (R)-japonilure, in the sensillar lymph to be 29.8 ms, whereas a behavioral antagonist has a half-life of 90.6 ms. The half-life of the sex pheromone and a behavioral antagonist estimated from the kinetic parameters obtained with recombinant PjapPDE were 30 and 81 ms, respectively.
It is an interesting feature of the Japanese beetle olfactory system that both sex pheromone and a behavioral antagonist are transported by the same pheromone-binding protein (6), detected by seemingly two different odorant receptors (6, 8), and inactivated by the same enzyme, PjapPDE. Although two odorant receptors seem to detect the sex pheromone and behavioral antagonist selectively, chiral discrimination of the enantiomers of japonilure by PjapPDE allows faster inactivation of the pheromone than the behavioral antagonist. Continuous resetting of the olfactory system is essential to report to the brain the detection of new signals encountered en route to a sex pheromone source. By contrast, ORNs reporting a “stop signal” may benefit from a slow recovery as the “old signal” would increase early rejection of a nonspecific female. Thus, the kinetic control of signal inactivation reported here is consistent with a peripheral mechanism underlining the physiological roles of enantiomers of japonilure in the chemical ecology of the Japanese beetle.
Materials and Methods
Isolation of PjapPDE.
Males of the Japanese beetle were collected in pheromone-baited traps in Ohio and Nebraska and kindly supplied by Mike Klein, and Fred Baxendale and Chelsey Wasen, respectively. Traps were emptied daily, beetles were frozen, packed in dry ice, and shipped overnight to Davis, CA, where they were stored at −80°C until use. To determine the site of PjapPDE expression, lamellae and flagella were collected independently to avoid cross-contamination. For protein isolation and identification, >100,000 antennae were collected with forceps under a microscope. Samples were homogenized in batches of 10,000–20,000 antennae with ice-cold 10 mM Tris·HCl (pH 8.0; hereafter Tris buffer) by using a 7-ml Dounce tissue grinder (Wheaton). After centrifugation (three times at 12,000 × g for 15 min) to precipitate debris, the supernatants were used for purification, native PAGE analysis, and/or kinetic studies. For protein isolation, the supernatants were loaded on a DEAE open column (Toyopearl 650S, 5-ml bed volume; Tosoh), which was flushed with 10 ml of Tris buffers containing increasing concentrations of NaCl (0–400 mM in 50 mM increments). The fractions were analyzed by native PAGE and visualized by the α/β-naphthyl acetate assay (18, 20, 21). Sensillar esterases, separated from integumental and other esterases, eluted with 100 and 150 mM NaCl. These fractions containing the antennae-specific esterase were loaded on a gel filtration column (Hiload Superdex 75; GE Healthcare) precalibrated with BSA, ovalbumin, chymotrypsinogen A, and ribonuclease, and eluted with 20 mM Tris·HCl (pH 8), 150 mM NaCl at 0.5 ml/min. The esterase(s) eluted from this column appeared in three fractions, with an estimated molecular mass of 59 kDa. The three fractions were combined, diluted with Tris buffer, and loaded on a high-performance ion exchange Mono Q column (HR10/10; GE Healthcare), which was eluted with a linear gradient of NaCl (0–400 mM at 2 ml/min for 40 min). The fraction containing the target band was desalted and concentrated with a centrifugal device (Microcon YM-30, nominal molecular mass, 30 kDa; Millipore), and analyzed by IEF (pH 3–7). Proteins were transferred to a poly(vinylidene difluoride) (PVDF) membrane, which was visualized with Coomassie blue. The N-terminal amino acid sequence of the five bands with pI values from 5.2 to 5.8 was obtained on a Procise protein sequencing system (Applied Biosystems) at the Molecular Structure Facility at the University of California, Davis.
cDNA Cloning and Sequence Analyses.
Two hundred frozen antennae were immediately homogenized in TRIzol (Invitrogen) to extract RNA. First-strand cDNA was synthesized from 1 μg of total RNA by a SMART RACE cDNA amplification kit (Clontech). Degenerate primers were designed on the basis of N-terminal amino acid sequence determined from the purified pheromone-degrading enzyme (NNPFPTLEI) and conserved region of insect esterases: PjapPDE-N, 5′-AA(C/T)AA(C/T)CC(A/C/G/T)TT(C/T)CC(A/C/G/T)AC(A/C/G/T)CT(A/C/G/T)GA(A/G)AT-3′; R-2, 5′-TG(A/G)TC(C/T)TTIA(A/G)ICC(A/G)TT(A/G)TTICC-3′ (I indicates inosine) (10). Seventy cycles of stepwise amplification program (94°C for 1 min, 40°C for 2 min, and 72°C for 3 min) were carried out. After gel purification, the 500 bp of PCR product was ligated at the EcoRV recognition site of pBluescript SK(+) (Stratagene) and sequenced at an automated DNA sequencing facility (Davis Sequencing). A whole cDNA fragment was amplified by using Advantage GC-2 polymerase mix (Clontech), and the gene-specific primers were designed on the basis of the 500 bp of partial cDNA fragment. The DNA sequence was determined by using 16 independent clones to exclude any artifact sequences derived by PCR. A putative signal sequence was identified by SignalP version 3.0 (www.cbs.dtu.dk) (22), and the predicted molecular mass and IEF point were calculated with PeptideMass (http://us.expasy.org) (23). N-linked glycosylation sites were searched with NetNGlyc 1.0 (www.cbs.dtu.dk/). Homology search and alignment of amino acid sequences were carried out with FASTA and ClustalW (www.ddbj.nig.ac.jp/) (24, 25).
RT-PCR.
Template cDNA pools for RT-PCR were prepared from antennae (described above), 10 hindlegs, and 1 thorax. We could not extract RNA from abdomen because of the high content of RNase. For detection of PjapPDE gene expression, the gene-specific primers PjapPDEf, 5′-CCACAAACAACCATGACGTTCCACGCCAGG-3′, and PjapPDEr, 5′-GGGCCTGCATAAAAGTGTCCACTTCCTAGC-3′, were used. The cDNA synthesized from 2.5 ng of total RNA equivalent was applied for each PCR. Twenty-five cycles of stepwise amplification program (94°C for 30 s, 55°C for 30 s, and 72°C for 1 min) were carried out. Detection of actin gene as a control was performed as described in ref. 10. PCR products were separated by agarose gel. Gel image was taken by Gel Print 2000i (BioPhotonics) and cropped by Photoshop 7.0 (Adobe).
Expression of Recombinant PjapPDE.
In an attempt to express PjapPDE, we tried 145 combinations of various expression conditions by using pET22-b(+) (Novagen) and pColdTF DNA (Takara) vectors, BL21(DE3), Rosetta2(DE3), BL21(DE3)pLysS, and Rosetta2(DE3)pLysS (Novagen) as host cells and various IPTG concentrations and incubation temperatures. For baculovirus expression, PjapPDE cDNA added with BamHI and XhoI recognition sites at the 5′ and 3′ ends, respectively, was ligated into the recombinant donor plasmid, pFastBac1 (Invitrogen). After confirmation of the DNA sequence, recombinant bacmid DNA was obtained according to protocols provided by Invitrogen. The confirmed bacmid DNA was transfected into Sf21 cells by using Cellfectin reagent (Invitrogen). The recombinant virus AcMNPV-PjapPDE was collected and used to inoculate Sf21 cells in Sf-900 II SFM medium (Invitrogen). After incubation at 28°C for 72 h, the medium was harvested and exchanged with 10 mM Tris·HCl (pH 8.0) by using a Microcon YM-10 filtration device (Millipore). Recombinant PjapPDE was purified as described above for the purification of the native enzyme.
Analytical Methods and Kinetic Studies.
LC-ESI/MS was performed with a LCMS-2010 (Shimadzu). Synthetic samples of (R)-japonilure (S)-japonilure, 4-(R)-hydroxy-5-tetradecenoic acid and 4-(S)-hydroxy-5-tetradecenoic acid were prepared as described in ref. 26. When analyzed in negative scan mode, these hydroxyacids gave essentially a base peak at m/z 241. The MS detector, operated in the selected ion (m/z 241) mode, with the curve desolvation line at 250°C, heat block at 200°C, nebulizer at 1 liter/min, and 0.1 ml/min, 50% methanol solution, gave a linear response, with very low threshold (Fig. S1). Reactions could be monitored by injections of crude mixtures because the background signal at m/z 241 generated by incubation of japonilure with reaction buffer only or even buffer alone (SI Fig. S2a) was very small and constant over time (SI Fig. S2b). To minimize problems with debris and allow direct injections, before the reactions the protein samples were filtered (HYCPTFE, 0.2 μm; Microcon). Reactions were carried out in glass vials deactivated by Silicote CL7 treatment (Kimble). To each reaction mixture (50 μl) consisting of 50 antennae-equivalent of native or recombinant PjapPDE in 20 mM Tris buffer was added 1 μl of ethanol solution to make concentrations of (R)- or (S)-japonilure ranging from 1 μM to 1.5 mM. To determine chiral discrimination accurately, the concentrations of the substrates were adjusted by gas chromatography. Thus, samples of (R)-japonilure and (S)-japonilure with the same nominal concentration were at least 99% accurate. Each reaction was repeated three to five times. The reactions were incubated at 25°C, and 1-μl aliquots were injected over time. Velocity was calculated by the production of hydroxyacid per time (n = 3–5). Kinetic parameters (Vmax, Km) were determined by Lineweaver–Burk plots (double reciprocal, 1/v versus 1/[S]). Because of the low yields of pure recombinant PjapPDE, it was not possible to determine the amount of enzyme per reaction. Thus, the concentrations of the samples were adjusted by native gels visualized with α/β-naphthyl acetate assay to have 50 antennae-equivalents per reaction.
Supplementary Material
Acknowledgments.
We thank Mike Klein (U.S. Department of Agriculture–Agricultural Research Service) and Fred Baxendale and Chelsey Wasen (University of Nebraska–Lincoln) for beetle collection; Brooke P. Pannell, Melissa C. Hardstone, Vicky P. Chiang, Angela M. Chen, Li Ning, Chunpei Wang, and Jesse G. Livingston (University of California, Davis, CA) for tissue collection; Robert Classon and Kara Merkle (Shimadzu) for valuable discussion on analytical method development; and Zain Syed, Karl-Ernst Kaissling, and S. George Kamita for critique of an earlier version of the manuscript. This work was supported by the National Research Initiative of the U.S. Department of Agriculture USDA-CSREES 2003-35302-13648 and National Science Foundation Grant 0234769.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY866482).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802610105/DCSupplemental.
References
- 1.Butenandt A, Beckmann R, Stamm D, Hecker E. On the sex pheromone of the silk moth Bombyx mori: Preparation in a pure state and constitution. Z Naturforsch B. 1959;14:283–284. [Google Scholar]
- 2.Silverstein RM, Rodin JO, Wood DL, Browne LE. Identification of two new terpene alcohols from frass produced by Ips confusus in ponderosa pine. Tetrahedron. 1996;22:1926–1936. [Google Scholar]
- 3.Seybold SJ. Role of chirality in olfactory-directed behavior: Aggregation of pine engraver beetles in the genus Ips (Coleoptera: Scarabaeidae) J Chem Ecol. 1993;19:1809–1831. doi: 10.1007/BF00982310. [DOI] [PubMed] [Google Scholar]
- 4.Tumlinson JH, Klein MG, Doolittle RE, Ladd TL, Proveaux AT. Identification of the female Japanese beetle sex pheromone: Inhibition of male response by an enantiomer. Science. 1997;197:789–792. doi: 10.1126/science.197.4305.789. [DOI] [PubMed] [Google Scholar]
- 5.Leal WS. Chemical communication in scarab beetles: Reciprocal behavioral agonist–antagonist activities of chiral pheromones. Proc Natl Acad Sci USA. 1996;93:12112–12115. doi: 10.1073/pnas.93.22.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wojtasek H, Hansson BS, Leal WS. Attracted or repelled? A matter of two neurons, one pheromone binding protein, and a chiral center. Biochem Biophys Res Commun. 1998;250:217–222. doi: 10.1006/bbrc.1998.9278. [DOI] [PubMed] [Google Scholar]
- 7.Kim J-Y, Leal WS. Ultrastructure of pheromone-detecting sensillum placodeum of the Japanese beetle, Popillia japonica Newmann (Coleoptera: Scarabaeidae) Arthropod Struct Dev. 2000;29:121–128. doi: 10.1016/s1467-8039(00)00022-0. [DOI] [PubMed] [Google Scholar]
- 8.Nikonov AA, Leal WS. Peripheral coding of sex pheromone and a behavioral antagonist in the Japanese beetle, Popillia japonica. J Chem Ecol. 2002;28:1075–1089. doi: 10.1023/a:1015274104626. [DOI] [PubMed] [Google Scholar]
- 9.Leal WS. In: Semiochemicals in Pest and Weed Control. Petroski RJ, Tellez MR, Behle RW, editors. Washington, DC: American Chemical Society; 2005. pp. 45–57. [Google Scholar]
- 10.Ishida Y, Leal WS. Cloning of putative odorant-degrading enzyme and integumental esterase cDNAs from the wild silkmoth, Antheraea polyphemus. Insect Biochem Mol Biol. 2002;32:1775–1780. doi: 10.1016/s0965-1748(02)00136-4. [DOI] [PubMed] [Google Scholar]
- 11.Cygler M, et al. Relationship between sequence conservation and three-dimensional structure in a large family of esterases, lipases, and related proteins. Protein Sci. 1993;2:366–382. doi: 10.1002/pro.5560020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishida Y, Leal WS. Rapid inactivation of a moth pheromone. Proc Natl Acad Sci USA. 2005;102:14075–14079. doi: 10.1073/pnas.0505340102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaissling K-E. Olfactory perireceptor and receptor events in moths: A kinetic model. Chem Senses. 2001;26:125–150. doi: 10.1093/chemse/26.2.125. [DOI] [PubMed] [Google Scholar]
- 14.Murlis J. In: Insect Pheromone Research: New Directions. Cardé RT, Minks AK, editors. New York: Chapman & Hall; 1997. pp. 221–247. [Google Scholar]
- 15.Murlis J, Willis MA, Cardé RT. Spatial and temporal structures of pheromone plumes in fields and forests. Physiol Entomol. 2000;25:211–222. [Google Scholar]
- 16.Vogt RG, Riddiford LM, Prestwich GD. Kinetic properties of a sex pheromone-degrading enzyme: The sensillar esterase of Antheraea polyphemus. Proc Natl Acad Sci USA. 1985;82:8827–8831. doi: 10.1073/pnas.82.24.8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keil TA. Reconstruction and morphometry of silkmoth olfactory hairs: A comparative study of sensilla trichodea on the antennae of male Antheraea polyphemus and Antheraea pernyi (Insecta: Lepidoptera) Zoomorphology. 1984;104:147–156. [Google Scholar]
- 18.Klein U. Sensillum-lymph proteins from antennal olfactory hairs of the moth Anthearea polyphemus (Saturniidae) Insect Biochem. 1987;17:1193–1204. [Google Scholar]
- 19.Meng LZ, Wu CH, Wicklein M, Kaissling K-E, Bestmann HJ. Number and sensitivity of three types of pheromone receptor cells in Antheraea pernyi and A polyphemus. J Comp Physiol A. 1989;165:139–146. [Google Scholar]
- 20.Shaw CR, Prasad R. Starch gel electrophoresis of enzymes: A complication of recipes. Biochem Genet. 1970;4:297–320. doi: 10.1007/BF00485780. [DOI] [PubMed] [Google Scholar]
- 21.Vogt RG. In: Pheromone Biochemistry. Prestwich GD, Blomquist GJ, editors. Orlando, FL: Academic; 1987. pp. 385–431. [Google Scholar]
- 22.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Wilkins MR, et al. Detailed peptide characterization using PEPTIDEMASS: A World-Wide-Web-accessible tool. Electrophoresis. 1997;18:403–408. doi: 10.1002/elps.1150180314. [DOI] [PubMed] [Google Scholar]
- 24.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson JD, Higgins DG, Gibson TJ. ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leal WS. Enantiomeric anosmia in scarab beetles. J Chem Ecol. 1999;25:1055–1066. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.