Abstract
Protein synthesis (PS) has been considered essential to sustain mammalian life, yet was found to be virtually arrested for weeks in brain and other organs of the hibernating ground squirrel, Spermophilus tridecemlineatus. PS, in vivo, was below the limit of autoradiographic detection in brain sections and, in brain extracts, was determined to be 0.04% of the average rate from active squirrels. Further, it was reduced 3-fold in cell-free extracts from hibernating brain at 37°C, eliminating hypothermia as the only cause for protein synthesis inhibition (active, 0.47 ± 0.08 pmol/mg protein per min; hibernator, 0.16 ± 0.05 pmol/mg protein per min, P < 0.001). PS suppression involved blocks of initiation and elongation, and its onset coincided with the early transition phase into hibernation. An increased monosome peak with moderate ribosomal disaggregation in polysome profiles and the greatly increased phosphorylation of eIF2α are both consistent with an initiation block in hibernators. The elongation block was demonstrated by a 3-fold increase in ribosomal mean transit times in cell-free extracts from hibernators (active, 2.4 ± 0.7 min; hibernator, 7.1 ± 1.4 min, P < 0.001). No abnormalities of ribosomal function or mRNA levels were detected. These findings implicate suppression of PS as a component of the regulated shutdown of cellular function that permits hibernating ground squirrels to tolerate “trickle” blood flow and reduced substrate and oxygen availability. Further study of the factors that control these phenomena may lead to identification of the molecular mechanisms that regulate this state.
Mammalian hibernation represents a state in which global physiologic functions are virtually arrested and delivery of glucose and oxygen is minimal, yet homeostatic control is maintained. The profound reduction of cerebral perfusion in hibernation (1) would lead to rapid autolysis of brain tissue (2) in the absence of adjustments that characterize hibernation, but has no adverse effects on hibernators. In fact, hippocampal slices from hibernating ground squirrels show increased tolerance to a superimposed insult of hypoxia and aglycemia even at 36°C (3). Surprisingly, the cellular mechanisms and signals that trigger and maintain this adaptation remain unknown. The identification of hibernation-specific changes in cellular functions and of the regulatory mechanisms employed by this tolerant state therefore may lead to the discovery of clinically effective measures that induce cellular resistance to insults such as brain ischemia and trauma in humans.
Whereas metabolic suppression (4–7) and membrane functions (8, 9) in hibernators as well as anoxia-tolerant species have been the focus of recent research, the regulation of another major cellular process, protein biosynthesis, has been less well explored. Previously, preservation of mammalian cell structure and function was considered to be critically dependent on the continuous formation of new proteins and peptides especially under circumstances of reduced cerebral perfusion, in which lack of recovery of protein synthesis (PS) has been viewed as a reliable predictor of cell death (10, 11). We have identified PS as a major cellular process that is actively regulated and rapidly arrested during hibernation by mechanisms that involve inhibition of protein initiation and elongation. This suppression is physiologic for the state yet incompatible with prevailing biologic concepts, and it appears to constitute a regulated step in the coordinated shutdown of cellular functions and associated neuroprotection in hibernation.
METHODS
Animal Preparation and Hibernaculum.
Thirteen-lined ground squirrels, Spermophilus tridecemlineatus, were captured by a United States Department of Agriculture-licensed trapper (TLS Research, Bartlett, IL). All experiments were approved by the National Institute of Neurological Disorders and Stroke Animal Care and Use Committee. Squirrels were chronically implanted with arterial and venous catheters and multichannel telemetry transmitters (TL10M3-F50-EET, Datasciences, Minneapolis) for long-term monitoring of the electrocardiogram and body temperature (1). The animals were housed in an environmental chamber kept at 5°C at 60% humidity and constant darkness to provide external cues conducive to hibernation. Immobility, a curled posture, and profound reductions in heart rate and body temperature characterized hibernation (1).
l-[1-14C]leucine Autoradiography.
Autoradiographic determination of the rate of protein synthesis in brain was performed as described previously (12, 13). Active, cold-adapted squirrels were transferred to tube restrainers. Hibernating animals were not touched during blood sampling and were monitored telemetrically. After intravenous injection of l-[1-14C]leucine (100 μCi/kg body weight in normal saline), timed arterial samples were taken for 2 hr to determine the time courses of labeled and unlabeled leucine concentrations in plasma. The animals were killed by a barbiturate overdose, and brains were removed for cryostat sectioning. Autoradiographic images were analyzed densitometrically (nih image). Rates of leucine incorporation into protein were calculated from local tissue concentrations of 14C-labeled protein, the integrated specific activity of the acid-soluble leucine in plasma, and λ, the fraction of unlabeled leucine in the tissue precursor pool that comes from plasma; the value of λ of 0.58 determined in the normal conscious rat was used (12, 13).
Protein Extraction from Whole Tissues and Separation by SDS/PAGE.
Organs were homogenized in 5% trichloroacetic acid (TCA). Pellets were washed five times in 5% TCA (0°C) and then suspended in hot TCA (5%, 90°C) for 15 min. After centrifugation, pellets were washed once in ethanol and then twice in ethanol-ether-chloroform (2:2:1 by volume) and twice in acetone. Dried pellets were hydrolyzed in 0.1 M NaOH for scintillation counting. Protein content was determined by the Bradford method (Pierce). For SDS/PAGE, l-[35S]methionine (2.5 mCi in normal saline) was injected i.v. in one active and one hibernating squirrel. The animals were killed 2 hr after injection, and proteins were extracted from brain, liver, and heart as described above. Discontinuous SDS/PAGE was carried out (14), applying 25 μg protein in 5 μl to each lane. After staining with Coomassie blue, gels were exposed on high-resolution phosphorimaging plates for 10 days and analyzed by a Fuji BAS 3000 imaging system (Fuji).
In Vitro Translational Assay.
Brain postmitochondrial supernatants (PMS) were prepared and adjusted for protein content (10 mg/ml) for initiation of in vitro translation as described previously (15). The reaction was started by the addition of 5 μCi/ml l-[3,4,5-3H(N)]leucine (specific activity, 180 Ci/mmol) at 37°C and stopped after 20–30 min by the addition of 10% TCA. The TCA-extracted protein pellet was hydrolyzed in 1 M NaOH for scintillation counting. Rates of leucine incorporation were adjusted for endogenous leucine concentrations determined by amino acid analysis (Model 7300, Beckman). Background was defined as the signal that remained after inhibition of protein synthesis by preparing the reaction mixture with 5 mM cycloheximide.
Quantitative Northern Hybridization.
Total RNA was extracted from active (n = 5) and hibernating (n = 3) brain extracts by the phenol/guanidinium thiocyanate-phenol method (STAT60, Tel-Test, Friendswood, TX) and analyzed by Northern blotting with probes to the mRNAs for cyclophilin, glyceraldehyde-3-phosphate dehydrogenase, calmodulin, and mitochondrial F1-ATPase subunits 6 and 8. Each of these mRNAs had been shown previously to be present at equivalent levels in RNA extracted directly from whole brains of hibernating and active ground squirrels (unpublished data).
Polysome Profiles.
Ribosomes were isolated by centrifugation of brain extracts through 2 M sucrose in hypotonic buffer (300,000 × g × 3 hr), and 3–4 OD260 units of purified ribosomes were centrifuged through a linear 15–55% (wt/vol) sucrose gradient (16). The absorbance profiles of the effluent fractions were measured at OD260, and relative concentrations of RNA were determined planimetrically.
eIF2α Phosphorylation.
Samples containing 50 μg protein were subjected to electrophoresis on 12.5% SDS-polyacrylamide gels followed by transfer to nitrocellulose (0.45 μm). Phosphorylated eIF2α was detected with a 1:1,000 dilution of rabbit polyclonal antibody specific for the phosphorylated form of eIF2α as described previously (17). Total eIF2α was detected with a 1:500 dilution of mouse mAb (18). Immunoreactive bands were detected by enhanced chemiluminescence (ECL, Amersham). To quantify the change in eIF2α phosphorylation, isoelectric focusing gels were used to separate phosphorylated eIF2α from the basal form as described previously (19). Crude protein extracts from active and hibernating squirrel brains were focused by using the same reagents and protocols established for analyzing yeast eIF2α (20). After focusing, eIF2α was detected by immunoblotting with eIF2α mAbs described previously (18).
Transit Time Determinations.
Protein elongation rates were determined as described previously (21). In vitro translation of PMS was carried out as above and stopped at 1-min intervals after pulse-labeling with l-[3,4,5-3H(N)]leucine by the addition of 5 mM cycloheximide and cooling to 4°C. Samples were divided for determination of radioactivity in the TCA-precipitable material from the total sample (completed and nascent chains) and from the supernatant only (soluble, completed chains) obtained after deposition of ribosomes by centrifugation. For determination of the size distribution of proteins, in vitro translation was stopped 20 min after labeling. After TCA extraction, SDS/PAGE was carried out (40 μg protein/lane) in 25 mM Tris/glycine (pH 8.3) polyacrylamide gels (4–20% linear gradient). Four lanes were pooled per determination and cut into 20 sequential pieces. Each piece was hydrolyzed with 1 M NaOH before scintillation counting. The molecular weight range for each gel piece was determined based on its position relative to standards.
Determination of the Presence of ADP-Ribosylated Elongation Factor-2 (eEF2).
The level of ADP-ribosylated eEF2 in both hibernating and active brains from ground squirrels was determined according to the method of Youle and Neville, Jr. (22). One gram of brain tissue was pulverized in the presence of liquid nitrogen and homogenized in 1 ml of homogenizing buffer [35 mM KHCO3/20 mM K2HPO4/4 mM MgCl2/25 mM KCl/2 μg/ml leupeptin/2 μg/ml aprotinin/50 μg/ml phenylmethylsulfonyl fluoride (PMSF), adjusted to pH 7.4 with HCl] with a motorized Teflon homogenizer for 15 strokes. The extract was centrifuged at 120,000 × g (4°C), and the supernatant fraction was collected. For removal of endogenous NAD, an aliquot (0.4 ml) of the supernatant fraction was applied to a Sephadex G-25 column (1.4 × 5 cm) equilibrated with 10 mM Tris⋅HCl, pH 8.0/1 mM EDTA/2 μg/ml leupeptin/2 μg/ml aprotinin/50 μg/ml PMSF. The eluted fraction containing the protein (0.5 ml) was used to assay for ADP-ribosylated eEF2.
A three-tube assay was used to analyze the level of ADP-ribosylated eEF2 (22). Each tube contained a final volume of 62 μl:50 μl of the protein fraction collected from Sephadex G-25, 20 mM DTT, and 1 μM [32P]NAD (2,000–20,000 cpm/pmol). To one tube, 0.1 μg/ml CRM107 [a mutant of diphtheria toxin (23)] was included, and to another tube, 0.1 μg/ml CRM 107 + 100 μM nicotinamide was included. The reactions were started with the addition of [32P]NAD and stopped by precipitation with 62 μl of 10% TCA after 30 min at 22°C. Samples were stored on ice for 2 hr or at 4°C overnight. The precipitated protein from each reaction was collected on Whatman GF/C glass-fiber filters, washed four times with 2 ml of 5% TCA, dried with 2 ml of methanol, and counted in a scintillation counter (Wallac 1409, Gaithersburg, MD).
RESULTS
Absence of Autoradiographic Evidence for Leucine Incorporation in Vivo in Brain from Hibernating Squirrels.
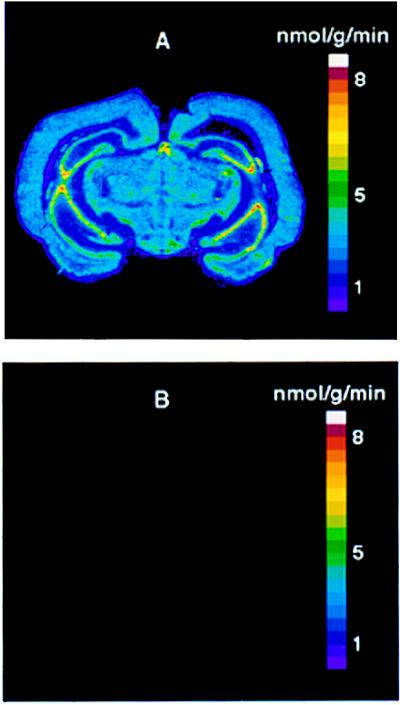
In an effort to identify brain regions that might be involved in the control of hibernation, we determined rates of PS with the quantitative autoradiographic l-[1-14C]leucine method (12, 13). In an active ground squirrel, the autoradiographic distribution and local rates of cerebral PS (Fig. 1A) were comparable to those of other rodents (12), ranging from 7.5 nmol leucine incorporation per g/min in the hippocampus to 1.5 nmol per g/min in the corpus callosum. During hibernation, no autoradiographic evidence of leucine incorporation was found in brain sections (4 months exposure on Eastman Kodak SB-5 film) (Fig. 1B). Undetectable incorporation of [14C]leucine into protein was confirmed in two additional hibernating squirrels.
Figure 1.
Color-coded transforms of representative autoradiograms, showing rates of cerebral PS in an active (A) and a hibernating (B) ground squirrel at the level of the arcuate nucleus. During hibernation, no autoradiographic evidence of leucine incorporation was detected.
Residual PS in Whole Brain, Heart, and Liver in Vivo.
We verified this autoradiographic result and determined the precise rates of cerebral PS in three additional hibernating and three active squirrels by extracting and counting brain protein after the [14C]leucine procedure. Analyses also were done on heart and liver of two hibernating and one active animal. One experiment was conducted during entrance into hibernation; body temperature in this animal decreased from 19.0°C to 7.5°C and heart rate dropped from 65 beats/min (bpm) to 24 bpm during this 2-hr experiment. Although cerebral PS during hibernation could not be detected autoradiographically, some residual level of [14C]leucine incorporation could be measured consistently in brain homogenate. PS rates in the hibernating squirrels were approximately 0.04% of average rates measured in active squirrels (Table 1). Importantly, during entrance into hibernation, while the declining body temperature averaged above steady-state levels in hibernation (6–7°C), the rate of PS already was as low as during full hibernation. This indicates that hypothermia was not the sole reason for the reduction in PS. Previous studies aimed at characterizing PS in hibernation have reported much more modest reductions (to 10% of active controls) in rates of amino acid incorporation (24, 25). However, in these prior studies the input function was not determined and no model for the behavior of the tracer amino acids was used, which compromises quantitative analysis.
Table 1.
In vivo rates of leucine incorporation into protein in active and hibernating ground squirrels and during entrance into hibernation (values are mean ± SD)
| Active | Hibernating, per nmol/100 mg protein per min | Entrance | |
|---|---|---|---|
| Brain | 3.0 ± 0.2 | 0.001 ± 0.001 | 0.001 |
| (n = 3) | (n = 3) | ||
| Liver | 6.4 | 0.006 ± 0.002 | |
| (n = 2) | |||
| Heart | 3.9 | 0.004 ± 0.002 | |
| (n = 2) |
Specific Species of Proteins That Account for the Residual Level of PS Could Not Be Identified.
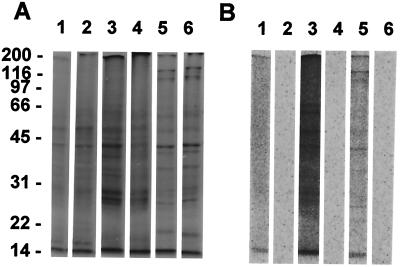
In an effort to determine whether the proteins synthesized during hibernation differ from those in active animals, protein extracts from brain, liver, and heart were subjected to SDS/PAGE. l-[35S]methionine was used as the tracer amino acid for these studies because its specific activity is 20 times higher than that of [14C]leucine and concentrations of individual, newly synthesized proteins had been noted to be too low to be detected by incorporation of 14C-labeled leucine. In active animals, protein extracts were separated into smears with multiple discrete bands that stained with Coomassie blue and corresponded to the autoradiographic image of the radioactively labeled proteins (Fig. 2). The pattern of Coomassie blue-stained protein from hibernating squirrel tissues was similar to that found in the active state, but produced no autoradiographic image. Any synthesis of specific proteins during hibernation is beneath the sensitivity of this method of detection.
Figure 2.
SDS/PAGE separation of proteins in brain, liver, and heart from a hibernating and an active squirrel. A shows the Coomassie-stained gel and B shows the autoradiogram produced by l-[35S]methionine incorporation. During hibernation, incorporation was below the detection limit of the PhosphorImager (1, brain/active; 2, brain/hibernating; 3, liver/active; 4, liver/hibernating; 5, heart/active; 6, heart/hibernating).
PS Is Suppressed in Vitro Independent of Temperature.
To test the hypothesis that suppression of PS during hibernation is not merely a passive thermodynamic phenomenon, we have studied PS at 37°C by means of an in vitro translational system in PMS prepared from brains of active and hibernating ground squirrels. Throughout 30 min of in vitro translation, much less l-[3,4,5-3H(N)]leucine was incorporated in PMS from hibernating brain as compared with active brain (Fig. 3). Incorporation could be blocked by cycloheximide and reduced to about half by a specific inhibitor of initiation, 7-methyl-GTP (26, 27) (data not shown), indicating that incorporation was indeed due to PS, with a significant contribution from in vitro initiation of translation. Quantitatively, in vitro rates of leucine incorporation at 37°C were about three times higher in active animals (0.47 ± 0.08 pmol/mg protein per min, n = 7) than in hibernators (0.16 ± 0.05 pmol/mg protein per min, n = 7, P < 0.001, Student’s t test).
Figure 3.
In vitro translation in PMS at 37°C. After introduction of l-[3,4,5-3H(N)]leucine into the medium, the reaction was allowed to proceed for 30 min. Incorporation of leucine into TCA-precipitable material was several times higher in PMS from active (Left) compared with PMS from hibernating (Right) brain (−CHX). Incorporation was blocked by 5 mM cycloheximide (+CHX).
Levels of Several Ubiquitous mRNAs Are Not Altered During Hibernation.
A temperature-independent reduction of PS could be the result of modifications of transcription, mRNA, translational factors, or ribosomes. Levels of mRNA in the in vitro translational system were compared by quantitative Northern blot analyses for several ubiquitous messages. Similar to whole brain (data not shown), the mRNA levels determined in PMS from three active and three hibernating brains did not differ (Table 2).
Table 2.
Ratios of four ubiquitous mRNA levels in PMS from active (NH) and hibernating (H) squirrels (n = 3 each; values are ratios NH/H) determined by Phosphor/Image analysis of Northern blots
| Calmodulin | Mitochondrial ATPase | Cyclophilin | GAPDH |
|---|---|---|---|
| 1.04 | 0.80 | 1.11 | 0.95 |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Hibernation Induces Moderate Ribosomal Disaggregation.
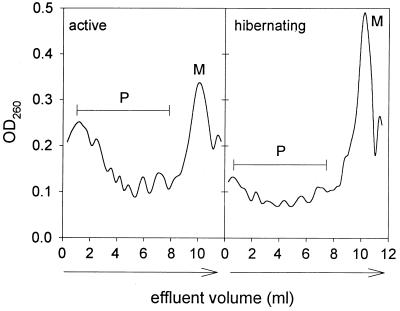
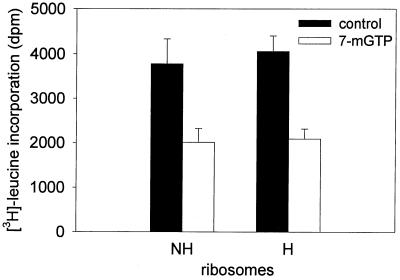
Since the aggregation state of ribosomes may serve as an indicator of the translational activity of the protein-synthesizing machinery, polysome profiles were prepared from PMS of hibernating and active brains. During hibernation, the monosomal peak was increased and the polysome fraction was decreased compared with profiles from active brains, indicating moderate ribosomal disaggregation (Fig. 4). Additional independent profiles confirmed a significant decrease in the polysome fraction during hibernation (48 ± 5% of total profile area, n = 6) compared with active brain (64 ± 8% of total profile area; n = 6, P < 0.002, Student’s t test). To test whether ribosomes from hibernating brain lack the ability to initiate, PMS from active brain was processed further to remove ribosomes and then used to assay the translational activity of ribosomes isolated from active and hibernating brain. No difference was seen in the incorporation of [3H]leucine by hibernating compared with active ribosomes (Fig. 5), either in the absence or presence of 7-methyl-GTP. This result indicates that the ribosomal component of the protein-synthesizing machinery was intact. The moderate ribosomal disaggregation during hibernation is compatible with the presence of a ribosomal inhibitor that interferes with the formation of the initiation complex.
Figure 4.
Analyses of representative ribosomal profiles in PMS from hibernating and active brains. PMS was analyzed by means of a linear 15–55% sucrose gradient. Note the moderate disaggregation of the polysomal fraction (P) and increase of the monosomal peak (M) in PMS from hibernating brain.
Figure 5.
In vitro translational capability of ribosomes isolated from hibernating and active brains. Ribosomes from hibernating (n = 3) and active (n = 3) brains were incubated in ribosome-depleted PMS from active brain. The ribosomes from the two sources showed comparable levels of leucine incorporation, both in the presence and absence of 1 mM 7-methyl-GTP. RNA content was adjusted to 1 OD260unit/assay. Bars represent the incorporation of [3H]leucine into protein (mean ± SD).
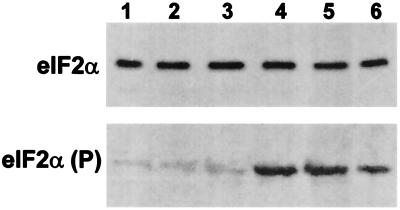
eIF2α Phosphorylation Is Greatly Increased During Hibernation.
Initiator methionyl-tRNA is introduced into the translation–initiation complex by eIF2, and phosphorylation of the α-subunit of eIF2 is known to inhibit global protein synthesis (28). Phosphorylation of serine 51 of the α-subunit generates a stable eIF2-GDP complex that binds the recycling protein, eIF2B, and prevents exchange of bound GDP for GTP, a necessary step for each round of translation initiation. On this basis, we analyzed eIF2α phosphorylation by Western blotting. As shown in Fig. 6, the total amount of eIF2α is unchanged, but the level of phosphorylation of eIF2α is greatly increased during hibernation. Isoelectric focusing gels were used to separate phosphorylated eIF2α from the basal form to quantify the change in eIF2α phosphorylation. Densitometric scanning and quantification of the results revealed that approximately 13% of the eIF2α in brains from hibernating squirrels was phosphorylated compared with less than 2% of the eIF2α in brains from active animals. Although the level of eIF2α phosphorylation observed in the brain tissue extracts from hibernating animals is consistent with a translation–initiation defect in these tissues (29) and this level of phosphorylation has been associated with substantial inhibition of translation initiation in several different cell types (30, 31), the observation that the molar ratio of eIF2 to eIF2B varies among tissues (32) dictates caution in attributing a substantial degree of the translation inhibition in the hibernating squirrels to eIF2α phosphorylation.
Figure 6.
Immunoblots of total (Upper) and phosphorylated (Lower) eIF2α in brain homogenates from active (lanes 1–3) and hibernating (lanes 4–6) squirrels.
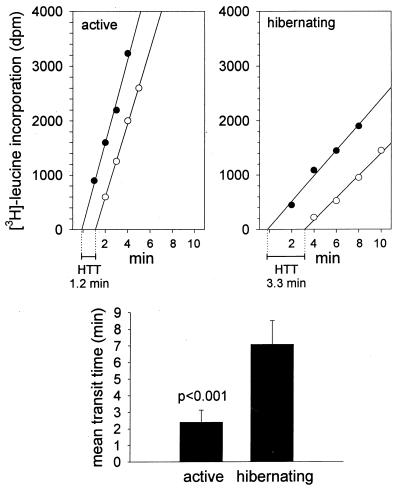
The Rate of Protein Elongation Is Slowed During Hibernation.
To specifically test for the presence of an elongation defect, mean translation rates (mean transit times) were determined in PMS from active and hibernating brain by measuring the flow of radioactive material from nascent polypeptide chains to completed protein. The measured variable, mean transit time, is independent of ribosome attachment, i.e., initiation rate. Mean transit times were increased about 3-fold in hibernating brain extracts (7.1 ± 1.4 min, n = 5) compared with active brains (2.4 ± 0.7 min, n = 5, P < 0.001, Student’s t test) (Fig. 7). These results indicate the presence of an elongation block in hibernation. Protein size distributions determined by SDS/PAGE of labeled protein from the hibernating and active brain extracts were similar, supporting the validity of the transit time comparisons (data not shown).
Figure 7.
Mean transit time determinations in PMS from active and hibernating squirrel brain. Incorporation of radioactivity in total (solid circles) and soluble (open circles) protein from a representative experiment is plotted against time (Upper). The difference between the time axis intercepts of the linear regression lines corresponds to a one-half transit time (HTT). (Lower) Ribosome transit times (mean ± SD) measured in extracts from five active and five hibernating squirrel brains were significantly different by Student’s t test.
ADP Ribosylation of eEF2.
Since the elongation rate of protein biosynthesis is decreased during hibernation, we looked at one of the key factors involved in the translocation process during polypeptide chain elongation, eEF2. eEF2 contains a unique posttranslationally modified histidine residue, named diphthamide, that is conserved throughout eukaryotic species, suggesting it has an essential role in cellular metabolism (33–35). ADP ribosylation of eEF2, specifically at the diphthamide residue, either endogenously (36, 37) or by bacterial toxins (38, 39), has been reported to cause inactivation of eEF2 with respect to its role in protein biosynthesis. With this connection, it seemed plausible that endogenous ADP ribosylation of eEF2 could be a molecular switch for controlling protein biosynthesis during variable metabolic states within the cell. A comparison between three sets of active versus hibernating extracts revealed no detectable ADP ribosylation of eEF2, however, and thus, less than 10%, if any, eEF2 is ADP-ribosylated during hibernation.
DISCUSSION
The principal finding of this study is that PS in the brain of hibernating ground squirrels is actively arrested for up to several weeks at a time without an absolute requirement for hypothermia and therefore may constitute an important component in the induction of cellular quiescence during hibernation. Active regulation of PS suppression in hibernation is demonstrated by the findings of increased phosphorylation of the initiation factor, eIF2α, and the persistence of PS suppression during translation in vitro at physiologic temperatures. Further, PS in vivo was maximally suppressed during entrance into hibernation while body temperature still averaged above steady-state temperature in hibernation. In addition, thermodynamic effects are unlikely to explain the approximate 2,500-fold reduction in PS. If Q10 effects (the fold-reduction in rate of biochemical reaction for each 10°C reduction in body temperature) regulated PS, the calculated Q10 during hibernation would approximate 13.6 and would be even higher during the entrance phase (16.3), whereas Q10 values normally range from 2 to 3 for most enzymes and tissues. In comparison, the rate of cerebral glucose metabolism during hibernation was reduced to a lesser degree, 1–2% of the values in active animals (total reduction of about 75-fold), corresponding to a Q10 of 4.3 (40).
The temperature-independent component of the PS reduction could result from modifications in transcription, mRNA, translational factors, or ribosomes. We have confirmed previous findings (41) of no differences between hibernating and active animals in total mRNA levels in brain and liver. This greatly reduces the likelihood that global suppression of PS occurs at the level of transcription. It does not rule out differential expression of specific mRNAs during hibernation, which has, in fact, been documented (42, 43). Furthermore, we have found no evidence that the function of the ribosomal machinery is perturbed during hibernation. The decreased polysome fraction in the polysome profile suggested that factors regulating the rate of initiation could be inhibited, and this was strongly supported by the large increase in phosphorylated eIF2α seen in hibernating animals. In addition, suppression of PS in hibernation occurs during elongation as indicated by markedly slowed protein elongation rates in vitro independent of temperature. Translational control of gene expression generally has been thought to occur at the level of initiation (28), but may also involve modifications of the elongation machinery. Phosphorylation of eEF2 can lead to such an arrest (44), which is usually only short-lived (minutes). eEF2 phosphorylation is postulated to be a separate signal-transduction pathway, the “translational arrest-dependent pathway,” to enable major changes in physiologic programs in response to extracellular signals such as hormones (45). Sustained suppression of PS by inhibition of eEF2 is usually lethal. For example, ADP ribosylation of eEF2 by diphtheria toxin freezes the translocation complex, and cell death follows within hours. Inhibition of elongation has also been implicated in the early stages of programmed cell death (45). It is likely that PS suppression is somehow linked to the regulation of protein degradation. Both actually could be regulated efficiently by modification of a single factor, eEF1A (previously termed eEF1α), which not only is required for polypeptide elongation but also is a cofactor for degradation of N-terminally blocked proteins (46). Such mechanisms have been shown to be operational in invertebrate quiescence, a state that resembles hibernation and is induced by anoxia (47). During this state, the ubiquitin-mediated proteolytic pathway is acutely blocked in response to anoxia, which preserves macromolecular integrity and possibly explains the tolerance of these organisms to oxygen deprivation.
Under normal circumstances PS is one of the cellular processes most sensitive to disruption. Even brief interruptions of cerebral perfusion suppress PS in brain, presumably because of inhibition of translation initiation (17, 48). In hibernation, a state with only trickle blood flow and massively reduced capacity to supply nutrients, PS is arrested by active regulation. While prolonged disturbances in PS after ischemia in nonhibernators reliably predict postischemic cell necrosis (10, 11), suppression of PS in hibernation is not only survived without ill effects but is even associated with tolerance to a superimposed insult in an in vitro model of cerebral ischemia (3). This extraordinary discrepancy may have its explanation in the fact that ischemia disrupts homeostatic control of all cellular functions, while in hibernation, a coordinated shutdown occurs that maintains homeostatic balance. Since suppression of PS in hibernation is a component of the adaptation of this tolerant state, study of its regulation would be expected to lead ultimately to identification of the signals that initiate and maintain the global shutdown of cellular functions. Strategies of reversible cellular arrest employed by hibernators in the future may be applied to the prevention of cell death as a consequence of various forms of cell injury.
Acknowledgments
This work has been supported in part by the Boston Neurosurgical Foundation (K.U.F.).
ABBREVIATIONS
- PS
protein synthesis
- PMS
postmitochondrial supernatants
- eEF2
elongation factor-2
References
- 1.Frerichs K U, Kennedy C, Sokoloff L, Hallenbeck J M. J Cereb Blood Flow Metab. 1994;14:193–205. doi: 10.1038/jcbfm.1994.26. [DOI] [PubMed] [Google Scholar]
- 2.Astrup J, Siesjo B K, Symon L. Stroke. 1981;12:723–725. doi: 10.1161/01.str.12.6.723. [DOI] [PubMed] [Google Scholar]
- 3.Frerichs K U, Hallenbeck J M. J Cereb Blood Flow Metab. 1998;18:168–175. doi: 10.1097/00004647-199802000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Hochachka P W, Buck L T, Doll C J, Land S C. Proc Natl Acad Sci USA. 1996;93:9493–9498. doi: 10.1073/pnas.93.18.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Storey K B. Comp Biochem Physiol. 1996;113B:23–35. doi: 10.1016/0305-0491(95)02043-8. [DOI] [PubMed] [Google Scholar]
- 6.Song X, Körtner G, Geiser F. J Comp Physiol B. 1995;165:291–297. doi: 10.1007/BF00367312. [DOI] [PubMed] [Google Scholar]
- 7.Storey K B, Storey J M. Q Rev Biol. 1990;65:145–174. doi: 10.1086/416717. [DOI] [PubMed] [Google Scholar]
- 8.Aloia R C, Raison J K. Biochim Biophys Acta. 1989;988:123–146. doi: 10.1016/0304-4157(89)90007-5. [DOI] [PubMed] [Google Scholar]
- 9.Wang L C H. In: Advances in Comparative Environmental Physiology. Gilles R, editor. Berlin: Springer; 1988. pp. 1–45. [Google Scholar]
- 10.Hossmann K-A. Progress Brain Res. 1993;96:161–177. doi: 10.1016/s0079-6123(08)63265-3. [DOI] [PubMed] [Google Scholar]
- 11.Widman R, Weber C, Bonnekoh P, Schlenker M, Hossmann K-A. J Cereb Blood Flow Metab. 1992;12:425–433. doi: 10.1038/jcbfm.1992.60. [DOI] [PubMed] [Google Scholar]
- 12.Smith C B. Neurochem Res. 1991;16:1037–1046. doi: 10.1007/BF00965848. [DOI] [PubMed] [Google Scholar]
- 13.Smith C B, Deibler G E, Eng N, Schmidt K, Sokoloff L. Proc Natl Acad Sci USA. 1988;85:9341–9345. doi: 10.1073/pnas.85.23.9341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson B L, Berry R W, Telsner A. Anal Biochem. 1983;132:365–375. doi: 10.1016/0003-2697(83)90022-2. [DOI] [PubMed] [Google Scholar]
- 15.Cosgrove J W, Rapoport S I. Neurochem Res. 1986;9:1289–1301. doi: 10.1007/BF00966123. [DOI] [PubMed] [Google Scholar]
- 16.Holbrook L, Brown I R. J Neurochem. 1976;27:77–82. doi: 10.1111/j.1471-4159.1976.tb01546.x. [DOI] [PubMed] [Google Scholar]
- 17.DeGracia D, Sullivan J, Neumar R, Alousi S, Hikade K, Pittman J, White B, Rafols J, Krause G. J Cereb Blood Flow Metab. 1997;17:1291–1302. doi: 10.1097/00004647-199712000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Scorsone K, Panniers R, Rowlands A, Henshaw E. J Biol Chem. 1987;262:14538–14543. [PubMed] [Google Scholar]
- 19.Dever T E, Sripriya R, McLachlin J R, Lu J, Fabian J R, Kimball S R, Miller L K. Proc Natl Acad Sci USA. 1998;95:4164–4169. doi: 10.1073/pnas.95.8.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dever T E, Feng L, Wek R C, Cigan A M, Donahue T F, Hinnebusch A G. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 21.Fan H, Penman S. J Mol Biol. 1970;50:655–670. doi: 10.1016/0022-2836(70)90091-4. [DOI] [PubMed] [Google Scholar]
- 22.Youle R J, Neville D M., Jr J Biol Chem. 1979;254:11089–11096. [PubMed] [Google Scholar]
- 23.Greenfield L, Johnson V G, Youle R J. Science. 1987;238:536–539. doi: 10.1126/science.3498987. [DOI] [PubMed] [Google Scholar]
- 24.Gulevsky A K, Grischenko V I, Zagnoiko V I, Shchenyavsky I I, Ilyasova E N. Cryobiology. 1992;29:679–684. doi: 10.1016/0011-2240(92)90071-9. [DOI] [PubMed] [Google Scholar]
- 25.Derij L, Shtark M B. Comp Biochem Physiol. 1985;80B:927–934. doi: 10.1016/0305-0491(85)90486-9. [DOI] [PubMed] [Google Scholar]
- 26.Hickey E D, Weber L A, Baglioni C. Proc Natl Acad Sci USA. 1976;73:19–23. doi: 10.1073/pnas.73.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lodish H F, Rose J K. J Biol Chem. 1977;252:1181–1188. [PubMed] [Google Scholar]
- 28.Merrick W C. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson R J. In: Translation in Eukaryotes. Trachsel H, editor. Boca Raton, FL: CRC; 1991. pp. 193–229. [Google Scholar]
- 30.Clemens M J, Galpine A, Austin S A, Panniers R, Henshaw E C, Duncan R, Hershey J W B, Pollard J W. J Biol Chem. 1987;262:767–771. [PubMed] [Google Scholar]
- 31.Barber G N, Jagus R, Meurs E F, Hovanessian A G, Katze M G. J Biol Chem. 1995;270:17423–17428. doi: 10.1074/jbc.270.29.17423. [DOI] [PubMed] [Google Scholar]
- 32.Oldfield S, Jones B L, Taton D, Proud C G. Eur J Biochem. 1994;221:399–410. doi: 10.1111/j.1432-1033.1994.tb18752.x. [DOI] [PubMed] [Google Scholar]
- 33.Van Ness B G, Howard J B, Bodley J W. J Biol Chem. 1980;255:10710–10716. [PubMed] [Google Scholar]
- 34.Brown B A, Bodley J W. FEBS Lett. 1979;103:253–255. doi: 10.1016/0014-5793(79)81339-3. [DOI] [PubMed] [Google Scholar]
- 35.Parentesis J P, Phan L D, Gleason W D, LaPorte D C, Livingston D M, Bodley J W. J Biol Chem. 1992;267:1190–1197. [PubMed] [Google Scholar]
- 36.Lee H, Iglewski W J. Proc Natl Acad Sci USA. 1984;81:2703–2707. doi: 10.1073/pnas.81.9.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fendrick J L, Iglewski W J. Proc Natl Acad Sci USA. 1989;86:554–557. doi: 10.1073/pnas.86.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pappenheimer A M., Jr Annu Rev Biochem. 1977;46:69–94. doi: 10.1146/annurev.bi.46.070177.000441. [DOI] [PubMed] [Google Scholar]
- 39.Iglewski B H, Liu P, Kabat D. Infect Immunol. 1977;15:138–144. doi: 10.1128/iai.15.1.138-144.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frerichs K U, Dienel G, Sokoloff L, Hallenbeck J M. Am J Physiol. 1995;268:R445–R453. doi: 10.1152/ajpregu.1995.268.2.R445. [DOI] [PubMed] [Google Scholar]
- 41.Srere H K, Belke D, Wang L C H, Martin S L. Am J Physiol. 1995;37:R1507–R1512. doi: 10.1152/ajpregu.1995.268.6.R1507. [DOI] [PubMed] [Google Scholar]
- 42.Srere H K, Wang L C H, Martin S L. Proc Natl Acad Sci USA. 1992;89:7119–7123. doi: 10.1073/pnas.89.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soukri A, Valverde F, Hafid N, Elkebbaj M S, Serrano A. Gene. 1996;181:139–145. doi: 10.1016/s0378-1119(96)00494-5. [DOI] [PubMed] [Google Scholar]
- 44.Ryazanov A G, Shestakova E A, Natapov P G. Nature (London) 1998;334:170–173. doi: 10.1038/334170a0. [DOI] [PubMed] [Google Scholar]
- 45.Ryazanov A G, Spirin A S. In: Translational Regulation of Gene Expression 2. Ilan J, editor. New York: Plenum; 1993. pp. 433–455. [Google Scholar]
- 46.Gonen H, Smith C E, Siegel N R, Kahana C, Merrick W C, Chakraburtty K, Schwartz A L, Ciechanover A. Proc Natl Acad Sci USA. 1994;91:7648–7652. doi: 10.1073/pnas.91.16.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anchordoguy T J, Hand S C. Am J Physiol. 1994;267:R895–R900. doi: 10.1152/ajpregu.1994.267.4.R895. [DOI] [PubMed] [Google Scholar]
- 48.DeGracia D J, Neumar R W, White B C, Krause G S. J Neurochem. 1996;67:2005–2012. doi: 10.1046/j.1471-4159.1996.67052005.x. [DOI] [PubMed] [Google Scholar]