Abstract
Endothelial cell (EC) apoptosis may play an important role in blood vessel development, homeostasis and remodelling. In support of this concept, EC apoptosis has been detected within remodelling vessels in vivo, and inactivation of EC apoptosis regulators has caused dramatic vascular phenotypes. EC apoptosis has also been associated with cardiovascular pathologies. Therefore, understanding the regulation of EC apoptosis, with the goal of intervening in this process, has become a current research focus. The protein-based signalling and cleavage cascades that regulate EC apoptosis are well known. However, the possibility that programmed transcriptome and glycome changes contribute to EC apoptosis has only recently been explored. Traditional bioinformatic techniques have allowed simultaneous study of thousands of molecular signals during the process of EC apoptosis. However, to progress further, we now need to understand the complex cause and effect relationships among these signals. In this article, we will first review current knowledge about the function and regulation of EC apoptosis including the roles of the proteome transcriptome and glycome. Then, we assess the potential for further bioinformatic analysis to advance our understanding of EC apoptosis, including the limitations of current technologies and the potential of emerging technologies such as gene regulatory networks.
Keywords: endothelial cell, apoptosis, transcriptome, gene network
1. Introduction
Apoptosis is the programmed suicide of a cell (Wyllie & Arends 2003). It serves to remove supernumerary cells during development and tissue remodelling (Meier et al. 2000); maintain homeostasis in adult tissues; remove self-reactive or non-reactive immune cells (Adams et al. 1999) and delete damaged, infected or transformed cells. It is an energy-requiring process, throughout which membrane integrity is maintained to avoid the initiation of an inflammatory response. This contrasts with necrosis in which cells die relatively passively following overwhelming damage, leading to loss of membrane integrity and to inflammation (Kerr et al. 1972).
Like other organs, blood vessels and the heart appear to employ apoptosis for remodelling during development and to maintain homeostasis during adulthood. Within blood vessels, vascular endothelial cells (ECs) play an especially important role in regulating overall vessel structure. Most ECs in adult blood vessels are relatively quiescent and resistant to apoptosis. However, they are thought to retain the latent capacity for proliferation and apoptosis to mediate angiogenesis and regression, respectively. In this review, we will discuss what is known about the function of EC apoptosis in health and cardiovascular disease. We will describe how this process appears to be controlled at the level of proteins (the proteome). Then, we will discuss the role that regulated RNA transcript abundance profiles (the transcriptome) and cell surface polysaccharide profiles (the glycome) may play in the EC apoptotic programme. To progress our knowledge of the regulation of EC apoptosis further, we now need to understand the cause and effect relationships between the molecular signals that control this process. In §10 of this review, we will assess whether a deeper analysis of the EC transcriptome using emerging technologies such as gene regulatory network analysis may help us understand the complex signals that control EC fate. Our conclusion is that these emerging technologies have great potential for EC research, especially when their results are integrated with those of cell biology studies, proteomics and in silico databases. However, it is critical that the cause and effect relationships predicted using technologies such as gene regulatory networks are thoroughly validated by laboratory experiments.
2. The function of EC apoptosis in normal physiology
(a) EC apoptosis may contribute to blood vessel remodelling
Several lines of evidence support the view that EC apoptosis plays an important role in vascular biology and pathology. Firstly, apoptosis can be detected during developmental vessel regression in the aortic arches (Fisher et al. 2000), the abdominal aorta (Cho et al. 1995) and the ductus arteriosus (Kim et al. 2000). EC apoptosis may play an especially important role in the eye, in development of the tunica vasculosa lentis of the ocular lens (Mitchell et al. 1998) and the pupillary membrane of the anterior chamber of the eye (Lang et al. 1994). Macrophages may promote this EC apoptosis (Lang et al. 1994), possibly through Wnt signalling (Lobov et al. 2005).
Apoptosis in these regressing vessels is thought to promote their remodelling to meet the changing requirements of the tissues they supply (Carmeliet 2000). Vessel regression may involve a classical series of events. For example, in one scenario following the inhibition of VEGF-A signalling, blood flow ceases, followed by EC apoptosis and loss of pericytes, leaving empty basement membrane sheaves that can act as scaffolds for future capillary re-growth (Baffert et al. 2006).
EC apoptosis appears to be less frequent in adult animals than during development. For example, EC apoptosis appears to be much less common in adult rat kidney than in the developing rat kidney (Fierlbeck et al. 2003). Nevertheless, adult ECs retain a latent capacity for apoptosis, and EC apoptosis is detected within specific adult tissues where substantial vessel remodelling occurs, such as healing wounds (Desmouliere et al. 1995; Greenhalgh 1998), regressing alveoli in the mammary gland after weaning (Matsumoto et al. 1992; Djonov et al. 2001), the developing placenta (Tertemiz et al. 2005) and during the cyclical remodelling of female reproductive tissues (Modlich et al. 1996; Niswender et al. 2000; Dickson et al. 2001; Gaytan et al. 2002). Further support for a role of EC apoptosis in vessel remodelling comes from in vitro studies. For example, incubation with pro-survival growth factors (Satake et al. 1998) or retroviral-mediated anti-apoptotic Bcl-2 expression (Pollman et al. 1999) reduces the incidence of EC apoptosis and inhibits the regression of vessel-like structures.
It is important to note that two studies have been unable to detect significant EC apoptosis within regressing tissues where blood vessel remodelling would be expected to occur (Hughes & Chang-Ling 2000; Arfuso & Meyer 2003). In addition, apoptosis was not observed in one in vitro study where vessels were disrupted by angiogenic inhibitors (Friis et al. 2006). However, the majority of studies suggest that EC apoptosis is frequently seen in remodelling vessels, implying that EC apoptosis plays a role in the remodelling process.
(b) EC apoptosis may contribute to blood vessel morphogenesis and lumen formation
In addition to participating in vessel regression, EC apoptosis may also contribute to angiogenesis and lumen formation. Several EC types spontaneously form vessel-like structures in vitro (vascular morphogenesis) when cultured on an extracellular matrix or with matrix-producing cells. Incubation of EC with anti-apoptotic caspase inhibitors or overexpression of the anti-apoptotic protein Bcl-2 appears to inhibit vascular morphogenesis in vitro (Segura et al. 2002 and M. Harris & C. Print 2003, unpublished results). It initially seems counterintuitive that EC apoptosis may contribute to vascular morphogenesis. However, it is possible that EC apoptosis contributes to this process by deleting wrongly placed EC as nascent vessels enlarge. Furthermore, the negatively charged membrane surface of apoptotic EC may promote angiogenic sprouting of adjacent vessels by causing localized plasma membrane hyperpolarization (Weihua et al. 2005). Angiogenesis may also be promoted through the engulfment of apoptotic EC debris by viable neighbouring EC, which may lead to the release of pro-angiogenic factors including VEGF-A (Golpon et al. 2004). However, since in vitro angiogenesis models only approximately recapitulate in vivo angiogenesis, these in vitro experiments should be interpreted with caution until they are confirmed by in vivo studies.
As well as supporting vessel regression and possibly promoting angiogenesis, EC apoptosis may in addition play a role in lumen formation. In support of this hypothesis, ECs undergoing apoptosis are seen in the centre of developing vascular structures (Duval et al. 2003), and newly formed lumen appear to contain post-apoptotic endothelial fragments (Bishop et al. 1999). Blocking apoptosis in the capillaries of the developing rat kidney by inhibiting the pro-apoptotic factor TGF-β1 appears to retard lumen formation (Fierlbeck et al. 2003). In addition, apoptotic EC can be detected during lumen formation in placental vessels in vivo (Tertemiz et al. 2005).
(c) Mice in which regulators of EC apoptosis are disrupted have vascular phenotypes
Additional evidence that EC apoptosis plays a role in vascular biology is provided by the inactivation of genes encoding regulators of EC apoptosis in mice. For example, the growth factors vascular endothelial growth factor-A (VEGF-A) and angiopoietin-1 (Ang-1) are important inhibitors of EC apoptosis. Inactivation of Ang-1 (Suri et al. 1996) causes severe vascular abnormalities early in development, and inactivation of VEGF-A (using either a genetic-based or protein inhibitor-based method) causes an increase in EC apoptosis and embryonic mortality (Gerber et al. 1999). An inherited mutation in the Tie2 receptor causes vascular malformations by inhibiting EC apoptosis through constitutive activation of p52 ShcA (Morris et al. 2006). Vascular endothelial (VE) cadherin is an EC–EC adhesion molecule that transduces anti-apoptoic signals into EC in conjunction with VEGF-A. Inactivation of VE-cadherin (Carmeliet et al. 1999) causes embryonic lethality due to failed vascular development. Disruption of gap junction formation by inactivation of connexin 43 alters coronary vessel development in association with altered apoptosis (Walker et al. 2005). In mice in which the anti-apoptotic gene Bcl-2 has been inactivated in the germ line retinal vascular development is retarded (Wang et al. 2005). Although these phenotypes imply an important role for EC apoptosis, since these signals also regulate EC proliferation, it is difficult to assess the proportion of each mutant phenotype that results specifically from the loss of EC apoptosis control. Therefore, more specific studies to assess the role of EC apoptosis are underway in our laboratory and others. In these ongoing studies, EC apoptosis is being inhibited directly by the EC-specific expression of anti-apoptotic transgenes. We hope that the results of these studies will reveal the definitive functions of EC apoptosis in vivo.
3. EC apoptosis in cardiovascular disease
Altered EC apoptosis is associated with several cardiovascular pathologies, in particular with atherosclerosis and thrombosis. EC apoptosis can be detected in atherosclerotic plaques (Mehta et al. 2002; Norata et al. 2002) and may provide an important step in the transition from a stable atherosclerotic plaque to a plaque with overlying thrombosis. The tissue environment of an atherosclerotic plaque may promote EC apoptosis. For example, oxidized LDL (ox-LDL), which is present in most atherosclerotic plaques, appears to downregulate EC expression of the anti-apoptotic protein cFLIP and subsequently activate Fas-mediated EC death in vitro (Sata & Walsh 1998). Ox-LDL also appears to activate the release of apoptosis-inducing factor (AIF) from coronary artery EC mitochondria. Suppression of AIF expression using antisense oligonucleotides suppresses ox-LDL-induced EC death (Zhang et al. 2004). In contrast, high-density lipoprotein (HDL) cholesterol, which is anti-atherogenic, appears to inhibit EC apoptosis in part by promoting the activity of nitric oxide (NO) synthase (Mineo et al. 2006). Oxidative stress has also been implicated in activating the apoptosis of EC in atherosclerosis (Valgimigli et al. 2003). The apoptosis of EC overlying atherosclerotic plaques may cause exposure of the underlying pro-thrombotic plaque tissue and subsequent thrombosis. For example, induction of EC apoptosis in vivo by the pro-apoptotic drug staurosporine promotes both EC apoptosis and local thrombosis. This chain of events can be broken by infusion of anti-apoptotic caspase inhibitors (Durand et al. 2004).
In addition, apoptotic EC are thought to become pro-coagulant by a process that does not require the exposure of underlying plaque tissues (Bombeli et al. 1997). The coagulation cascade initiator tissue factor (TF, thromboplastin) may provide an especially important link between EC apoptosis and thrombosis. TF activity is increased by phosphatidylserine (PS) exposure on the surface of EC during apoptosis (Tedgui & Mallat 2003). Women with systemic lupus erythematosus (SLE) are at increased risk of atherosclerosis and superimposed thrombosis. This may be due in part to an increased incidence of EC apoptosis and increased TF activity, since elevated numbers of PS-expressing post-apoptotic EC are detected in the blood of SLE patients.
In atherosclerotic plaques, vascular smooth muscle cells (VSMC) become activated, migrate into the tunica intima, survive and proliferate. This proliferation may be promoted in part by factors derived from apoptotic EC overlying the atherosclerotic plaque. Raymond et al. (2004) suggest that proteolytic activity initiated by caspases from apoptotic EC causes degradation of proteoglycans, which release unidentified factors that activate VSMC ERK phosphorylation and Bcl-xL expression. Other cardiovascular pathologies with which EC apoptosis has been associated include vasculitis (Woywodt et al. 2003) and transplant rejection (Krupnick et al. 2002; Hall & Jevnikar 2003). EC apoptosis is also associated with inflammatory disorders (Winn & Harlan 2005) and non-cardiovascular pathologies such as neoplasia, reviewed by Carmeliet & Jain (2000), which are beyond the scope of this review.
4. The control of EC apoptosis by protein-based signalling and cleavage cascades
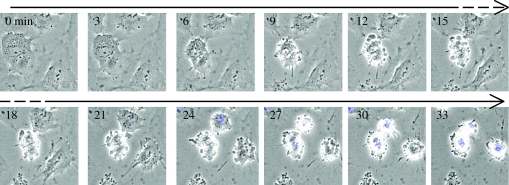
Whether an EC lives or whether it dies by apoptosis is determined by the interplay of numerous signals. It is the integration of all these signals, rather than any individual signal, that ultimately determines an EC's fate. The signals that regulate EC apoptosis are closely integrated with the signals that regulate other activities such as cell division, stress responses, activation and migration (Blagosklonny 2003). The most widely studied mechanisms that regulate EC apoptosis are in the realm of proteins (the proteome). These include regulated protein translation, phosphorylation, re-localization, cleavage and oligomerization. We have reviewed these mechanisms in detail previously (Duval et al. 2003), and they can be broadly divided into those that regulate (i) the mitochondrial and (ii) the death receptor apoptosis pathways. When triggered, these two pathways converge to activate the ‘executioner’ caspases 3, 6 and 7 and nuclease enzymes that, along with other enzymes such as cathepsins (Madge et al. 2003) and calpains (Porn-Ares et al. 2003), cleave structural proteins, metabolic enzymes and nucleic acids. This results in a series of biochemical and morphological changes including (in approximate order of appearance) loss of cell–cell and cell–matrix contact, cytoplasmic retraction, re-localization of cytochrome c from mitochondria to cytoplasm, activation of caspases, phosphatidyl serine exposure on the plasma membrane, membrane blebbing and chromatin condensation (Granville et al. 1999). This process can be seen in the time-lapse sequence in figure 1 (Johnson et al. 2004). The end result is post-apoptotic EC corpses that retain their membrane integrity and are either dispersed in the bloodstream or occasionally phagocytosed by neighbouring cells. Interestingly, components of EC corpses that are dispersed in the bloodstream may be taken up by endothelial progenitors and promote their proliferation and differentiation (Hristov et al. 2004).
Figure 1.
HUVEC were cultured to 70% confluence in their optimal medium, washed and their media replaced with a low serum (2% charcoal-stripped FCS) medium. Their morphology was followed over time in the presence of Hoechst 33342 and their death imaged by time-lapse epifluorescence microscopy. The following series of events was observed: (i) loss of contact with matrix and adjacent cells, (ii) retraction of cytoplasm, (iii) membrane blebbing and (iv) nuclear condensation (seen as blue coloration representing the density of Hoechst fluorescence surpassing a pre-set threshold for display). Times are in minutes (adapted from Johnson et al. 2004).
The mitochondrial apoptosis pathway is initiated by intracellular damage or by the loss of extracellular survival signals, for example, by growth factor deprivation when vessels become occluded during the process of regression (Lang & Bishop 1993; Meeson et al. 1999). This activates caspase 9 and releases pro-apoptotic molecules such as AIF from the mitochondria, thus initiating the apoptotic programme described above.
A plethora of extracellular survival signals normally suppress the mitochondrial apoptosis pathway. These include integrin-mediated cell adhesion to basement membrane (Meredith et al. 1993; Re et al. 1994; Frisch et al. 1996), adhesion to adjacent cells mediated by VE-cadherin (Carmeliet et al. 1999) and platelet EC adhesion molecule-1 (PECAM-1; Gao et al. 2003; Limaye et al. 2005), haemodynamic shear forces from blood flow (Meeson et al. 1996), vascular EC-specific phosphotyrosine phosphatase (VE-PTP; Baumer et al. 2006), VEGF-A (Lobov et al. 2002), interleukin (IL)-8 (Li et al. 2003), basic fibroblast growth factor (FGF; Fuks et al. 1994) and survival signals derived from pericytes or VSMC (Benjamin et al. 1998). These extracellular survival signals are transduced into the cytoplasm where they activate intracellular signalling intermediates, including pro- and anti-apoptotic members of the Bcl-2 family. Overexpression of anti-apoptotic Bcl-2 or Bcl-xL proteins inhibits EC apoptosis (Schechner et al. 2000; Zheng et al. 2000), while antisense oligonucleotide-mediated inhibition of the pro-survival Bcl-xL protein induces EC apoptosis in vitro (Ackermann et al. 1999). The radio-protective drug amifostine may act in part by elevating the expression of anti-apoptotic members of the Bcl-2 family (Khodarev et al. 2004). Interestingly, immediately after birth, the ratio of pro-apoptotic Bax to pro-survival Bcl-2 is elevated in regressing vessels (such as the ductus arteriosus) relative to non-regressing vessels (Kim et al. 2000). A central hub in the inhibition of EC apoptosis is the kinase AKT. For example, inactivation of AKT in mice markedly alters blood vessel development, possibly by reducing the expression of thrombospondin (TSP)-1 and -2 (Chen et al. 2005).
The death receptor apoptosis pathway is activated when tumour necrosis factor (TNF)-family proteins (Slowik et al. 1997) such as TNFα, Fas ligand and vascular endothelial growth inhibitor (VEGI; Yu et al. 2001) bind to TNF-family ‘death receptors’ on the EC surface. Death receptors activate cytoplasmic adapter proteins, which in turn activate caspase 8 and the p38 mitogen-activated protein kinase (MAPK) pathway (Cardier & Erickson-Miller 2002; Pru et al. 2003). Although ECs express death receptors on their surface (Richardson et al. 1994; Sata et al. 2002), TNFα and Fas ligand only activate apoptosis in a subset of EC (Biancone et al. 1997; Sata et al. 2002; Filippatos et al. 2004). The apparent resistance of many ECs to death receptor-induced apoptosis may be in part due to their expression of the caspase 8-antagonist cFLIP (Aoudjit & Vuori 2001; Bannerman et al. 2001). It may also be due to a second action of the TNF-receptor family—activation of the transcription factor NFκB (Yasumoto et al. 1992; Rath & Aggarwal 1999) which upregulates the expression of RNAs encoding apoptosis inhibitors (Bach et al. 1997) including A1 (Duriez et al. 2000), A20 and xIAP (Stehlik et al. 1998). There are additional ‘death ligands’ for ECs that are not members of the TNF superfamily. For example, adult capillary and venous EC, but not arterial EC, are sensitive to bone morphogenetic protein 4 (BMP4)-induced apoptosis. The relatively higher resistance of arterial EC to apoptosis may be due in part to their expression of inhibitory (I) Smads. After the knockdown of I-Smad expression using short interfering RNA, the resistant arterial EC become sensitive to BMP4-induced death. In contrast, ectopic expression of I-Smads in BMP4-sensitive cells suppressed BMP4-induced death. Furthermore, intravenous administration of BMP4 into mice caused haemorrhage of capillary EC in brain and lung (Kiyono & Shibuya 2006).
5. Could changes to the transcriptome and glycome potentiate EC apoptosis?
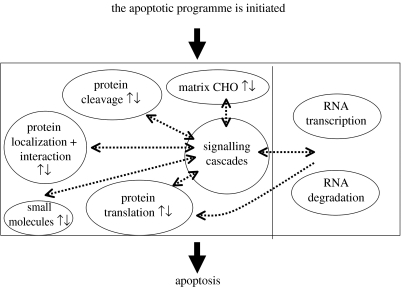
In addition to the extensively studied protein-based events described above, EC apoptosis may also be regulated by altered RNA transcript abundance profiles (an altered transcriptome), altered cell surface polysaccharide profiles (an altered glycome) or by altered concentrations of intracellular small molecules. The potential interplay between the intracellular events that form the apoptotic programme is shown schematically in figure 2.
Figure 2.
A schematic of the intracellular participants in the apoptotic programme. RNA transcriptome-associated events are shown on the right and other events are shown on the left. CHO denotes carbohydrates.
To map the transcriptome and glycome changes that accompany EC apoptosis, our laboratory recently modelled the apoptosis of EC that may occur during vessel regression by culturing primary human umbilical vein EC (HUVEC) in conditions of partial survival factor deprivation (SFD). Time-lapse microscopy of the last events in this process is shown in figure 1. Within 1 h of the onset of SFD, HUVEC showed signs of stress. After 28 h, progression through the cell cycle had ceased and only approximately 60% of original cell numbers remained. At this time, approximately 85–90% of the cultures consisted of stressed cells that, without intervention, were destined to die over the next 3 days and approximately 10–15% of the cultures consisted of cells actively undergoing apoptosis. Since almost all post-apoptotic cells appeared to detach and float into the medium within 3 h of death, the remaining adherent cultures, which were subsequently analysed, contained very few post-apoptotic corpses. To determine whether SFD-induced EC apoptosis was associated with transcriptome changes, we used Affymetrix human U95A gene arrays to compare an abundance of 12 600 transcripts in HUVEC cultured in their optimal medium with the corresponding abundance in HUVEC after 28 and 48 h of SFD. Each experiment was replicated five times, each time using primary HUVEC derived from a different individual. We found that most of the 12 600 transcripts were not significantly affected by SFD. However, Bayesian Student's t-tests identified 171 transcripts that were consistently upregulated and 495 downregulated in all five experiments (p≤0.01). Independent component analysis (ICA; a machine learning method for the analysis of noisy data that makes very few statistical assumptions) (Saidi et al. 2004) confirmed the selection of regulated transcripts made by the Bayesian Student's t-tests (Johnson et al. 2004).
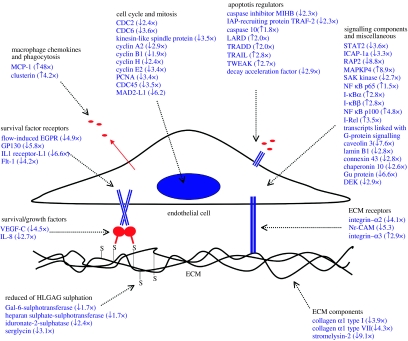
Interestingly, very few of the apoptosis-associated transcriptome changes indicated a protective stress response. Instead, most changes appeared to be either directly pro-apoptotic or to indirectly prime cells for future apoptosis. They are summarized in figure 3, some are of special interest. For example, transcripts encoding the death receptor LARD (DR3) and the death receptor ligands Trail and Tweak were upregulated. RNAs encoding survival signals and their receptors, such as VEGF-C, IL-8, flow-induced G-protein-coupled receptor, GP130, IL1 receptor component-L1 and Flt-1, were downregulated. Transcripts encoding intracellular inhibitors of apoptosis were also downregulated including cIAP1, TRAF-2, STAT2 and the integrin-associated kinase ICAP-1a. Bax, a pro-apoptotic member of the Bcl-2 family, was upregulated. The final stage of the apoptotic programme is engulfment of apoptotic corpses by phagocytes. Transcript abundance changes may assist this process, since RNAs encoding the chemokine monocyte chemoattractant protein-1 (MCP-1) and clusterin (apolipoprotein J, thought to promote the uptake of apoptotic bodies by non-professional phagocytes) were also upregulated.
Figure 3.
Summary of the regulated transcript abundance and carbohydrate sulphation changes that may play a role in preparing EC for apoptosis (adapted from Johnson et al. 2004). Abbreviations: ECM, extracellular matrix; HLGAG, heparin-like glycosaminoglycan.
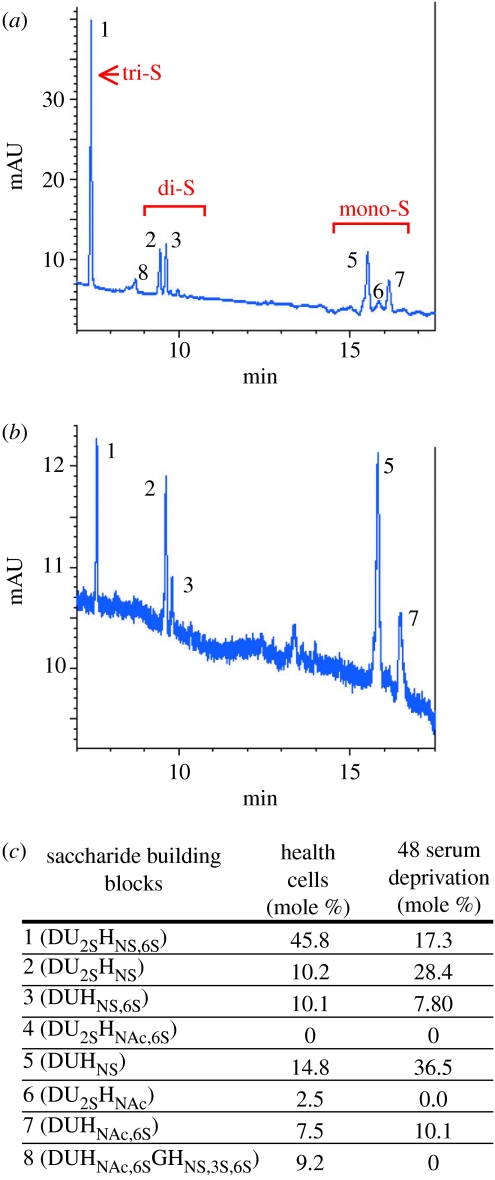
We also observed changes in the abundance of RNAs encoding enzymes that determine the structure of heparin-like cell surface glycosaminoglycans (HSGAG). Highly sulphated HSGAG are required to present extracellular matrix-binding survival factors, such as FGF and VEGF-A, to their receptors on the EC surface (Park et al. 1993; Venkataraman et al. 1999). After 28 h apoptosis, the RNA encoding N-deacetylase N-sulphotransferase (which promotes HSGAG sulphation) was downregulated, the RNA encoding iduronate-2-sulphatase (which removes sulphate groups from HSGAG) was upregulated and serglycin (a proteoglycan core protein secreted by EC) was downregulated. In line with these transcript abundance changes, when we analysed HSGAG profiles, we found that during EC apoptosis there was a relative loss of tri-sulphated disaccharides and a relative increase in mono-sulphated disaccharides (figure 4). This apoptosis-associated reduction in HSGAG sulphation may further reduce the ability of stressed or apoptotic EC to respond to survival factors such as VEGF-A. In agreement with this hypothesis, we found that the ability of EC to mount a response to VEGF-A (as measured by the degree of VEGF-induced transcriptome change) was dramatically reduced following SFD (Johnson et al. 2004).
Figure 4.
Compositional analysis of heparin sulphates isolated from control (grown in optimal media) and 48 h SFD HUVEC cultures. (a) CE electropherogram depicting the profile of cell surface HSGAGs from healthy ECs and (b) from SFD cells. There is a loss of the highly sulphated disaccharides (peak 1) and an increase in mono-sulphated disaccharides (peaks 5 and 7). (c) The relative contribution of each HSGAG to the total pool investigated is shown for four separate experiments, each using a different set of three pooled HUVEC isolates (adapted from Johnson et al. 2004).
Based on these results, we suggest that the cell biology of stress/deprivation-induced EC apoptosis is underpinned by synergy between previously described protein-based changes and the changes to the transcriptome and glycome identified in this study. This provides a new paradigm for the regulation of EC apoptosis, and the possibility that a small number of the apoptosis-associated transcriptome and glycome changes identified by this study may provide future drug targets for the regulation of EC apoptosis.
6. What is the time course of transcriptome change during EC apoptosis?
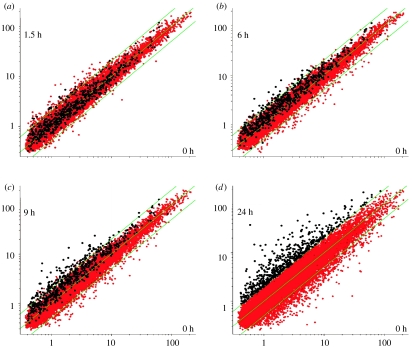
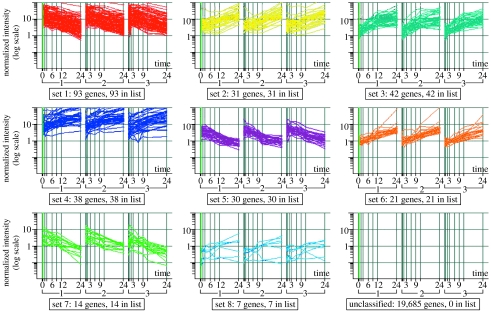
To understand the kinetics of transcriptome change during EC apoptosis, we have performed a time course experiment. HUVEC (a pooled culture from 10 donors) were exposed to SFD conditions identical to those used in the study described above. RNA was prepared from the cultures before the induction of apoptosis (time 0) and after 0.5, 1.5, 3, 6, 9, 12 and 24 h, hybridized to CodeLink Human Uniset 20K gene arrays. This experiment was repeated three times independently. The regulation of transcripts during EC apoptosis can be visualized in the scatterplots shown in figure 5a–d. We then selected a subset of 276 transcripts that were consistently regulated over the time course of EC apoptosis for further analysis. To do this, we excluded all transcripts from our analysis that were not regulated by Z≧1.5σ at 3 or more adjacent time points in all three experimental replicates (see Appendix for methods). These data are available in file 1 in the electronic supplementary material. From these 276 transcripts, k-means clustering was used to select eight groups of transcripts (in addition to an unclassified group) with similar time-course profiles (figure 6). From these profiles and from the scatterplots shown in figure 5a–d, it can be seen that a number of transcripts start to be regulated form 1.5 h and that, in general, the rate of change appears to low after 12 h. When we plotted the individual transcripts identified in the previous study, we noted that, in general, the transcripts encoding growth factors (such as Ang-2 and IL-8) were among the earliest transcripts to be regulated, while transcripts associated with the cell cycle (such as cyclins A2, H, and E, CDC6, CDC28 and kinesin-like spindle protein) were regulated later (table 1). The regulation of many of these transcripts following 28 h SFD has been validated by our previous Affymetrix study. These data suggest that some functional classes follow common patterns of regulation during the SFD-induced apoptosis of EC cultures. More sophisticated analysis (see below) suggests common upstream regulators of these transcripts.
Figure 5.
A time course of transcriptome change during EC apoptosis. Scatterplots show each transcript represented by a dot, with abundance at time 0 shown on each x-axis and abundance at other time points shown on the y-axes: (a) 1.5 h, (b) 6 h, (c) 9 h and (d) 24 h SFD. Those transcripts that are not regulated remain approximately on the diagonal. Transcripts that appeared to be upregulated when cultures were exposed to SFD conditions for 24 h compared with healthy cultures at time 0 are highlighted in white. The gradual upregulation of these transcripts over time can be seen.
Figure 6.
k-means clustering revealed eight groups of transcripts (from a shortlist of 685 highly regulated transcripts) that followed distinct temporal patterns of regulation.
Table 1.
The kinetics of SFD-dependent regulation of transcript abundance. Values represent median fold change (relative to 0 h) of normalized transcript abundance of the triplicate time-course data. Negative values represent downregulation. Positive values represent upregulation.
| 0.5 h | 1.5 h | 3 h | 6 h | 9 h | 12 h | 24 h | |
|---|---|---|---|---|---|---|---|
| cell cycle and mitosis | |||||||
| CCNA2 | −1.03 | 1.01 | −1.08 | −1.19 | −1.21 | −1.67 | −3.62 |
| CCNE | 1.01 | 1.03 | −1.05 | −1.32 | −1.91 | −3.29 | −4.25 |
| CCNH | 1.02 | 1.05 | −1.07 | −1.41 | −1.43 | −1.68 | −1.69 |
| CDC6 | 1.20 | 1.42 | 1.30 | −1.03 | −1.43 | −2.27 | −4.11 |
| CDC28 | −1.19 | −1.23 | −1.16 | −1.12 | −1.21 | −1.17 | −2.57 |
| KNSL1 | −1.04 | −0.04 | 1.04 | 1.14 | 1.04 | −1.29 | −3.49 |
| apoptosis regulators | |||||||
| TRAIL | −1.03 | 1.87 | 1.74 | 2.91 | 3.28 | 4.59 | 11.62 |
| LARD | −1.03 | −1.09 | 1.25 | 2.35 | 2.68 | 3.17 | 2.56 |
| growth factors | |||||||
| ANGPT2 | −1.08 | 1.02 | −1.64 | −2.42 | −3.11 | −4.49 | −1.93 |
| IL-8 | −1.51 | −4.29 | −3.92 | −3.30 | −1.55 | 1.07 | 1.04 |
| signalling components | |||||||
| RELB | 1.02 | 1.20 | 1.02 | 1.38 | 3.17 | 5.73 | 3.02 |
| chaperonin 10 | 1.03 | −1.02 | −1.03 | −1.09 | −1.26 | −1.51 | −2.45 |
7. Programmed changes to the transcriptome may potentiate apoptosis in other cell types
Contribution of the transcriptome and glycome to the apoptotic programme is not unique to EC. It has been known for some time that regulation of RNA transcript abundance may turn the apoptotic programme on or off during development. For example, ecdysone-dependent expression of RNAs encoding the caspases DRONC (Dorstyn et al. 1999) and DRICE (Kilpatrick et al. 2005) appears to prime Drosophila tissues for histolysis at specific developmental stages. Interestingly, the strategy of regulated transcript abundance activating the apoptotic programme during development may also be employed in plants. In the tissues of barley undergoing developmental cell death, RNAs encoding plant homologues of the endonucleases that a cleave DNA during mammalian apoptosis are upregulated (Zaina et al. 2003).
Further support for the hypothesis that transcriptome change may play a role in apoptotic programmes comes from experiments in which apoptosis of some cell types can be blocked by inhibiting RNA synthesis (Galli et al. 1995; Schulz et al. 1996). In addition, several studies have identified programmed changes in RNA transcript abundance as cultured cells undergo apoptosis. For example, infection with group A Streptococcus pyogenes induces apoptosis in some human epithelial cells. Microarray analysis suggests that this apoptosis is associated with altered gene expression including induction of RNA transcripts encoding caspases and pro-apoptotic members of the Bcl-2 family (Nakagawa et al. 2004). Ionizing radiation-induced apoptosis of lymphoblastoid cells is accompanied by the induction of RNAs encoding pro-apoptotic proteins such as Fas and Bak (Akerman et al. 2005). 5-Fluorouracil-induced apoptosis of colon carcinoma cells is accompanied by a programme of altered gene expression including the induction of RNAs encoding p53, Fas and TNF receptor-2 (Zhang et al. 2003). Apoptosis of human lung cancer cells is associated with the upregulation of a number of genes including pro-apoptotic members of the Bcl-2 family (Robinson et al. 2003). The dexamethasone-induced apoptosis of lymphoid cells is also accompanied by alterations in gene expression including the induction of BimEL, a pro-apoptotic member of the Bcl-2 family (Medh et al. 2003). In neurons, a set of RNA transcripts appears to be upregulated during apoptosis including those encoding caspase 3, the death receptor DR6 (Chiang et al. 2001), Id2 (Gleichmann et al. 2002) and the pro-apoptotic Bcl-2 family member Bax (Wullner et al. 1998).
As would be expected, different cell lineages appear to regulate different cohorts of RNA transcripts during their apoptotic programmes. However, some apoptosis-associated RNA changes are conserved across different cell lineages (including EC), such as the induction of transcripts encoding pro-apoptotic members of the Bcl-2 family. Taken together, these data suggest that in many cell types the altered abundance, during or immediately before apoptosis, of transcripts encoding apoptotic regulators or apoptotic machinery may play a ‘feed-forward’ role to assist the apoptotic programme.
8. What are the limitations of traditional gene array bioinformatic techniques for the study of EC apoptosis?
As discussed above, traditional bioinformatic techniques have revealed interesting new paradigms for the control of EC apoptosis. However, they do have real limitations. While these limitations should not deter prospective EC researchers from using transcriptome profiling studies, they require consideration.
Owing to the false discovery introduced by noise or individual differences (Schoenfeld et al. 2004), in addition to any annotation or contamination errors inherent in the gene array platform itself, a proportion of the results of every gene array experiment will be incorrect. Researchers and clinicians using microarray techniques must accept this low rate of error. However, accepting a low rate of error is not as difficult as it sounds. After all, deciding on an acceptable probability of error forms the basis of most statistical analyses of scientific experiments.
The results obtained when the same RNA is analysed using different gene array platforms are not always concordant (Jordan 2004). This has shaken the confidence of some researchers in gene array technology (Tan et al. 2003). This problem is illustrated by the microarray data we describe in this paper. Our previous study of the effect of 28 h SFD on HUVEC used Affymetrix U95A genechips and highlighted 79 regulated transcripts (listed in file 3 in the electronic supplementary material of Johnson et al. 2004). The CodeLink genechip time-course study presented in this paper used the same cell type and SFD conditions and contained a 24 h time point. We compared the transcripts apparently regulated by 28 h SFD in the Affymetrix study with the transcripts apparently regulated by 24 h SFD in the CodeLink study. Of the 79 transcripts highlighted by the Affymetrix study, 48 could be identified by reference to Unigene IDs in the CodeLink gene arrays, but only 30 of these appeared to be regulated by 1.5-fold or more in a congruent direction to the Affymetrix study. Although the lack of full concordance is concerning, some of the differences between the results of our two studies may be due to experimental differences. Other differences may be due to occasional misannotation of one or both gene array platforms, since it was recently shown that matching of gene array probes between platforms by actual sequences rather than by Unigene IDs dramatically improved the apparent concordance (Mecham et al. 2004). This microarray cross-platform discordance problem urgently requires further study. The safest approach when possible may be to focus on results that have been confirmed by two independent gene array platforms or by one platform and quantitative polymerase chain reactions.
Finally, there is an important but hidden limitation of traditional bioinformatic techniques. Although techniques such as clustering (Eisen et al. 1998), ICA (Saidi et al. 2004) and self organizing maps (Tamayo et al. 1999) have been extremely valuable in revealing hidden patterns of transcript co-regulation, in general they are not good at revealing the causal relationships among molecular signals within cells. However, it is the combination of the cause and effect relationships between all the signals operating in a cell, rather than the isolated actions of individual signals, that drive and regulate processes such as apoptosis.
9. The contribution and limitations of the proteome
An obvious limitation of gene array data is that it does not give direct information about proteins or post-transcriptional regulation. Although the use of gene array technology has helped to enrich our knowledge of the key regulatory genes involved in processes such as EC apoptosis, understanding regulation at the level of the proteome will be of more direct value. Large amounts of useful information have already emerged from proteomic studies of EC (Bruneel et al. 2003; Kamino et al. 2003; Nylund & Leszczynski 2004; Sprenger et al. 2004; Yu et al. 2004). Notable among these is a subtractive proteomics study that identified tumour-associated EC surface proteins, which may be targets for anti-tumour therapy. In fact, radio-immunotherapy directed against one of the EC surface proteins identified, annexin A1, is effective in vivo (Oh et al. 2004). Another notable study from the same research group illustrated the differences between in vitro and in vivo lung microvascular ECs, and highlighted the need for replicated in vivo confirmation of in vitro results (Durr et al. 2004). A study of the proteomic changes that occur during etoposide-induced EC apoptosis has suggested that multiple protein-based signalling cascades, including the mitochondrial and the ER stress pathways, may play a role (Bruneel et al. 2005).
The information derived from these proteomics studies could not have been derived from transcriptome studies, since it is very difficult to correlate gene expression levels with protein abundance (Gygi et al. 1999; Lian et al. 2002). Although technologies such as protein microarrays are rapidly developing and now enable relatively specific and sensitive analysis of proteins (Letarte et al. 2005), we believe that at present transcriptome studies still remain extremely viable. The best approach in the future may be to combine the results of proteomics with that of transcriptome analysis using systems biology techniques (see below).
10. Inference of causal relationships from transcriptome data
Methods to infer cause and effect relationships based on transcriptome data are emerging. Of these methods, some researchers believe the technique of gene regulatory network analysis may hold particular promise (Brazhnik et al. 2002; Schlitt & Brazma 2005). Practitioners of gene regulatory network transcriptome analysis claim to produce ‘circuit diagrams’ that show the hierarchical networks of cause and effect transcription control relationships that underlie a biological process like EC apoptosis or interventions like drug treatment. These gene regulatory networks are specific to the cell type under study. Unlike the traditional post-translational signalling network charts that most biologists are used to seeing, the nodes on gene regulatory network graphs represent transcripts rather than proteins. The links between the nodes (called edges) represent putative cause and effect relationships between transcripts, where the abundance of one transcript, through one or more intervening protein-based steps, can regulate the abundance of a second transcript. Therefore, although proteins are not explicitly shown in gene regulatory network graphs, they provide the hidden engine that underlies the network structure.
Several mathematical methods have been used to infer gene regulatory networks such as differential equation models (Chen et al. 1999; de Hoon et al. 2003), state space models (Rangel et al. 2004), Boolean network models (Akutsu et al. 1998; Shmulevich et al. 2002) and Bayesian network models (Friedman et al. 2000; Imoto et al. 2002). In some situations, a combination of these techniques is helpful. For example, Boolean and Bayesian network algorithms can be combined (Imoto et al. 2003b) to infer gene networks for response to a drug—the Boolean network approach is used initially to identify the set of transcripts regulated in abundance by the drug, and a Bayesian network approach is then used to map the upstream regulators of the drug-affected genes, which provide potential ‘druggable’ targets. This combination of approaches has been used successfully in yeast (Aburatani et al. 2003) to determine that the protein regulated by the oral antifungal drug griseofulvin is CIK1 (Savoie et al. 2003).
Some features of gene regulatory network inference techniques appear especially important when inferring networks from microarray data. For example, transcripts are not always regulated in a linear manner; therefore, capturing nonlinear relationships between transcripts can greatly increase the power of network analysis. This can be achieved by the use of non-parametric additive regression models (Imoto et al. 2002). In addition, most gene expression data contain many outlying data points and show heteroscedasticity (the dispersion of distributions differs between transcripts). Therefore, modelling which takes the effects of outliers and heteroscedasticity into account is especially well suited to inferring networks from microarray data (Imoto et al. 2003a). Like all bioinformatics techniques, both the quality and quantity of the available data determine the statistical power of gene network techniques.
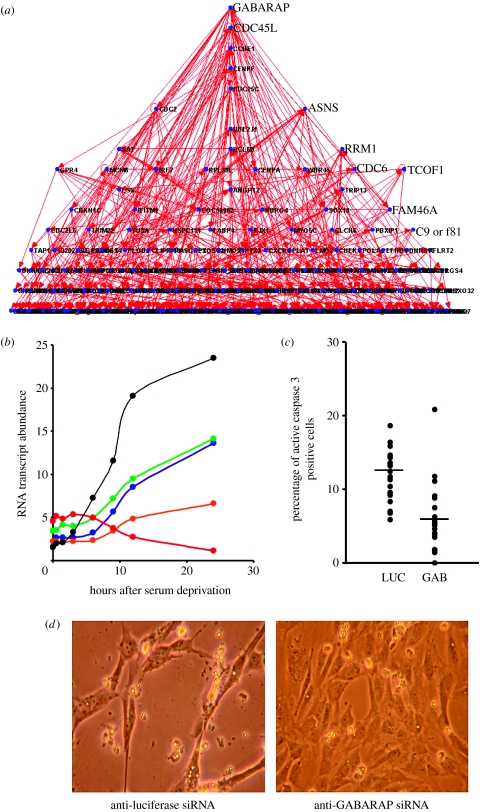
To illustrate the principles of applying gene regulatory networks to endothelial biology, we have used the dynamic Bayesian network inference method (Kim et al. 2003) to generate a gene network based on the median of the HUVEC apoptosis time-course gene array data which was described above. A graph representing this network is shown in figure 7a. In this network, the regulation of abundance of most parents and their multiple children over the apoptosis time course appears to be correlated, with the up- or downregulation of the putative children often lagging behind that of their parents (figure 7b and file 2 in the electronic supplementary material). In this gene network, the parent node with the most children is GABA receptor-associated protein (GABARAP). According to this network, GABARAP is potentially acting as a novel transcriptional ‘hub’ during the process of EC apoptosis. Interestingly, GABARAP has recently been shown in other laboratories to induce apoptosis by interacting with DEAD box polypeptide 47 (Lee et al. 2005) and appears to be encoded by a tumour suppressor gene mutated in breast cancer (Klebig et al. 2005). To illustrate the potential function of GABARAP in the regulation of EC apoptosis, we knocked down the expression of GABARAP and luciferase (a negative control) in HUVEC using siRNA. RT-PCR analysis indicated that 24 h after anti-GABARAP siRNA treatment, the abundance of GABARAP mRNA was reduced to less than 20% of its former levels (data not shown). siRNA-mediated reduction of GABARAP expression appeared to inhibit HUVEC apoptosis. The median incidence of apoptosis induced in HUVEC by 24 h of SFD (performed as in Johnson et al. 2004) was 11.7% in cells treated with anti-luciferase siRNA (control) but only 5.8% in cells treated by anti-GABARAP siRNA (figure 7c). Analysis of this data using the Mann–Whitney two-tailed test, a non-parametric statistical method, gave a statistically significant result of U=426.5 and p≤0.001. In addition, the morphological changes induced in HUVEC by 24 h of SFD appeared to be reduced by the inhibition of GABARAP expression (figure 7d). This suggests that GABARAP may indeed be a regulator of EC apoptosis, as is suggested by its prominent position in the gene network. Another node that appears to act as transcription hub during apoptosis is interferon regulatory factor-7 (IRF7), a transcription factor known to be regulated by the TNF and NFκB families. Interestingly, the TNF family member TRAIL and the NFκB family member RelB lie upstream of IRF7 in our gene network. The complete parent–child relationships for this gene network are given in file 2 in the electronic supplementary material. Web-based tools such as Cell Illustrator (http://www.cellillustrator.com) and Cytoscape (http:///www.cytoscape.org) can be used by the reader to explore this putative network and the gene-to-gene relationships it symbolizes.
Figure 7.
An example of a dynamic time-course gene regulatory network. (a) A graph representing a dynamic Bayesian gene network generated from the median of triplicated apoptosis time-course data. Dots represent transcripts (‘nodes’) and lines between the dots represent potential cause and effect interactions (‘edges’). (b) A graph showing apoptosis time-course gene array transcript abundance data for a parent RNA transcript in the network (CDKN1C, black) and its putative children (C1QTNF5, green; AKR1C3, blue; MLF1, orange; SUV39H1, red). (c) Reduction of GABARAP mRNA abundance inhibits HUVEC apoptosis. Incidence of apoptosis induced by 24 h of SFD in anti-GABARAP siRNA-transfected HUVEC (GAB) and anti-luciferae siRNA-transfected HUVEC (LUC, control) were assessed by immunocytochemical detection of caspase 3 cleavage. Horizontal bars indicate median values. (d) Images of HUVEC cultures deprived of serum for 24 h after treatment with anti-luciferase siRNA (control) and anti-GABARAP siRNA. Fields are 225×300 μM in size.
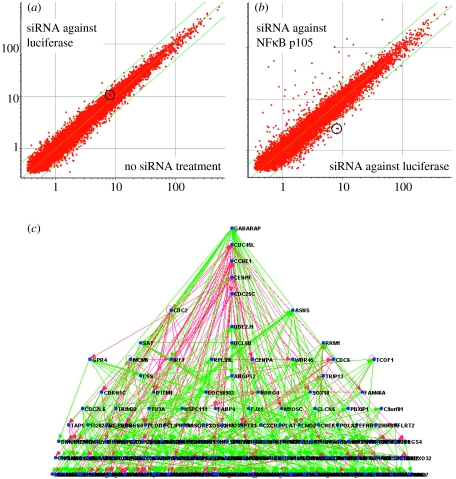
As a further illustration of gene network analysis techniques, we also generated a second gene network from the same apoptosis time-course gene array data. This network differs from the one shown in figure 7a, from which it was derived by the incorporation of new gene array data from eight siRNA disruptant experiments. This new gene array data was used as a Bayesian prior and is provided as file 3 in the electronic supplementary material. In these eight disruptant experiments, specific RNAs related to cell cycle control (CDC45L, CCNE1, CENPA, CENPF, CDC2, CDC25C, MCM6 and CDC6) were reduced in abundance by more than 65% in HUVEC using siRNA pools. Gene array analysis showed that these siRNA treatments caused large numbers of transcripts to be regulated (presumably as a consequence of the targeted RNA downregulation, or possibly in some cases also as a consequence of unexpected off-target siRNA effects). An example of the effects of siRNA treatment on the HUVEC transcriptome is shown in figure 8a,b. The putative time-course gene network modified by the incorporation of this disruptant information as a Bayesian prior is shown in figure 8c and its complete parent–child relationships are listed in file 4 in the electronic supplementary material. Of the 488 edges contained in the modified gene network shown in figure 8c, 338 are shared with the unmodified gene network shown in figure 7a. Of the 150 edges found in the modified but not the unmodified networks, 94 have as parents one of the eight RNAs that were targeted in the siRNA disruptant experiments. These particular edges appear to have inherited cause and effect information from the disruptant gene array data, since they all correctly predict the effects on children of disrupting parents by siRNA treatment (data not shown). Therefore, the inclusion of disruptant data as a Bayesian prior appears to enhance the ability of dynamic time-course gene networks to predict specific directional relationships between transcripts. It seems possible that the addition of data from biological database annotations and proteomics experiments could further enhance the predictive power of this type of gene network. We and other researchers are generating larger-scale gene regulatory networks using gene array data from hundreds of disruptant experiments, which will be combined with time-course data using Bayesian techniques, as illustrated above (Imoto et al. 2006). We believe these large networks will provide valuable hypotheses about functional interactions within ECs, which can then be tested experimentally.
Figure 8.
An example of a dynamic time-course gene regulatory network modified by the inclusion of prior information from siRNA disruptant experiments. (a) A scatterplot comparing transcript abundance in untransfected HUVEC (x-axis) with transcript abundance in mock HUVEC transfected with control siRNAs directed against luciferase (y-axis)—little regulation of transcript abundance appears to have occurred. (b) A scatterplot comparing transcript abundance in HUVEC transfected with siRNAs directed against luciferase (x-axis) with transcript abundance in HUVEC transfected with siRNAs directed against NFκB p105 (y-axis)—NFκB p105 (circled) was downregulated more than fourfold and a large number of additional transcripts were also regulated in abundance. (c) A graph representing an apoptosis time-course gene network generated that had been modified by incorporating, as a Bayesian prior, gene array data from eight siRNA disruptant experiments, similar to those shown in (a,b). Edges in common with the network shown in figure 7a are green, and edges present in this network but not in the network shown in figure 7a are pink.
Therefore, gene regulatory network technologies, along with other techniques for inferring cause and effect relationships in cells from transcriptome data, promise a great deal for vascular biology research and for accelerating the rational design of new cardiovascular medications. They will be especially valuable if their results are used to compliment those of traditional cell biology studies and EC proteomic studies currently underway. Cautious integration of the cause and effect relationships that are predicted by gene networks into systems biology databases (Aggarwal & Lee 2003) may be especially useful. A study showing the usefulness of this approach modelled a yeast galactose utilization pathway by analysing experimental perturbations using information from microarrays, proteomics and computer databases (Ideker et al. 2001). Another example is a study that defined pathways that control endoderm and mesoderm specification during sea urchin development. In this study, the results of perturbation analyses were combined with information from computer databases, cis-regulatory analysis and molecular embryology (Davidson et al. 2002). In Professor Eugene Butcher's laboratory, systems biology approaches using small amounts of data from cultured ECs have already revealed important new information about the role of NFκB and Ras EC signalling pathways (Plavec et al. 2004). The addition of data from gene regulatory networks may dramatically increase the power of this type of systems biology analysis.
However, a critical challenge for technologies such as gene regulatory networks is that the predictions they make must be extensively tested using traditional ‘wet’ laboratory experiments. This will require clear communication and extensive cooperation between bioinformaticians and biologists/clinicians. Hopefully, the emerging generation of vascular biologists fluent in both bioinformatics and wet laboratory experimentation will be able to meet this challenge.
11. Conclusions
EC apoptosis appears to play an essential role in blood vessel development and remodelling and has been associated with cardiovascular pathologies. Therefore, understanding the regulation of EC apoptosis with the goal of intervening in this process has become a current research focus. Much is known about the protein-based signalling and cleavage cascades that regulate EC apoptosis. However, the potential role of pro-apoptotic transcriptome and glycome changes in the orchestration of EC death has only recently been recognized. For a complete understanding of EC apoptosis, we now need to define the complex relationships between the molecular signals that operate in these cells. New technologies such as gene regulatory networks that promise to infer cause and effect relationships from EC transcriptome data provide powerful tools, especially when integrated with information from proteomic and cell biology studies and with computer databases. However, the predictions made by all of these techniques must be extensively validated by laboratory experiments. This will require close cooperation between bioinformaticians and experimental biologists and is likely to be driven by a new generation of cross-disciplinary scientists fluent in the languages of both fields.
Appendix: material and methods
HUVEC were isolated from umbilical cords by collagenase digestion and cultured at 37°C/5% CO2 in basal culture medium supplemented with a proprietary mixture of heparin, hydrocortisone, epidermal growth factor, FGF and 2% foetal calf serum (FCS; EGM-2, Cambrex, Workingham, UK). HUVEC from 10 individuals were pooled, plated at 4.8×105 cells in 25 cm2 flasks and allowed to recover for 24 h, at which time they were approximately 60% confluent. Supplemented medium was then removed and replaced with basal culture medium containing only 2% charcoal stripped FCS (Invitrogen, Paisley, UK). At time points 0, 0.5, 1.5, 3, 6, 9, 12 and 24 h, total RNA was prepared using Trizol reagent (Invitrogen) and assessed using an Agilent 2100 bioanalyser. Biotin-labelled complex cRNAs were prepared and hybridized to CodeLink UniSet Human 20K gene chips according to the manufacturer's protocols (GE Healthcare, Amersham, UK). To generate triplicate data (of biological replicates), the protocol was repeated two further times using HUVEC from 10 different individuals for each pool. The quality of the expression data from all gene chips was confirmed using CodeLink expression analysis Software (v. 4.1). To ensure that expression levels were comparable between the arrays, the data was normalized using intensity-dependent scaling as described in Schoenfeld et al. (2004). To knock down, the abundance of RNA transcripts in HUVEC siRNA ‘smartpools’ from Dharmacon Inc. (Lafayette, Colorado, USA) were transfected into HUVEC using the siFectamine transfection reagent (ICVEC, London, UK) used according to the manufacturers instructions. The incidence of HUVEC apoptosis was assessed by cytospinning HUVEC onto glass slides followed by immunohistochemical staining using an anti-active caspase 3 antibody (rabbit polyclonal, Promega, Southampton, UK, 1 : 250 dilution as described in Johnson et al. (2004)). For each experimental condition, the staining of more than 1000 cells was counted.
Data were log2-transformed and log ratios between each time point and the 0 h control calculated. In addition, the ratio between each time point and 24 h was also calculated to capture regulation at both ends of the time course. For each transcript at each time point, Z-scores were calculated. Log2 ratios of individual transcripts were subtracted from the mean of log2 ratios for that time point and then divided by the standard deviation of log2 ratios for that time point. For each time-course replicate, transcripts that had ‘good’- or ‘low’-quality flags (based on CodeLink expression analysis software results) and a Z-score of −2≤Z≤+2 at three or more adjacent time points were selected for further study. Transcripts that contained ‘good flags only’ and had a Z-score of −1.5≤Z≤+1.5 at three or more adjacent time points were also selected. Those regulated transcripts concordant between the triplicate time-course data were selected for the network generation. In addition, transcripts that were known to be involved in apoptosis and were significantly regulated in a previous Affymetrix apoptosis study (Johnson et al. 2004), that were also regulated by greater than 1.5-fold in the triplicate CodeLink data were included. This gave a list of 276 transcripts for apoptosis network building.
Dynamic Bayesian apoptosis time-course networks were generated from the list of 276 apoptosis regulated transcripts, as described by Kim et al. (2003). Data generated from eight siRNA knock down arrays were used to make an array prior to the apoptosis time-course network. The eight knockdown arrays were selected from a list of 270 different knockdown arrays, based on the following criteria: (i) the transcript knocked down is in the list of 276 transcripts regulated in the apoptosis time-course triplicate data (as explained above) and (ii) a large number of the regulated genes resulting from the knock down were also contained in the 276 list of apoptosis-regulated transcripts. Gene networks, both with and without this prior information, were inferred from the apoptosis data as described (Kim et al. 2003). Comparisons of the gene networks generated with and without knockdown prior information, and their visualization and data mining, were carried out using the software iNet (now known as Cell Illustrator) which can be downloaded from http://www.cellillustrator.com
Acknowledgments
We wish to thank Mrs Deborah Sanders and Mrs Sally Humphreys for assistance with HUVEC culture, as well as Prof. Satoru Kuhara for discussions about gene regulatory networks. Prof. Stephen Smith and Dr Shiladitya Sengupta co-supervised some the microarray and glycomic studies described in this review, which were funded by The Biotechnology and Biological Sciences Research Council, The Wellcome Trust and by a research collaboration agreement between Cambridge University and the biotechnology company GNI, Ltd. Computation time was provided by the Super Computer System, Human Genome Centre, Institute of Medical Science, University of Tokyo.
Footnotes
One contribution of 21 to a Theme Issue ‘Bioengineering the heart’.
Supplementary Material
The triplicated apoptosis timecuorse Codelink Human Uniset I gene array data for the transcripts discussed here are shown. Column 1 shows the probe ID. The remaining columns show normalised transcript abundance. Column headers begin with the time after apoptosis induction (hrs or min) and end with the replicate number
Edges of the Bayesian gene regulatory network inferred without the use of priors are listed. Parent Codelink probe ID is in column 1, Parent entrez ID in column 2, Parent official gene symbol in column 3, Child Codelink probe ID in column 4, Child entrez ID in column 5 and Child official gene symbol in column 6.
This file shows the Codelink Human Uniset I gene array data from siRNA experiments that were used as a Bayesian prior for the gene networks shown in Figure 8. Column 1 shows the probe ID. The remaining columns show normalised transcript abundance following 24hrs incubation with siRNA duplexes. The official gene symbols of the transcripts targeted by the siRNA duplexes are shown in the column headers.
Edges of the Bayesian gene regulatory network inferred with the use of priors are listed. Parent Codelink probe ID is in column 1, Parent entrez ID in column 2, Parent official gene symbol in column 3, Child Codelink probe ID in column 4, Child entrez ID in column 5 and Child official gene symbol in column 6
References
- Aburatani S, et al. Discovery of novel transcription control relationships with gene regulatory networks generated from multiple-disruption full genome expression libraries. DNA Res. 2003;10:1–8. doi: 10.1093/dnares/10.1.1. doi:10.1093/dnares/10.1.1 [DOI] [PubMed] [Google Scholar]
- Ackermann E.J, Taylor J.K, Narayana R, Bennett C.F. The role of antiapoptotic Bcl-2 family members in endothelial apoptosis elucidated with antisense oligonucleotides. J. Biol. Chem. 1999;274:11 245–11 252. doi: 10.1074/jbc.274.16.11245. doi:10.1074/jbc.274.16.11245 [DOI] [PubMed] [Google Scholar]
- Adams J.M, et al. Control of apoptosis in hematopoietic cells by the Bcl-2 family of proteins. Cold Spring Harb. Symp. Quant. Biol. 1999;64:351–358. doi: 10.1101/sqb.1999.64.351. doi:10.1101/sqb.1999.64.351 [DOI] [PubMed] [Google Scholar]
- Aggarwal K, Lee K.H. Functional genomics and proteomics as a foundation for systems biology. Brief Funct. Genomics Proteomics. 2003;2:175–184. doi: 10.1093/bfgp/2.3.175. doi:10.1093/bfgp/2.3.175 [DOI] [PubMed] [Google Scholar]
- Akerman G.S, et al. Alterations in gene expression profiles and the DNA-damage response in ionizing radiation-exposed TK6 cells. Environ. Mol. Mutagen. 2005;45:188–205. doi: 10.1002/em.20091. doi:10.1002/em.20091 [DOI] [PubMed] [Google Scholar]
- Akutsu T, Kuhara S, Maruyama O, Miyano S. A system for identifying genetic networks from gene expression patterns produced by gene disruptions and overexpressions. Genome Inform. Ser. Workshop Genome Inform. 1998;9:151–160. [PubMed] [Google Scholar]
- Aoudjit F, Vuori K. Matrix attachment regulates Fas-induced apoptosis in endothelial cells: a role for c-flip and implications for anoikis. J. Cell Biol. 2001;152:633–643. doi: 10.1083/jcb.152.3.633. doi:10.1083/jcb.152.3.633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfuso F, Meyer G.T. Apoptosis does not affect the vasculature of the corpus luteum of pregnancy in the rat. Apoptosis. 2003;8:665–671. doi: 10.1023/A:1026283414726. doi:10.1023/A:1026283414726 [DOI] [PubMed] [Google Scholar]
- Bach F.H, Hancock W.W, Ferran C. Protective genes expressed in endothelial cells: a regulatory response to injury. Immunol. Today. 1997;18:483–486. doi: 10.1016/s0167-5699(97)01129-8. doi:10.1016/S0167-5699(97)01129-8 [DOI] [PubMed] [Google Scholar]
- Baffert F, Le T, Sennino B, Thurston G, Kuo C.J, Hu-Lowe D, McDonald D.M. Cellular changes in normal blood capillaries undergoing regression after inhibition of VEGF signaling. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H547–H559. doi: 10.1152/ajpheart.00616.2005. doi:10.1152/ajpheart.00616.2005 [DOI] [PubMed] [Google Scholar]
- Bannerman D.D, Tupper J.C, Ricketts W.A, Bennett C.F, Winn R.K, Harlan J.M. A constitutive cytoprotective pathway protects endothelial cells from lipopolysaccharide-induced apoptosis. J. Biol. Chem. 2001;276:14 924–14 932. doi: 10.1074/jbc.M100819200. doi:10.1074/jbc.M100819200 [DOI] [PubMed] [Google Scholar]
- Baumer S, et al. Vascular endothelial cell-specific phosphotyrosine phosphatase (VE-PTP) activity is required for blood vessel development. Blood. 2006;107:4754–4762. doi: 10.1182/blood-2006-01-0141. doi:10.1182/blood-2006-01-0141 [DOI] [PubMed] [Google Scholar]
- Benjamin L.E, Hemo I, Keshet E. A plasticity window for blood vessel remodelling is defined by pericyte coverage of the preformed endothelial network and is regulated by PDGF- B and VEGF. Development. 1998;125:1591–1598. doi: 10.1242/dev.125.9.1591. [DOI] [PubMed] [Google Scholar]
- Biancone L, Martino A.D, Orlandi V, Conaldi P.G, Toniolo A, Camussi G. Development of inflammatory angiogenesis by local stimulation of Fas in vivo. J. Exp. Med. 1997;186:147–152. doi: 10.1084/jem.186.1.147. doi:10.1084/jem.186.1.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop E.T, Bell G.T, Bloor S, Broom I.J, Hendry N.F.K, Wheatley D.N. An in vitro model of angiogenesis: basic features. Angiogenesis. 1999;3:335–344. doi: 10.1023/a:1026546219962. doi:10.1023/A:1026546219962 [DOI] [PubMed] [Google Scholar]
- Blagosklonny M.V. Apoptosis, proliferation, differentiation: in search of the order. Semin. Cancer Biol. 2003;13:97–105. doi: 10.1016/s1044-579x(02)00127-x. doi:10.1016/S1044-579X(02)00127-X [DOI] [PubMed] [Google Scholar]
- Bombeli T, Karsan A, Tait J.F, Harlan J.M. Apoptotic vascular endothelial cells become procoagulant. Blood. 1997;89:2429–2442. [PubMed] [Google Scholar]
- Brazhnik P, de la Fuente A, Mendes P. Gene networks: how to put the function in genomics. Trends Biotechnol. 2002;20:467–472. doi: 10.1016/s0167-7799(02)02053-x. doi:10.1016/S0167-7799(02)02053-X [DOI] [PubMed] [Google Scholar]
- Bruneel A, Labas V, Mailloux A, Sharma S, Vinh J, Vaubourdolle M, Baudin B. Proteomic study of human umbilical vein endothelial cells in culture. Proteomics. 2003;3:714–723. doi: 10.1002/pmic.200300409. doi:10.1002/pmic.200300409 [DOI] [PubMed] [Google Scholar]
- Bruneel A, Labas V, Mailloux A, Sharma S, Royer N, Vinh J, Pernet P, Vaubourdolle M, Baudin B. Proteomics of human umbilical vein endothelial cells applied to etoposide-induced apoptosis. Proteomics. 2005;5:3876–3884. doi: 10.1002/pmic.200401239. doi:10.1002/pmic.200401239 [DOI] [PubMed] [Google Scholar]
- Cardier J.E, Erickson-Miller C.L. Fas (CD95)- and tumor necrosis factor-mediated apoptosis in liver endothelial cells: role of caspase-3 and the p38 MAPK. Microvasc. Res. 2002;63:10–18. doi: 10.1006/mvre.2001.2360. doi:10.1006/mvre.2001.2360 [DOI] [PubMed] [Google Scholar]
- Carmeliet P. Mechanisms of angiogenesis and arteriogenesis. Nat. Med. 2000;6:389–395. doi: 10.1038/74651. doi:10.1038/74651 [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Jain R.K. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. doi:10.1038/35025220 [DOI] [PubMed] [Google Scholar]
- Carmeliet P, et al. Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell. 1999;98:147–157. doi: 10.1016/s0092-8674(00)81010-7. doi:10.1016/S0092-8674(00)81010-7 [DOI] [PubMed] [Google Scholar]
- Chen T, He H.L, Church G.M. Modeling gene expression with differential equations. Pac. Symp. Biocomput. 1999;99:29–40. [PubMed] [Google Scholar]
- Chen J, Somanath P.R, Razorenova O, Chen W.S, Hay N, Bornstein P, Byzova T.V. Akt1 regulates pathological angiogenesis, vascular maturation and permeability in vivo. Nat. Med. 2005;11:1188–1196. doi: 10.1038/nm1307. doi:10.1038/nm1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang L.W, Grenier J.M, Ettwiller L, Jenkins L.P, Ficenec D, Martin J, Jin F, DiStefano P.S, Wood A. An orchestrated gene expression component of neuronal programmed cell death revealed by cDNA array analysis. Proc. Natl Acad. Sci. USA. 2001;98:2814–2819. doi: 10.1073/pnas.051630598. doi:10.1073/pnas.051630598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A, Courtman D.W, Langille B.L. Apoptosis (programmed cell death) in arteries of the neonatal lamb. Circ. Res. 1995;76:168–175. doi: 10.1161/01.res.76.2.168. [DOI] [PubMed] [Google Scholar]
- Davidson E.H, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. doi:10.1126/science.1069883 [DOI] [PubMed] [Google Scholar]
- de Hoon M.J, Imoto S, Kobayashi K, Ogasawara N, Miyano S. Inferring gene regulatory networks from time-ordered gene expression data of Bacillus subtilis using differential equations. Pac. Symp. Biocomput. 2003;8:17–28. [PubMed] [Google Scholar]
- Desmouliere A, Redard M, Darby I, Gabbiani G. Apoptosis mediates the decrease in cellularity during the transition between granulation tissue and scar. Am. J. Pathol. 1995;146:56–66. [PMC free article] [PubMed] [Google Scholar]
- Dickson S.E, Bicknell R, Fraser H.M. Mid-luteal angiogenesis and function in the primate is dependent on vascular endothelial growth factor. J. Endocrinol. 2001;168:409–416. doi: 10.1677/joe.0.1680409. doi:10.1677/joe.0.1680409 [DOI] [PubMed] [Google Scholar]
- Djonov V, Andres A.C, Ziemiecki A. Vascular remodelling during the normal and malignant life cycle of the mammary gland. Microsc. Res. Tech. 2001;52:182–189. doi: 10.1002/1097-0029(20010115)52:2<182::AID-JEMT1004>3.0.CO;2-M. doi:10.1002/1097-0029(20010115)52:2<182::AID-JEMT1004>3.0.CO;2-M [DOI] [PubMed] [Google Scholar]
- Dorstyn L, Colussi P.A, Quinn L.M, Richardson H, Kumar S. DRONC, an ecdysone-inducible Drosophila caspase. Proc. Natl Acad. Sci. USA. 1999;96:4307–4312. doi: 10.1073/pnas.96.8.4307. doi:10.1073/pnas.96.8.4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E, et al. In vivo induction of endothelial apoptosis leads to vessel thrombosis and endothelial denudation: a clue to the understanding of the mechanisms of thrombotic plaque erosion. Circulation. 2004;109:2503–2506. doi: 10.1161/01.CIR.0000130172.62481.90. doi:10.1161/01.CIR.0000130172.62481.90 [DOI] [PubMed] [Google Scholar]
- Duriez P.J, Wong F, Dorovini-Zis K, Shahidi R, Karsan A. A1 functions at the mitochondria to delay endothelial apoptosis in response to tumor necrosis factor. J. Biol. Chem. 2000;275:18 099–18 107. doi: 10.1074/jbc.M908925199. doi:10.1074/jbc.M908925199 [DOI] [PubMed] [Google Scholar]
- Durr E, Yu J, Krasinska K.M, Carver L.A, Yates J.R, Testa J.E, Oh P, Schnitzer J.E. Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat. Biotechnol. 2004;22:985–992. doi: 10.1038/nbt993. doi:10.1038/nbt993 [DOI] [PubMed] [Google Scholar]
- Duval H, Harris M, Li J, Johnson N, Print C. New insights into the function and regulation of endothelial cell apoptosis. Angiogenesis. 2003;6:171–183. doi: 10.1023/B:AGEN.0000021390.09275.bc. doi:10.1023/B:AGEN.0000021390.09275.bc [DOI] [PubMed] [Google Scholar]
- Eisen M.B, Spellman P.T, Brown P.O, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14 863–14 868. doi: 10.1073/pnas.95.25.14863. doi:10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierlbeck W, Liu A, Coyle R, Ballermann B.J. Endothelial cell apoptosis during glomerular capillary lumen formation in vivo. J. Am. Soc. Nephrol. 2003;14:1349–1354. doi: 10.1097/01.asn.0000061779.70530.06. doi:10.1097/01.ASN.0000061779.70530.06 [DOI] [PubMed] [Google Scholar]
- Filippatos G, Ang E, Gidea C, Dincer E, Wang R, Uhal B.D. Fas induces apoptosis in human coronary artery endothelial cells in vitro. BMC Cell Biol. 2004;5:6. doi: 10.1186/1471-2121-5-6. doi:10.1186/1471-2121-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S.A, Langille B.L, Srivastava D. Apoptosis during cardiovascular development. Circ. Res. 2000;87:856–864. doi: 10.1161/01.res.87.10.856. [DOI] [PubMed] [Google Scholar]
- Friedman N, Linial M, Nachman I, Pe'er D. Using Bayesian networks to analyze expression data. J. Comput. Biol. 2000;7:601–620. doi: 10.1089/106652700750050961. doi:10.1089/106652700750050961 [DOI] [PubMed] [Google Scholar]
- Friis T, Hansen A.B, Houen G, Engel A.M. Influence of angiogenesis inhibitors on endothelial cell morphology in vitro. APMIS. 2006;114:211–224. doi: 10.1111/j.1600-0463.2006.apm_189.x. doi:10.1111/j.1600-0463.2006.apm_189.x [DOI] [PubMed] [Google Scholar]
- Frisch S.M, Vuori K, Ruoslahti E, Chan-Hui P.Y. Control of adhesion-dependent cell survival by focal adhesion kinase. J. Cell Biol. 1996;134:793–799. doi: 10.1083/jcb.134.3.793. doi:10.1083/jcb.134.3.793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks Z, Persaud R.S, Alfieri A, McLoughlin M, Ehleiter D, Schwartz J.L, Seddon A.P, Cordon-Cardo C, Haimovitz-Friedman A. Basic fibroblast growth factor protects endothelial cells against radiation-induced programmed cell death in vitro and in vivo. Cancer Res. 1994;54:2582–2590. [PubMed] [Google Scholar]
- Galli C, Meucci O, Scorziello A, Werge T.M, Calissano P, Schettini G. Apoptosis in cerebellar granule cells is blocked by high KCl, forskolin, and IGF-1 through distinct mechanisms of action: the involvement of intracellular calcium and RNA synthesis. J. Neurosci. 1995;15:1172–1179. doi: 10.1523/JNEUROSCI.15-02-01172.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Sun W, Christofidou-Solomidou M, Sawada M, Newman D.K, Bergom C, Albelda S.M, Matsuyama S, Newman P.J. PECAM-1 functions as a specific and potent inhibitor of mitochondrial-dependent apoptosis. Blood. 2003;102:169–179. doi: 10.1182/blood-2003-01-0003. doi:10.1182/blood-2003-01-0003 [DOI] [PubMed] [Google Scholar]
- Gaytan F, Morales C, Bellido C, Sanchez-Criado J.E. Selective apoptosis of luteal endothelial cells in dexamethasone-treated rats leads to ischemic necrosis of luteal tissue. Biol. Reprod. 2002;66:232–240. doi: 10.1095/biolreprod66.1.232. doi:10.1095/biolreprod66.1.232 [DOI] [PubMed] [Google Scholar]
- Gerber H.P, Vu T.H, Ryan A.M, Kowalski J, Werb Z, Ferrara N. VEGF couples hypertrophic cartilage remodeling, ossification and angiogenesis during endochondral bone formation. Nat. Med. 1999;5:623–628. doi: 10.1038/9467. doi:10.1038/9467 [DOI] [PubMed] [Google Scholar]
- Gleichmann M, Buchheim G, El-Bizri H, Yokota Y, Klockgether T, Kugler S, Bahr M, Weller M, Schulz J.B. Identification of inhibitor-of-differentiation 2 (Id2) as a modulator of neuronal apoptosis. J. Neurochem. 2002;80:755–762. doi: 10.1046/j.0022-3042.2002.00760.x. doi:10.1046/j.0022-3042.2002.00760.x [DOI] [PubMed] [Google Scholar]
- Golpon H.A, Fadok V.A, Taraseviciene-Stewart L, Scerbavicius R, Sauer C, Welte T, Henson P.M, Voelkel N.F. Life after corpse engulfment: phagocytosis of apoptotic cells leads to VEGF secretion and cell growth. FASEB J. 2004;18:1716–1718. doi: 10.1096/fj.04-1853fje. [DOI] [PubMed] [Google Scholar]
- Granville D.J, Shaw J.R, Leong S, Carthy C.M, Margaron P, Hunt D.W, McManus B.M. Release of cytochrome c, Bax migration, Bid cleavage, and activation of caspases 2, 3, 6, 7, 8, and 9 during endothelial cell apoptosis. Am. J. Pathol. 1999;155:1021–1025. doi: 10.1016/S0002-9440(10)65202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh D.G. The role of apoptosis in wound healing. Int. J. Biochem. Cell Biol. 1998;30:1019–1030. doi: 10.1016/s1357-2725(98)00058-2. doi:10.1016/S1357-2725(98)00058-2 [DOI] [PubMed] [Google Scholar]
- Gygi S.P, Rochon Y, Franza B.R, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A.V, Jevnikar A.M. Significance of endothelial cell survival programs for renal transplantation. Am. J. Kidney Dis. 2003;41:1140–1154. doi: 10.1016/s0272-6386(03)00345-7. doi:10.1016/S0272-6386(03)00345-7 [DOI] [PubMed] [Google Scholar]
- Hristov M, Erl W, Linder S, Weber P.C. Apoptotic bodies from endothelial cells enhance the number and initiate the differentiation of human endothelial progenitor cells in vitro. Blood. 2004;104:2761–2766. doi: 10.1182/blood-2003-10-3614. doi:10.1182/blood-2003-10-3614 [DOI] [PubMed] [Google Scholar]
- Hughes S, Chang-Ling T. Roles of endothelial cell migration and apoptosis in vascular remodeling during development of the central nervous system. Microcirculation. 2000;7:317–333. doi:10.1038/sj.mn.7300119 [PubMed] [Google Scholar]
- Ideker T, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. doi:10.1126/science.292.5518.929 [DOI] [PubMed] [Google Scholar]
- Imoto S, Goto T, Miyano S. Estimation of genetic networks and functional structures between genes by using Bayesian networks and nonparametric regression. Pac. Symp. Biocomput. 2002;7:175–186. [PubMed] [Google Scholar]
- Imoto S, Kim S, Goto T, Miyano S, Aburatani S, Tashiro K, Kuhara S. Bayesian network and nonparametric heteroscedastic regression for nonlinear modeling of genetic network. J. Bioinform Comput. Biol. 2003a;1:231–252. doi: 10.1142/s0219720003000071. doi:10.1142/S0219720003000071 [DOI] [PubMed] [Google Scholar]
- Imoto S, Savoie C.J, Aburatani S, Kim S, Tashiro K, Kuhara S, Miyano S. Use of gene networks for identifying and validating drug targets. J. Bioinform. Comput. Biol. 2003b;1:459–474. doi: 10.1142/s0219720003000290. doi:10.1142/S0219720003000290 [DOI] [PubMed] [Google Scholar]
- Imoto S. Computational strategy for discovering druggable gene networks from genome-wide RNA expression profiles. Pac. Symp. Biocomput. 2006;11:559–571. [PubMed] [Google Scholar]
- Johnson N.A, et al. Endothelial cells preparing to die by apoptosis initiate a program of transcriptome and glycome regulation. FASEB J. 2004;18:188–190. doi: 10.1096/fj.03-0097fje. [DOI] [PubMed] [Google Scholar]
- Jordan B.R. How consistent are expression chip platforms? Bioessays. 2004;26:1236–1242. doi: 10.1002/bies.20128. doi:10.1002/bies.20128 [DOI] [PubMed] [Google Scholar]
- Kamino H, Hiratsuka M, Toda T, Nishigaki R, Osaki M, Ito H, Inoue T, Oshimura M. Searching for genes involved in arteriosclerosis: proteomic analysis of cultured human umbilical vein endothelial cells undergoing replicative senescence. Cell Struct. Funct. 2003;28:495–503. doi: 10.1247/csf.28.495. doi:10.1247/csf.28.495 [DOI] [PubMed] [Google Scholar]
- Kerr J.F, Wyllie A.H, Currie A.R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodarev N.N, Kataoka Y, Murley J.S, Weichselbaum R.R, Grdina D.J. Interaction of amifostine and ionizing radiation on transcriptional patterns of apoptotic genes expressed in human microvascular endothelial cells (HMEC) Int. J. Radiat. Oncol. Biol. Phys. 2004;60:553–563. doi: 10.1016/j.ijrobp.2004.04.060. doi:10.1016/j.ijrobp.2004.04.060 [DOI] [PubMed] [Google Scholar]
- Kilpatrick Z.E, Cakouros D, Kumar S. Ecdysone-mediated upregulation of the effector caspase DRICE is required for hormone-dependent apoptosis in Drosophila cells. J. Biol. Chem. 2005;25:11 981–11 986. doi: 10.1074/jbc.M413971200. doi:10.1074/jbc.M413971200 [DOI] [PubMed] [Google Scholar]
- Kim H.S, Hwang K.K, Seo J.W, Kim S.Y, Oh B.H, Lee M.M, Park Y.B. Apoptosis and regulation of Bax and Bcl-X proteins during human neonatal vascular remodeling. Arterioscler. Thromb. Vasc. Biol. 2000;20:957–963. doi: 10.1161/01.atv.20.4.957. [DOI] [PubMed] [Google Scholar]
- Kim S.Y, Imoto S, Miyano S. Inferring gene networks from time series microarray data using dynamic Bayesian networks. Brief. Bioinform. 2003;4:228–235. doi: 10.1093/bib/4.3.228. doi:10.1093/bib/4.3.228 [DOI] [PubMed] [Google Scholar]
- Kiyono M, Shibuya M. Inhibitory Smad transcription factors protect arterial endothelial cells from apoptosis induced by BMP4. Oncogene. 2006;25:7131–7137. doi: 10.1038/sj.onc.1209700. doi:10.1038/sj.onc.1209700 [DOI] [PubMed] [Google Scholar]
- Klebig C, Seitz S, Arnold W, Deutschmann N, Pacyna-Gengelbach M, Scherneck S, Petersen I. Characterization of γ-aminobutyric acid type A receptor-associated protein, a novel tumor suppressor, showing reduced expression in breast cancer. Cancer Res. 2005;65:394–400. [PubMed] [Google Scholar]
- Krupnick A.S, et al. Mechanism of T cell-mediated endothelial apoptosis. Transplantation. 2002;74:871–876. doi: 10.1097/00007890-200209270-00022. doi:10.1097/00007890-200209270-00022 [DOI] [PubMed] [Google Scholar]
- Lang R.A, Bishop J.M. Macrophages are required for cell death and tissue remodeling in the developing mouse eye. Cell. 1993;74:453–462. doi: 10.1016/0092-8674(93)80047-i. doi:10.1016/0092-8674(93)80047-I [DOI] [PubMed] [Google Scholar]
- Lang R, Lustig M, Francois F, Sellinger M, Plesken H. Apoptosis during macrophage-dependent ocular tissue remodelling. Development. 1994;120:3395–3403. doi: 10.1242/dev.120.12.3395. [DOI] [PubMed] [Google Scholar]
- Lee J.H, Rho S.B, Chun T. GABAA receptor-associated protein (GABARAP) induces apoptosis by interacting with DEAD (Asp-Glu-Ala-Asp/His) box polypeptide 47 (DDX 47) Biotechnol. Lett. 2005;27:623–628. doi: 10.1007/s10529-005-3628-2. doi:10.1007/s10529-005-3628-2 [DOI] [PubMed] [Google Scholar]
- Letarte M, Voulgaraki D, Hatherley D, Foster-Cuevas M, Saunders N.J, Barclay N. Analysis of leukocyte membrane protein interactions using protein microarrays. BMC Biochem. 2005;6:2. doi: 10.1186/1471-2091-6-2. doi:10.1186/1471-2091-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Dubey S, Varney M.L, Dave B.J, Singh R.K. IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J. Immunol. 2003;170:3369–3376. doi: 10.4049/jimmunol.170.6.3369. [DOI] [PubMed] [Google Scholar]
- Lian Z, Kluger Y, Greenbaum D.S, Tuck D, Gerstein M, Berliner N, Weissman S.M, Newburger P.E. Genomic and proteomic analysis of the myeloid differentiation program: global analysis of gene expression during induced differentiation in the MPRO cell line. Blood. 2002;100:3209–3220. doi: 10.1182/blood-2002-03-0850. doi:10.1182/blood-2002-03-0850 [DOI] [PubMed] [Google Scholar]
- Limaye V.S, Li X, Hahn C, Xia P, Berndt M.C, Vadas M.A, Gamble J.R. Sphingosine kinase-1 enhances endothelial cell survival through a PECAM-1-dependent activation of PI-3K/Akt and regulation of Bcl-2 family members. Blood. 2005;105:3169–3177. doi: 10.1182/blood-2004-02-0452. doi:10.1182/blood-2004-02-0452 [DOI] [PubMed] [Google Scholar]
- Lobov I.B, Brooks P.C, Lang R.A. Angiopoietin-2 displays VEGF-dependent modulation of capillary structure and endothelial cell survival in vivo. Proc. Natl Acad. Sci. USA. 2002;99:11 205–11 210. doi: 10.1073/pnas.172161899. doi:10.1073/pnas.172161899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobov I.B, et al. WNT7b mediates macrophage-induced programmed cell death in patterning of the vasculature. Nature. 2005;437:417–421. doi: 10.1038/nature03928. doi:10.1038/nature03928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madge L.A, Li J.H, Choi J, Pober J.S. Inhibition of phosphatidylinositol 3-kinase sensitizes vascular endothelial cells to cytokine-initiated cathepsin-dependent apoptosis. J. Biol. Chem. 2003;278:21 295–21 306. doi: 10.1074/jbc.M212837200. doi:10.1074/jbc.M212837200 [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Nishinakagawa H, Kurohmaru M, Hayashi Y, Otsuka J. Pregnancy and lactation affect the microvasculature of the mammary gland in mice. J. Vet. Med. Sci. 1992;54:937–943. doi: 10.1292/jvms.54.937. [DOI] [PubMed] [Google Scholar]
- Mecham B.H, et al. Sequence-matched probes produce increased cross-platform consistency and more reproducible biological results in microarray-based gene expression measurements. Nucleic Acids Res. 2004;32:e74. doi: 10.1093/nar/gnh071. doi:10.1093/nar/gnh071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medh R.D, Webb M.S, Miller A.L, Johnson B.H, Fofanov Y, Li T, Wood T.G, Luxon B.A, Thompson E.B. Gene expression profile of human lymphoid CEM cells sensitive and resistant to glucocorticoid-evoked apoptosis. Genomics. 2003;81:543–555. doi: 10.1016/s0888-7543(03)00045-4. doi:10.1016/S0888-7543(03)00045-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeson A, Palmer M, Calfon M, Lang R. A relationship between apoptosis and flow during programmed capillary regression is revealed by vital analysis. Development. 1996;122:3929–3938. doi: 10.1242/dev.122.12.3929. [DOI] [PubMed] [Google Scholar]
- Meeson A.P, Argilla M, Ko K, Witte L, Lang R.A. VEGF deprivation-induced apoptosis is a component of programmed capillary regression. Development. 1999;126:1407–1415. doi: 10.1242/dev.126.7.1407. [DOI] [PubMed] [Google Scholar]
- Mehta U, Kang B.P, Bansal G, Bansal M.P. Studies of apoptosis and bcl-2 in experimental atherosclerosis in rabbit and influence of selenium supplementation. Gen. Physiol. Biophys. 2002;21:15–29. [PubMed] [Google Scholar]
- Meier P, Finch A, Evan G. Apoptosis in development. Nature. 2000;407:796–801. doi: 10.1038/35037734. doi:10.1038/35037734 [DOI] [PubMed] [Google Scholar]
- Meredith J.E, Jr, Fazeli B, Schwartz M.A. The extracellular matrix as a cell survival factor. Mol. Biol. Cell. 1993;4:953–961. doi: 10.1091/mbc.4.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineo C, Deguchi H, Griffin J.H, Shaul P.W. Endothelial and antithrombotic actions of HDL. Circ. Res. 2006;98:1352–1364. doi: 10.1161/01.RES.0000225982.01988.93. doi:10.1161/01.RES.0000225982.01988.93 [DOI] [PubMed] [Google Scholar]
- Mitchell C.A, Risau W, Drexler H.C. Regression of vessels in the tunica vasculosa lentis is initiated by coordinated endothelial apoptosis: a role for vascular endothelial growth factor as a survival factor for endothelium. Dev. Dyn. 1998;213:322–333. doi: 10.1002/(SICI)1097-0177(199811)213:3<322::AID-AJA8>3.0.CO;2-E. doi:10.1002/(SICI)1097-0177(199811)213:3<322::AID-AJA8>3.0.CO;2-E [DOI] [PubMed] [Google Scholar]
- Modlich U, Kaup F.J, Augustin H.G. Cyclic angiogenesis and blood vessel regression in the ovary: blood vessel regression during luteolysis involves endothelial cell detachment and vessel occlusion. Lab Invest. 1996;74:771–780. [PubMed] [Google Scholar]
- Morris P.N, Dunmore B.J, Brindle N.P. Mutant Tie2 causing venous malformation signals through Shc. Biochem. Biophys. Res. Commun. 2006;346:335–338. doi: 10.1016/j.bbrc.2006.05.128. doi:10.1016/j.bbrc.2006.05.128 [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Nakata M, Kawabata S, Hamada S. Transcriptome analysis and gene expression profiles of early apoptosis-related genes in Streptococcus pyogenes-infected epithelial cells. Cell Microbiol. 2004;6:939–952. doi: 10.1111/j.1462-5822.2004.00412.x. doi:10.1111/j.1462-5822.2004.00412.x [DOI] [PubMed] [Google Scholar]
- Niswender G.D, Juengel J.L, Silva P.J, Rollyson M.K, McIntush E.W. Mechanisms controlling the function and life span of the corpus luteum. Physiol. Rev. 2000;80:1–29. doi: 10.1152/physrev.2000.80.1.1. [DOI] [PubMed] [Google Scholar]
- Norata G.D, Tonti L, Roma P, Catapano A.L. Apoptosis and proliferation of endothelial cells in early atherosclerotic lesions: possible role of oxidised LDL. Nutr. Metab. Cardiovasc. Dis. 2002;12:297–305. [PubMed] [Google Scholar]
- Nylund R, Leszczynski D. Proteomics analysis of human endothelial cell line EA.hy926 after exposure to GSM 900 radiation. Proteomics. 2004;4:1359–1365. doi: 10.1002/pmic.200300773. doi:10.1002/pmic.200300773 [DOI] [PubMed] [Google Scholar]
- Oh P, Li Y, Yu J, Durr E, Krasinska K.M, Carver L.A, Testa J.E, Schnitzer J.E. Subtractive proteomic mapping of the endothelial surface in lung and solid tumours for tissue-specific therapy. Nature. 2004;429:629–635. doi: 10.1038/nature02580. doi:10.1038/nature02580 [DOI] [PubMed] [Google Scholar]
- Park J.E, Keller G.A, Ferrara N. The vascular endothelial growth factor (VEGF) isoforms: differential deposition into the subepithelial extracellular matrix and bioactivity of extracellular matrix-bound VEGF. Mol. Biol. Cell. 1993;4:1317–1326. doi: 10.1091/mbc.4.12.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plavec I, et al. Method for analyzing signaling networks in complex cellular systems. Proc. Natl Acad. Sci. USA. 2004;101:1223–1228. doi: 10.1073/pnas.0308221100. doi:10.1073/pnas.0308221100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollman M.J, Naumovski L, Gibbons G.H. Endothelial cell apoptosis in capillary network remodeling. J. Cell Physiol. 1999;178:359–370. doi: 10.1002/(SICI)1097-4652(199903)178:3<359::AID-JCP10>3.0.CO;2-O. doi:10.1002/(SICI)1097-4652(199903)178:3<359::AID-JCP10>3.0.CO;2-O [DOI] [PubMed] [Google Scholar]
- Porn-Ares M.I, Saido T.C, Andersson T, Ares M.P. Oxidized low-density lipoprotein induces calpain-dependent cell death and ubiquitination of caspase 3 in HMEC-1 endothelial cells. Biochem. J. 2003;374:403–411. doi: 10.1042/BJ20021955. doi:10.1042/BJ20021955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pru J.K, Lynch M.P, Davis J.S, Rueda B.R. Signaling mechanisms in tumor necrosis factor alpha-induced death of microvascular endothelial cells of the corpus luteum. Reprod. Biol. Endocrinol. 2003;1:17. doi: 10.1186/1477-7827-1-17. doi:10.1186/1477-7827-1-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel C, Angus J, Ghahramani Z, Lioumi M, Sotheran E, Gaiba A, Wild D.L, Falciani F. Modeling T-cell activation using gene expression profiling and state-space models. Bioinformatics. 2004;20:1361–1372. doi: 10.1093/bioinformatics/bth093. doi:10.1093/bioinformatics/bth093 [DOI] [PubMed] [Google Scholar]
- Rath P.C, Aggarwal B.B. TNF-induced signaling in apoptosis. J. Clin. Immunol. 1999;19:350–364. doi: 10.1023/a:1020546615229. doi:10.1023/A:1020546615229 [DOI] [PubMed] [Google Scholar]
- Raymond M.A, Desormeaux A, Laplante P, Vigneault N, Filep J.G, Landry K, Pshezhetsky A.V, Hebert M.J. Apoptosis of endothelial cells triggers a caspase-dependent anti-apoptotic paracrine loop active on VSMC. FASEB J. 2004;18:705–707. doi: 10.1096/fj.03-0573fje. [DOI] [PubMed] [Google Scholar]
- Re F, Zanetti A, Sironi M, Polentarutti N, Lanfrancone L, Dejana E, Colotta F. Inhibition of anchorage-dependent cell spreading triggers apoptosis in cultured human endothelial cells. J. Cell Biol. 1994;127:537–546. doi: 10.1083/jcb.127.2.537. doi:10.1083/jcb.127.2.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B.C, Lalwani N.D, Johnson K.J, Marks R.M. Fas ligation triggers apoptosis in macrophages but not endothelial cells. Eur. J. Immunol. 1994;24:2640–2645. doi: 10.1002/eji.1830241111. doi:10.1002/eji.1830241111 [DOI] [PubMed] [Google Scholar]
- Robinson M, Jiang P, Cui J, Li J, Wang Y, Swaroop M, Madore S, Lawrence T.S, Sun Y. Global genechip profiling to identify genes responsive to p53-induced growth arrest and apoptosis in human lung carcinoma cells. Cancer Biol. Ther. 2003;2:406–415. doi: 10.4161/cbt.2.4.437. [DOI] [PubMed] [Google Scholar]
- Saidi S.A, Holland C.M, Kreil D.P, MacKay D.J, Charnock-Jones D.S, Print C.G, Smith S.K. Independent component analysis of microarray data in the study of endometrial cancer. Oncogene. 2004;23:6677–6683. doi: 10.1038/sj.onc.1207562. doi:10.1038/sj.onc.1207562 [DOI] [PubMed] [Google Scholar]
- Sata M, Walsh K. Endothelial cell apoptosis induced by oxidized LDL is associated with the down-regulation of the cellular caspase inhibitor FLIP. J. Biol. Chem. 1998;273:33 103–33 106. doi: 10.1074/jbc.273.50.33103. doi:10.1074/jbc.273.50.33103 [DOI] [PubMed] [Google Scholar]
- Sata M, Hirata Y, Nagai R. Role of Fas/Fas ligand interaction in ischemia-induced collateral vessel growth. Hypertens. Res. 2002;25:577–582. doi: 10.1291/hypres.25.577. doi:10.1291/hypres.25.577 [DOI] [PubMed] [Google Scholar]
- Satake S, Kuzuya M, Ramos M.A, Kanda S, Iguchi A. Angiogenic stimuli are essential for survival of vascular endothelial cells in three-dimensional collagen lattice. Biochem. Biophys. Res. Commun. 1998;244:642–646. doi: 10.1006/bbrc.1998.8313. doi:10.1006/bbrc.1998.8313 [DOI] [PubMed] [Google Scholar]
- Savoie C.J, Aburatani S, Watanabe S, Eguchi Y, Muta S, Imoto S, Miyano S, Kuhara S, Tashiro K. Use of gene networks from full genome microarray libraries to identify functionally relevant drug-affected genes and gene regulation cascades. DNA Res. 2003;10:19–25. doi: 10.1093/dnares/10.1.19. doi:10.1093/dnares/10.1.19 [DOI] [PubMed] [Google Scholar]
- Schechner J.S, et al. In vivo formation of complex microvessels lined by human endothelial cells in an immunodeficient mouse. Proc. Natl Acad. Sci. USA. 2000;97:9191–9196. doi: 10.1073/pnas.150242297. doi:10.1073/pnas.150242297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitt T, Brazma A. Modelling gene networks at different organisational levels. FEBS Lett. 2005;579:1859–1866. doi: 10.1016/j.febslet.2005.01.073. doi:10.1016/j.febslet.2005.01.073 [DOI] [PubMed] [Google Scholar]
- Schoenfeld J, et al. Bioinformatic analysis of primary endothelial cell gene array data illustrated by the analysis of transcriptome changes in endothelial cells exposed to VEGF-A and PlGF. Angiogenesis. 2004;7:143–156. doi: 10.1007/s10456-004-1677-0. doi:10.1007/s10456-004-1677-0 [DOI] [PubMed] [Google Scholar]
- Schulz J.B, Weller M, Klockgether T. Potassium deprivation-induced apoptosis of cerebellar granule neurons: a sequential requirement for new mRNA and protein synthesis, ICE-like protease activity, and reactive oxygen species. J. Neurosci. 1996;16:4696–4706. doi: 10.1523/JNEUROSCI.16-15-04696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura I, et al. Inhibition of programmed cell death impairs in vitro vascular-like structure formation and reduces in vivo angiogenesis. FASEB J. 2002;16:833–841. doi: 10.1096/fj.01-0819com. doi:10.1096/fj.01-0819com [DOI] [PubMed] [Google Scholar]
- Shmulevich I, Dougherty E.R, Kim S, Zhang W. Probabilistic Boolean networks: a rule-based uncertainty model for gene regulatory networks. Bioinformatics. 2002;18:261–274. doi: 10.1093/bioinformatics/18.2.261. doi:10.1093/bioinformatics/18.2.261 [DOI] [PubMed] [Google Scholar]
- Slowik M.R, Min W, Ardito T, Karsan A, Kashgarian M, Pober J.S. Evidence that tumor necrosis factor triggers apoptosis in human endothelial cells by interleukin-1-converting enzyme-like protease- dependent and -independent pathways. Lab Invest. 1997;77:257–267. [PubMed] [Google Scholar]
- Sprenger R.R, Speijer D, Back J.W, De Koster C.G, Pannekoek H, Horrevoets A.J. Comparative proteomics of human endothelial cell caveolae and rafts using two-dimensional gel electrophoresis and mass spectrometry. Electrophoresis. 2004;25:156–172. doi: 10.1002/elps.200305675. doi:10.1002/elps.200305675 [DOI] [PubMed] [Google Scholar]
- Stehlik C, de Martin R, Kumabashiri I, Schmid J.A, Binder B.R, Lipp J. Nuclear factor (NF)-kappaB-regulated X-chromosome-linked iap gene expression protects endothelial cells from tumor necrosis factor alpha- induced apoptosis. J. Exp. Med. 1998;188:211–216. doi: 10.1084/jem.188.1.211. doi:10.1084/jem.188.1.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri C, Jones P.F, Patan S, Bartunkova S, Maisonpierre P.C, Davis S, Sato T.N, Yancopoulos G.D. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 1996;87:1171–1180. doi: 10.1016/s0092-8674(00)81813-9. doi:10.1016/S0092-8674(00)81813-9 [DOI] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander E.S, Golub T.R. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl Acad. Sci. USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. doi:10.1073/pnas.96.6.2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P.K, Downey T.J, Spitznagel E.L, Jr, Xu P, Fu D, Dimitrov D.S, Lempicki R.A, Raaka B.M, Cam M.C. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res. 2003;31:5676–5684. doi: 10.1093/nar/gkg763. doi:10.1093/nar/gkg763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedgui A, Mallat Z. Apoptosis, a major determinant of atherothrombosis. Arch. Mal. Coeur Vaiss. 2003;96:671–675. [PubMed] [Google Scholar]
- Tertemiz F, Kayisli U.A, Arici A, Demir R. Apoptosis contributes to vascular lumen formation and vascular branching in human placental vasculogenesis. Biol. Reprod. 2005;72:727–735. doi: 10.1095/biolreprod.104.034975. doi:10.1095/biolreprod.104.034975 [DOI] [PubMed] [Google Scholar]
- Valgimigli M, Merli E, Malagutti P, Soukhomovskaia O, Cicchitelli G, Macri G, Ferrari R. Endothelial dysfunction in acute and chronic coronary syndromes: evidence for a pathogenetic role of oxidative stress. Arch. Biochem. Biophys. 2003;420:255–261. doi: 10.1016/j.abb.2003.07.006. doi:10.1016/j.abb.2003.07.006 [DOI] [PubMed] [Google Scholar]
- Venkataraman G, Shriver Z, Davis J.C, Sasisekharan R. Fibroblast growth factors 1 and 2 are distinct in oligomerization in the presence of heparin-like glycosaminoglycans. Proc. Natl Acad. Sci. USA. 1999;96:1892–1897. doi: 10.1073/pnas.96.5.1892. doi:10.1073/pnas.96.5.1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D.L, Vacha S.J, Kirby M.L, Lo C.W. Connexin43 deficiency causes dysregulation of coronary vasculogenesis. Dev. Biol. 2005;284:479–498. doi: 10.1016/j.ydbio.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Wang S, Sorenson C.M, Sheibani N. Attenuation of retinal vascular development and neovascularization during oxygen-induced ischemic retinopathy in Bcl-2-/- mice. Dev. Biol. 2005;279:205–219. doi: 10.1016/j.ydbio.2004.12.017. doi:10.1016/j.ydbio.2004.12.017 [DOI] [PubMed] [Google Scholar]
- Weihua Z, Tsan R, Schroit A.J, Fidler I.J. Apoptotic cells initiate endothelial cell sprouting via electrostatic signaling. Cancer Res. 2005;65:11 529–11 535. doi: 10.1158/0008-5472.CAN-05-2718. doi:10.1158/0008-5472.CAN-05-2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn R.K, Harlan J.M. The role of endothelial cell apoptosis in inflammatory and immune diseases. J. Thromb. Haemost. 2005;3:1815–1824. doi: 10.1111/j.1538-7836.2005.01378.x. doi:10.1111/j.1538-7836.2005.01378.x [DOI] [PubMed] [Google Scholar]
- Woywodt A, Streiber F, de Groot K, Regelsberger H, Haller H, Haubitz M. Circulating endothelial cells as markers for ANCA-associated small-vessel vasculitis. Lancet. 2003;361:206–210. doi: 10.1016/S0140-6736(03)12269-6. doi:10.1016/S0140-6736(03)12269-6 [DOI] [PubMed] [Google Scholar]
- Wullner U, Weller M, Schulz J.B, Krajewski S, Reed J.C, Klockgether T. Bcl-2, Bax and Bcl-x expression in neuronal apoptosis: a study of mutant weaver and lurcher mice. Acta Neuropathol. (Berl.) 1998;96:233–238. doi: 10.1007/s004010050889. doi:10.1007/s004010050889 [DOI] [PubMed] [Google Scholar]
- Wyllie A.H, Arends M.J. Apoptosis in health and disease. In: Warrell D.A, Cox T.M, Firth J.D, Benz E.J, editors. Oxford textbook of medicine. 4th edn. Oxford University Press; Oxford, UK: 2003. 119–130. [Google Scholar]
- Yasumoto K, Okamoto S, Mukaida N, Murakami S, Mai M, Matsushima K. Tumor necrosis factor alpha and interferon gamma synergistically induce interleukin 8 production in a human gastric cancer cell line through acting concurrently on AP-1 and NF-kB-like binding sites of the interleukin 8 gene. J. Biol. Chem. 1992;267:22 506–22 511. [PubMed] [Google Scholar]
- Yu J, Tian S, Metheny-Barlow L, Chew L.J, Hayes A.J, Pan H, Yu G.L, Li L.Y. Modulation of endothelial cell growth arrest and apoptosis by vascular endothelial growth inhibitor. Circ. Res. 2001;89:1161–1167. doi: 10.1161/hh2401.101909. [DOI] [PubMed] [Google Scholar]
- Yu M, Chen D.M, Hu G, Wang H. Proteomic response analysis of endothelial cells of human coronary artery to stimulation with carbachol. Acta Pharmacol. Sin. 2004;25:1124–1130. [PubMed] [Google Scholar]
- Zaina G, Morassutti C, De Amicis F, Fogher C, Marchetti S. Endonuclease genes up-regulated in tissues undergoing programmed cell death are expressed during male gametogenesis in barley. Gene. 2003;315:43–50. doi: 10.1016/s0378-1119(03)00820-5. doi:10.1016/S0378-1119(03)00820-5 [DOI] [PubMed] [Google Scholar]
- Zhang W, Ramdas L, Shen W, Song S.W, Hu L, Hamilton S.R. Apoptotic response to 5-fluorouracil treatment is mediated by reduced polyamines, non-autocrine Fas ligand and induced tumor necrosis factor receptor 2. Cancer Biol. Ther. 2003;2:572–578. doi: 10.4161/cbt.2.5.532. [DOI] [PubMed] [Google Scholar]
- Zhang W, Li D, Mehta J.L. Role of AIF in human coronary artery endothelial cell apoptosis. Am. J. Physiol. Heart Circ. Physiol. 2004;286:H354–H358. doi: 10.1152/ajpheart.00579.2003. doi:10.1152/ajpheart.00579.2003 [DOI] [PubMed] [Google Scholar]
- Zheng L, Dengler T.J, Kluger M.S, Madge L.A, Schechner J.S, Maher S.E, Pober J.S, Bothwell A.L. Cytoprotection of human umbilical vein endothelial cells against apoptosis and CTL-mediated lysis provided by caspase-resistant Bcl-2 without alterations in growth or activation responses. J. Immunol. 2000;164:4665–4671. doi: 10.4049/jimmunol.164.9.4665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The triplicated apoptosis timecuorse Codelink Human Uniset I gene array data for the transcripts discussed here are shown. Column 1 shows the probe ID. The remaining columns show normalised transcript abundance. Column headers begin with the time after apoptosis induction (hrs or min) and end with the replicate number
Edges of the Bayesian gene regulatory network inferred without the use of priors are listed. Parent Codelink probe ID is in column 1, Parent entrez ID in column 2, Parent official gene symbol in column 3, Child Codelink probe ID in column 4, Child entrez ID in column 5 and Child official gene symbol in column 6.
This file shows the Codelink Human Uniset I gene array data from siRNA experiments that were used as a Bayesian prior for the gene networks shown in Figure 8. Column 1 shows the probe ID. The remaining columns show normalised transcript abundance following 24hrs incubation with siRNA duplexes. The official gene symbols of the transcripts targeted by the siRNA duplexes are shown in the column headers.
Edges of the Bayesian gene regulatory network inferred with the use of priors are listed. Parent Codelink probe ID is in column 1, Parent entrez ID in column 2, Parent official gene symbol in column 3, Child Codelink probe ID in column 4, Child entrez ID in column 5 and Child official gene symbol in column 6