Abstract
Feedback loops play an important role in determining the dynamics of biological networks. To study the role of negative feedback loops, this article introduces the notion of distance-to-positive-feedback which, in essence, captures the number of independent negative feedback loops in the network, a property inherent in the network topology. Through a computational study using Boolean networks, it is shown that distance-to-positive-feedback has a strong influence on network dynamics and correlates very well with the number and length of limit cycles in the phase space of the network. To be precise, it is shown that, as the number of independent negative feedback loops increases, the number (length) of limit cycles tends to decrease (increase). These conclusions are consistent with the fact that certain natural biological networks exhibit generally regular behavior and have fewer negative feedback loops than randomized networks with the same number of nodes and same connectivity.
INTRODUCTION
An understanding of the design principles of biochemical networks such as gene regulatory, metabolic, or intracellular signaling networks is a central concern of systems biology. In particular, the intricate interplay between network topology and resulting dynamics is crucial to our understanding of such networks, as is their presumed modular structure. Features that relate network topology to dynamics may be considered robust in the sense that their influence does not depend on detailed quantitative features such as exact flux rates. A topological feature of central interest in this context is the existence of positive and negative feedback loops. There is broad consensus that feedback loops have a decisive effect on dynamics, which has been studied extensively through the analysis of mathematical network models, both continuous and discrete. Indeed, it has long been appreciated by biologists that positive and negative feedback loops play a central role in controlling the dynamics of a wide range of biological systems. Thomas et al. (1) conjectured that positive feedback loops are necessary for multistationarity, whereas negative feedback loops are necessary for the existence of periodic behaviors. Proofs for different partial cases of these conjectures have been given (see (2–5), D. Angeli, M. Hirsch, and E. Sontag, unpublished). Moreover, it has often been pointed out (see, for instance, (7)) that an abundance of loops, and specifically negative loops (8,9), should result in longer cycles and thus more “chaotic” behavior in the network. Our results provide strong evidence in support of this.
We focus here on Boolean network models, a popular model type for biochemical networks, initially introduced by Kauffman (10). In particular, we study Boolean network models in which each directed edge can be characterized as either an inhibition or an activation. In Boolean models of biological networks, each variable can only attain two values (0/1 or on/off). These values represent whether a gene is being expressed, or the concentration of a protein is above a certain threshold, at time t. When detailed information on kinetic rates of protein-DNA or protein-protein interactions is lacking, and especially if regulatory relationships are strongly sigmoidal, such models are useful in theoretical analysis, because they serve to focus attention on the basic dynamical characteristics while ignoring specifics of reaction mechanisms (see (11–14)).
Boolean networks constructed from monotone Boolean functions (i.e., each node or gate computes a function which is increasing on all arguments) are of particular interest, and have been studied extensively, in the electronic circuit design and pattern recognition literature (15,16), as well as in the computer science literature (see, e.g., (17–19) for recent references). For Boolean and all other finite iterated systems, all trajectories must either settle into equilibria or into periodic orbits, whether the system is made up of monotone functions or not, but monotone networks have always somewhat shorter cycles. This is because periodic orbits must be antichains, i.e., no two different states can be compared (see (15,20)). An upper bound may be obtained by appealing to Sperner's Theorem (21): Boolean systems on n variables can have orbits of period up to 2n, but monotone systems cannot have orbits of size larger than 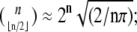 these are all classical facts in Boolean circuit design (15). It is also known that the upper bound is tight (15), in the sense that it is possible to construct Boolean systems on n variables, made up of monotone functions, for which orbits of the maximal size
these are all classical facts in Boolean circuit design (15). It is also known that the upper bound is tight (15), in the sense that it is possible to construct Boolean systems on n variables, made up of monotone functions, for which orbits of the maximal size 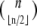 given by Sperner's Theorem exist. This number is still exponential in n. However, anecdotal experience suggests that monotone systems, constructed according to reasonable interconnection topologies and/or using restricted classes of gate functions, tend to exhibit shorter orbits (22,23). One may ask if the architecture of the network, that is, the structure of its dependency (also called interconnection) graph, helps insure shorter orbits. In this direction, Aracena et al. (17) showed that on certain graphs, called caterpillars, monotone networks can only have cycles of length of at most two in their phase space.
given by Sperner's Theorem exist. This number is still exponential in n. However, anecdotal experience suggests that monotone systems, constructed according to reasonable interconnection topologies and/or using restricted classes of gate functions, tend to exhibit shorter orbits (22,23). One may ask if the architecture of the network, that is, the structure of its dependency (also called interconnection) graph, helps insure shorter orbits. In this direction, Aracena et al. (17) showed that on certain graphs, called caterpillars, monotone networks can only have cycles of length of at most two in their phase space.
This article asks the even more general question of whether networks that are not necessarily made up from monotone functions, but which are close-to-monotone (in a sense to be made precise, roughly meaning that there are few independent negative loops), have shorter cycles than networks which are relatively farther to monotone.
In Sontag (8), we conjectured that smaller distance-to-monotone should correlate with more ordered (i.e., less chaotic) behavior, for random Boolean networks. A partial confirmation of this conjecture was provided in Kwon and Cho (9), where the relationship between the dynamics of random Boolean networks and the ratio of negative/positive feedback loops was investigated, albeit only for the special case of small Kauffman-type NK and NE networks, and with the additional restriction that all nodes have the same function chosen from AND, OR, or UNBIAS. Based on computer simulations, the authors of Kwon and Cho (9) found a positive (negative) correlation between the ratio of fixed points (other limit cycles) and the ratio of positive feedback loops. Observe that this differs from our conjecture in two fundamental ways:
Our measure of disorder is related to the number of independent negative loops, rather than their absolute number.
We do not consider that the number of positive loops should be part of this measure: a large number of negative loops will tend to produce large periodic orbits, even if the negative/ positive ratio is small due to a larger number of positive loops.
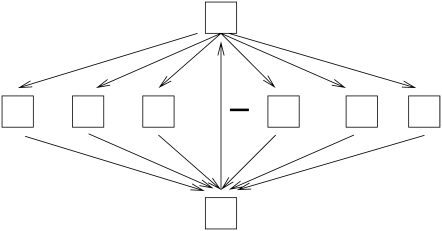
Thus, in the spirit of the conjecture in Sontag (8), the current article has as its goal an experimental study (as opposed to a theoretical analysis) of the effect of independent negative feedback loops on network dynamics, based on an appropriately defined measure of distance to positive-feedback. We study the effect of this distance on features of the network dynamics, namely the number and length of limit cycles. Rather than focusing on the number of negative feedback loops in the network as the characteristic feature of a network, we focus on the number of switches of the activation/inhibition character of edges that need to be made to obtain a network that has only positive feedback loops. We relate this measure to the cycle structure of the phase space of the network. It is worth emphasizing that the absolute number of negative feedback loops and the distance to positive feedback are not correlated in any direct way, as it is easy to construct networks with a fixed distance to positive feedback that have arbitrarily many negative feedback loops (see Fig. 1).
FIGURE 1.
A graph that has an arbitrary number of negative loops, as many as the number of nodes in the second layer, but its PF-distance is 1: to avoid negative feedback, it suffices to switch the sign of the single (negative) arrow from the bottom to the top node. All unlabeled arrows are positive.
Motivations
There are three different motivations for posing the question that we ask in this article. The first is that most biological networks appear to have highly regular dynamical behavior, settling upon simple periodic orbits or steady states. The second motivation is that it appears that real biological networks such as gene regulatory networks and protein signaling networks are indeed close to monotone (24–26). Thus, one may ask if being close to monotone correlates in some way with shorter cycles. Unfortunately, as mentioned above, one can build networks that are monotone yet exhibit exponentially long orbits. This suggests that one way to formulate the problem is through a statistical exploration of graph topologies, and that is what we do here.
A third motivation arises from the study of systems with continuous variables, which arguably provide more accurate models of biochemical networks. There is a rich theory of continuous-variable monotone (to be more precise, cooperative) systems. These are systems defined by the property that an inequality a(0) < b(0) in initial conditions propagates in time so that the inequality a(t) < b(t) remains true for all future times t > 0. Note that this is entirely analogous to the Boolean case, when one makes the obvious definition that two Boolean vectors satisfy the inequality a = (a1, …, an) ≤ b = (b1, …, bn) if ai ≤ bi for each i = 1, …, n (setting 0 < 1). Monotone continuous systems have convergent behavior. For example, in continuous-time (ordinary differential models), they cannot admit any possible stable oscillations (27–29), and, when there is only one steady state, every bounded solution converges to this unique steady state (monostability), see (30). When, instead, there are multiple steady states, the Hirsch Generic Convergence Theorem is the fundamental result (20,29,31,32); it states, under an additional technical assumption (strong monotonicity) that generic bounded solutions must converge to the set of steady states. Biological applications of these theorems include positive gene feedback circuits (20) as well as single phosphorylation/dephosphorylation loops (33) and double phosphorylation/dephosphorylation loops under appropriate assumptions on kinetic constants (34). For discrete-time strongly monotone systems, generically also stable oscillations are allowed besides convergence to equilibria, but no more complicated behavior. In neither case, discrete-time or continuous-time continuous monotone systems, does one observe chaotic behavior. It is an open question whether continuous systems that are in some sense close to being monotone have more regular behavior, in a statistical sense, than systems that are far from being monotone, just as for the Boolean analog considered in this article. The Boolean case is more amenable to computational exploration than continuous-variable systems, however. Since long orbits in discrete systems may be viewed as an analog of chaotic behavior, we focus on lengths of orbits.
One can proceed in several ways to define precisely the meaning of distance to positive feedback. One associates to a network made of unate (definition below) gate functions a signed graph whose edges have signs (positive or negative) that indicate how each variable affects each other variable (activation or inhibition). The first definition, explored in the literature (8,24,25,35,36), starts from the observation that in a network with all monotone node functions there are no negative undirected cycles. Conversely, if the dependency graph has no undirected negative parity cycles (a sign-consistent graph), then a change of coordinates (globally replacing a subset of the variables by their complements) renders the overall system monotone. Thus, asking what is the smallest number of sign-flips needed to render a graph sign-consistent is one way to define distance to monotone. This approach makes contact with areas of statistical physics (the number in question amounts to the ground energy of an associated Ising spin-glass model), as well as with the general theory of graph-balancing for signed graphs (37) that originated in Harary (38). It is also consistent with the generally accepted meaning of monotone-with-respect-to-some-orthant-order in the ordinary differential equation literature as a system that is cooperative under some inversion of variables.
A second, and different, definition starts from the fact that a network with all monotone node functions has, in particular, no negative-sign directed loops. For a strongly connected graph, the property that no directed negative cycles exist is equivalent to the property that no undirected negative cycles exist. However, for nonstrongly connected graphs, the properties are not the same. Thus, this second property is weaker. The second property is closer to what biologists and engineers mean by not having negative feedbacks in a system, and hence is perhaps more natural for applications. In addition, it is intuitively clear that negative feedbacks should be correlated to possible oscillatory behavior. (This is basically Thomas' conjecture. See (25) for precise statements for continuous-time systems; interestingly, published proofs of Thomas' conjecture use the first definition, because they appeal to results from monotone dynamical systems.) Thus, one could also define distance to monotone as the smallest number of sign-flips needed to render a graph free of negative directed loops. To avoid confusion, we will call this notion, which is the one studied in this article, distance-to-positive-feedback (PF-distance).
THEORY
Distance-to-positive-feedback
We give here the basic definitions of the concepts relevant to the study.
Definition 1
Let k = {0, 1} be the field with two elements. We order the two elements as 0 < 1. This ordering can be extended to a partial ordering on kn by comparing vectors coordinatewise in the lexicographic ordering.
A Boolean function h: kn→k is monotone if, whenever a ≤ b coordinatewise, for a, b ∈ kn, then h(a) ≤ h(b).
- A Boolean function h is unate if, whenever xi appears in h, the following holds:
- Either
- (a) For all a1, …, ai–1, ai+1, …, an ∈ k,
- h(a1, …, ai–1, 0, ai+1, …, an) ≤ h(a1, …, ai–1, 1, ai+1, …, an),
- Or
- (b) For all a1, …, ai–1, ai+1, …, an ∈ k,
- h(a1, …, ai–1, 0, ai+1, …, an) ≥ h(a1, …, ai–1, 1, ai+1, …, an).
The definition of unate function is equivalent to requiring that whenever ai appears in h, then it appears either everywhere as ai or everywhere as ¬ai: = 1 + ai.
Let f be a Boolean network with variables x1, …, xn, and coordinate functions f1, …, fn. That is, f = (f1,…,fn): kn → kn. We can associate to f its dependency graph 𝒟(f): The vertices are v1, …, vn, corresponding to the variables x1, …, xn, and there is an edge vi → vj if and only if there exists a1, …, ai–1, ai+1, …, an ∈ k such that fj(a1, …, ai–1, 0, ai+1, …, an) ≠ fj(a1, …, ai–1, 1, ai+1, …, an). If all coordinate functions fi of f are unate, then the dependency graph of f is a signed graph. Namely, we associate to an edge vi → vj, a plus sign (+) if fj preserves the ordering as in 2(a) of Definition 1 and a minus sign (−) if it reverses the ordering as in 2(b) of Definition 1. For later use we observe that this graph (as any directed graph) can be decomposed into a collection of strongly connected components, with edges between strongly connected components going one way but not the other. (Recall that a strongly connected directed graph is one in which any two vertices are connected by a directed path.) That is, the graph can be represented by a partially ordered set in which the strongly connected components make up the elements and the edge direction between components determines the order in the partially ordered set.
Definition 2
Let f be a Boolean network with unate Boolean functions and 𝒟(f) be its signed dependency graph. Then
The graph 𝒟(f) is a positive-feedback (PF) if it does not contain any odd parity directed cycles. (The parity of a directed cycle is the product of the signs of all the edges in the cycle.) In this case, f is called a positive-feedback (PF) network.
The PF-distance of f is the smallest number of signs that need to be changed in the dependency graph to obtain a PF network. We denote this number by |𝒟(f)| or simply | f |.
Notice that for a given directed graph G, different assignments of sign to the edges produce graphs with varying PF-distance. In particular, there is a maximal PF-distance that a given graph topology can support.
The dynamics of f are presented in a directed graph, called the phase space of f, which has the 2n elements of kn as a vertex set, and there is an edge  if f(a) = b. It is straightforward to see that each component of the phase space has the structure of a directed cycle, i.e., a limit cycle, with a directed tree feeding into each node of the limit cycle. The elements of these trees are called transient states.
if f(a) = b. It is straightforward to see that each component of the phase space has the structure of a directed cycle, i.e., a limit cycle, with a directed tree feeding into each node of the limit cycle. The elements of these trees are called transient states.
In this article, we relate the dynamics of a Boolean network to its PF-distance. The following is a motivational example that explains the main results.
Example
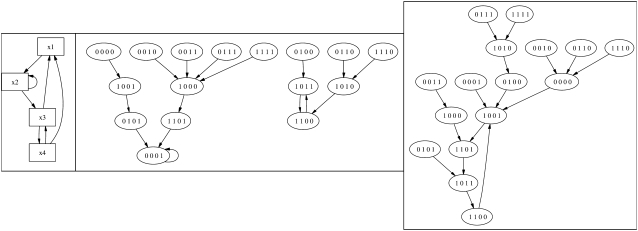
Let G be the directed graph depicted in Fig. 2 (left). It is easy to check that the maximal PF-distance of G is 3. Let f = (x3 ∨ ¬x4, i1 ∧ x2, x2 ∧ ¬x4, ¬x3): {0, 1}4→{0, 1}4 and g = (¬x3 ∨ x4, x1 ∧ ¬x2, x2 ∧ x4, ¬x3): {0, 1}4→{0, 1}4. It is clear that f and g are sign-modifications of the same PF network (x3 ∨ x4, x1 ∧ x2, x2 ∧ x4, x3); in particular, they have the same (unsigned) dependency graph. However, the PF-distance of f is 0 while it is 3 for g. The phase space of f is depicted in Fig. 2 (middle) and that of g is on the right. Notice that f has two limit cycles of lengths 1 and 2, respectively, while g has only one limit cycle of length 4.
FIGURE 2.
The dependency graph (left), the phase space of f (middle), and the phase space of g (right) from the Example in text. These graphs were generated using DVD (40).
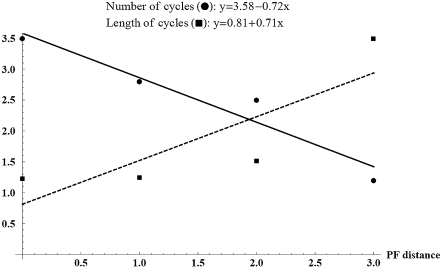
For each distance 0 ≤ d ≤ 3, we analyze the dynamics of 10 random PF networks and their sign modifications of distance d on the directed graph in Fig. 2 (left). The average of the numbers (lengths) of limit cycles is computed as in Table 1. The best fit-line of the averages of the number (length) of limit cycles is computed and its slope is reported as in Fig. 3. The details of this analysis are provided in the Supplementary Material, Data S1.
TABLE 1.
The average of the numbers (lengths) of limit cycles of the networks from the Example in text
| D | Average number | Average length |
|---|---|---|
| 0 | 3.5 | 1.23 |
| 1 | 2.80 | 1.25 |
| 2 | 2.50 | 1.52 |
| 3 | 1.20 | 3.50 |
FIGURE 3.
The best fit-line of the averages of the numbers (lengths) of limit cycles from Table 1.
Note that the purpose of the experiment is to determine a trend in the number and length of cycles as the PF-distance increases. It is thus appropriate to use a straight line to approximate the data points, even though this might not at all be the best possible approximation. But the slope of the line of best fit incorporates that trend adequately. We have repeated the experiment in the Example above for many different graphs and observed that the slope of the best fit-line of the length (respectively, number) of limit cycles is positive (respectively, negative) most of the time. In the next section we present the details of the experiments and the algorithms used in the computations.
METHODS
The main results of this article relate the PF-distance of Boolean networks with the number and length of their limit cycles. Specifically, our hypothesis is that, for Boolean networks consisting of unate functions, as the PF-distance increases, the total number of limit cycles decreases on average and their average length increases. This is equivalent to saying that for most or all experiments this slope is negative for the number of limit cycles and is positive for their length.
To test this hypothesis we analyzed the dynamics of >6,000,000 Boolean networks arranged in ∼130,000 experiments.
Random generation of unate functions
We generated a total of >130,000 random directed graphs, where each graph has 5,7,10,15, 20, or 100 nodes, with maximum in-degree 5 for each node. The graphs were generated as random adjacency matrices, with the restriction that each row has at least one “1” and at most five “1”s. For each directed graph G, we generated 10 Boolean networks with unate functions and dependency graph G, by using the following fact.
Lemma
A Boolean function f of n variables is unate if and only if it is of the form f(x) = g(x + s), where g is a monotone Boolean function of n variables and s ∈ kn and “+” denotes addition modulo 2.
Proof
If f is unate then each variable xi appears in f always as xi or always as ¬xi. Suppose that all xi appear without negations. Then f is constructed using ∧ and ∨ . Hence f is monotone. Otherwise, let s ∈ kn be the vector whose ith entry is 1 if and only if xi appears as ¬xi in f. Then g(x) = f(x + s) is a monotone function and f(x) = g(x + s). The converse is clear.
So to generate unate functions it is sufficient to generate monotone functions. We generated the set Mi of monotone functions in i variables by exhaustive search for i = 1, …, 5. (For example, M5 has 6894 elements.) Unate functions for a given signed dependency graph can then be generated by choosing random functions from Mi and random vectors s ∈ kn. The nonzero entries in s for a given node correspond to the incoming edges with negative sign in the dependency graph. Using this process we generated Boolean networks with unate Boolean functions.
We then carry out the following experiment.
The Experiment
Let G be a random unsigned directed graph on n nodes with a maximal PF-distance t, and let D ≤ t. Consider 10 unate Boolean networks chosen at random with G as their dependency graph.
- For 1 ≤ d ≤ D, let Gd be a signed graph of G of distance d.
- (a) For each network f of the 10 networks,
- i. Let g be a modified network of f such that 𝒟(g) = Gd; the signed dependency graph of g is Gd.
- ii. Compute the number and length of all limit cycles in the phase space of g.
- (b) Compute the average number N (respectively, average length L) of limit cycles in the phase spaces of the g values.
Compute the slope sN (respectively, sL) of the best fit-line of the N values (respectively, L values).
The output of a single experiment consists of the two nonnegative integers sN and sL.
Computation of PF-distance
Let f be a Boolean network with unate Boolean functions and let | f | be its PF-distance. The proofs of the following facts are straightforward.
Suppose the dependency graph of f has a negative feedback loop at a vertex. Let f′ be the Boolean network obtained by changing a single sign to make the loop positive. Then | f | = | f ′| + 1.
- Let H1, …, Hs be the strongly connected components of the dependency graph 𝒟(f). Then
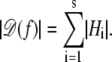
The algorithm for computing |Hi| now follows.
Algorithm: Distance to PF
Input. A signed, directed, and strongly connected graph G.
Output. |G|; the PF-distance of G.
Let d = 0.
Step 1. Let G1, …, Gr be the collection of all signed graphs obtained by making exactly d sign changes in G.
Step 2. For
If Gi is PF, then RETURN |G| = d.
Step 3. Otherwise, d: = d + 1, Go to Step 1 above.
In Step 2 above, to check whether a strongly connected graph is PF, it is equivalent to check whether it has any (undirected) negative cycles, which can easily be done in many different ways, see, e.g., Sontag (25). This algorithm must terminate, since G has finitely many edges and hence the PF-distance of G is finite.
If G has m directed edges, then there are 2m possible sign assignments. However, to compute the maximal PF-distance, one does not need to find the PF-distance of such possible assignments; see Data S1 for the algorithm we used to compute the maximal PF-distance.
We have coded these algorithms into a Mathematica code that we use to carry out our experiment. Once the number of nodes n, the maximal in-degree k of each node, and the maximal considered distance D are fixed, a random directed graph is generated and the experiment is carried out as shown above. Although we will not analyze the complexity here, it is clear that, as the value of n, k, or D increases, the time needed to do the experiment increases exponentially. In Table 2 we report the average CPU time (minutes) for some of the experiments where two of the variables are fixed.
TABLE 2.
The average CPU time (seconds) as we increase the number of nodes n, the maximum in-degree of each node k, and the maximal-considered distance D
|
k = 2 and D = 4
|
n = 5 and D = Max.
|
n = 10 and k = 5
|
|||
|---|---|---|---|---|---|
| n | Average CPU | k | Average CPU | D | Average CPU |
| 5 | 0.5951 | 2 | 0.6256 | 5 | 56.779 |
| 7 | 2.842 | 3 | 1.525 | 6 | 82.718 |
| 10 | 32.892 | 4 | 7.4078 | 7 | 101.35 |
| 15 | 2445.3 | 5 | 208.39 | 8 | 204.45 |
RESULTS
As mentioned in the previous section, we have carried out ∼130,000 such experiments for networks whose number of nodes ranges from 5 to 100. Table 3 summarizes the outcome of these experiments and represents the main result of the article. The rightmost two columns list the percentage of experiments for each network size that conform to our main hypothesis. As can be seen, the table confirms the hypothesis strongly, in particular for networks of smaller size. To explain the drop in the percentage of networks that conform to the hypothesis for larger networks, we need to consider some details of the experiments.
TABLE 3.
The percentage of experiments that conform to the hypotheses
| n | Number of experiments | Average number | Average length |
|---|---|---|---|
| 5 | 117,000 | 99.75 | 99.83 |
| 7 | 5000 | 97.82 | 99.92 |
| 10 | 6000 | 95.70 | 99.58 |
| 15 | 2921 | 95.72 | 98.25 |
| 20 | 331 | 90.03 | 94.86 |
| 100 | 659 | 77.39 | 93.93 |
To begin with, we need to observe that a computationally expensive part of an experiment is the computation of the maximal PF-distance which a given directed graph topology can support, since this is done essentially by an exhaustive test of all possible sign distributions on the edges. This computation becomes prohibitive for even modest-size graphs, with, e.g., 10 nodes. So unlike in the Example above, for networks on more than five nodes, we only considered PF-distances that are less than or equal to the number of nodes in the network. (See Methods for a detailed description of the experiment.) In fact, for graphs with 20 (respectively, 100) nodes, all considered networks have PF-distance ≤5 (respectively, 10). That is, our experiments reflect only the relationship between dynamics and PF-distance for networks that are very close to positive feedback, relative to their actual PF-distance.
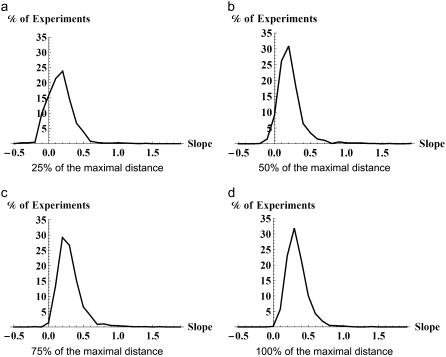
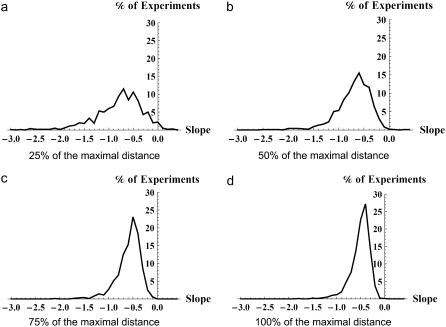
For networks on five nodes, we carried out 4000 experiments by varying the PF-distance considered in the computations. Table 4 shows the number of experiments that do not conform to our hypothesis as we vary the considered PF-distance. A more instructive way to visualize the effect of restricting the range of PF-distance relative to maximal PF-distance on the likelihood of experiments validating the conjecture is in the form of a histogram. In Figs. 4 and 5, the horizontal axis represents the slope of the lines of best fit and the vertical axis represents the percentage of experiments that confirm our hypothesis. Fig. 4 shows the results of the 4000 experiments on five-node networks. The histograms show the results when PF-distance of the network, respectively, is 25%, 50%, 75%, and 100% of the maximal PF-distance. It can be seen as the allowed range of PF-distance approaches the maximal distance, almost all experiments show positive slope of the best-fit line, thereby conforming to the conjecture. Similar results for the average number of limit cycles are shown in Fig. 5, demonstrating that if the PF-distance of networks is allowed the whole possible range, almost all the experiments conform to our hypothesis as we already noticed in Table 4.
TABLE 4.
The number of experiments that did not conform to our hypothesis for five-node networks; we considered PF-distance 25%, 50%, 75%, and 100% of the maximal distance (for each d, we considered 1000 experiments)
| D | Average number | Average length | Medium number | Medium length |
|---|---|---|---|---|
| 25% | 26 | 114 | 29 | 542 |
| 50% | 4 | 16 | 18 | 59 |
| 75% | 0 | 1 | 6 | 3 |
| 100% | 1 | 0 | 2 | 0 |
FIGURE 4.
Five-node networks. Histogram of slopes of best-fit lines to the average length of limit cycles (horizontal axis) versus percentage of experiments with a given slope (vertical axis). The panels from left to right include networks with increasing PF-distance, with 25%, 50%, 75%, and 100% of the maximal distance.
FIGURE 5.
Five-node networks. Histogram of slopes of best-fit lines to the average number of limit cycles (horizontal axis) versus percentage of experiments with a given slope (vertical axis). The panels from left to right include networks with increasing PF-distance, with 25%, 50%, 75%, and 100% of the maximal distance.
We have carried out similar computations for networks with seven (5000 experiments) and 10 (6000 experiments) nodes; see Data S1 for details. The results there are not quite as clear as for five-node networks, primarily because all computations were done with PF-distance ≤5, due to the computational complexity involved. For instance, for networks with 10 nodes (and up to four incoming edges per node), 31 out of 1000 experiments did not conform to our hypothesis for networks with PF-distance up to five.
In summary, the extensive computations confirm our hypothesis that, as the PF-distance increases, the total number of limit cycles decreases on average and their average length increases. Furthermore, the slopes of the best-fit lines increasingly conform to our hypothesis the closer the PF-distance of the networks comes to the maximum PF-distance of the network topology.
For details of our analysis of the five-node networks and all other considered networks, see Data S1.
DISCUSSION
Negative feedback loops in biological networks play a crucial role in controlling network dynamics. The new measure of distance-to-positive-feedback (PF-distance) introduced in this article is designed to capture the notion of independent feedback loops. We have shown that PF-distance correlates very well with the average number and length of limit cycles in networks, key measures of network dynamics. By analyzing the dynamics of >6,000,000 Boolean networks, we have provided evidence that networks with a larger number of independent negative feedback loops tend to have longer limit cycles and thus may exhibit more random or chaotic behavior. Furthermore, the number of limit cycles tends to decrease as the number of independent negative feedback loops increases.
In general, the problem of computing the PF-distance of a network is NP-complete, as MAX-CUT can be mapped into it as a special case (see the literature (24,25,35) for a discussion for the analogous problem of distance to monotone). The question of computing distance to monotone has been the subject of a few recent articles (24,35,36). The first two of these proposed a randomized algorithm based on a semidefinite programming relaxation, while the last one suggested an efficient deterministic algorithm for graphs with small distance to monotone. Since a strongly connected component of a graph is monotone if and only if it has the PF property, methods for computing PF distance for large graphs may be developed by similar techniques. Work along these lines is in progress.
One may speculate that the regular behavior observed in biological networks is due, in some measure, to their presumably small distance to positive feedback. There are as of yet too few large networks known with information on directionality and signs of interactions, to be able to draw statistically meaningful conclusions. However, one may make some preliminary statements. It was shown in Prill et al. (7) that for five networks (Escherichia coli transcription, Saccharomyces cerevisiae transcription, STKE signaling, Drosophila transcription, and a Caenorhabditis elegans neuron), feedback loops are statistically highly underrepresented among other motifs. No sign information was available for some of these networks, but it has been observed that negative (i.e., incoherent) cycles are less abundant than positive (i.e., coherent) ones in certain networks (39), and this will in turn bias feedback loops to be negative, when posttranscriptional modifications are added to the model. Similarly, models of a segment polarity network in Drosophila and of an S. cerevisiae gene network were shown to be close to monotone (no negative undirected cycles) compared to random graphs (24,25,35), and a similar statement of small PF distance (now including feedback loops as well as feedforward loops) was made for a CA-1 neuron signaling network and an E. coli and S. cerevisiae network (26), although this study was restricted to small loops.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
The computational results presented here were in part obtained using Virginia Tech's Advanced Research Computing (http://www.arc.vt.edu) 128p SGI Altix 3700 (Inferno2) system.
E.S. was supported in part by National Science Foundation grant No. DMS-0614371. A.V.-C., R.L., and A.S.J. were supported partially by National Science Foundation grant No. DMS-0511441.
The four authors contributed equally to this article.
Editor: Costas D. Maranas.
References
- 1.Thomas, R., D. Thieffry, and M. Kaufman. 1995. Dynamical behavior of biological regulatory networks. I. Biological role of feedback loops and practical use of the concept of the loop-characteristic state. Bull. Math. Biol. 57:247–276. [DOI] [PubMed] [Google Scholar]
- 2.Soulé, C. 2003. Graphic requirements for multistationarity. Complexus. 1:123–133. [Google Scholar]
- 3.Plahte, E., T. Mestl, and S. Omholt. 1995. Feedback loops, stability and multistationarity in dynamical systems. J. Biol. Syst. 3:409–413. [Google Scholar]
- 4.Gauzé, J. 1998. Positive and negative circuits in dynamical systems. J. Biol. Syst. 6:11–15. [Google Scholar]
- 5.Cinquin, O., and J. Demongeot. 2002. Positive and negative feedback: striking a balance between necessary antagonists. J. Theor. Biol. 216:229–241. [DOI] [PubMed] [Google Scholar]
- 6.Reference deleted in proof.
- 7.Prill, R., P. Iglesias, and A. Levchenko. 2005. Dynamic properties of network motifs contribute to biological network organization. PLoS Biol. 3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sontag, E. 2005. Molecular systems biology and control. Eur. J. Control. 11:396–435. [Google Scholar]
- 9.Kwon, Y.-K., and K.-H. Cho. 2007. Boolean dynamics of biological networks with multiple coupled feedback loops. Biophys. J. 92:2975–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kauffman, S. 1969. Metabolic stability and epigenesis in randomly constructed genetic nets. J. Theor. Biol. 22:437–467. [DOI] [PubMed] [Google Scholar]
- 11.Kauffman, S. 1969. Homeostasis and differentiation in random genetic control networks. Nature. 224:177–178. [DOI] [PubMed] [Google Scholar]
- 12.Kauffman, S., and K. Glass. 1973. The logical analysis of continuous, nonlinear biochemical control networks. J. Theor. Biol. 39:103–129. [DOI] [PubMed] [Google Scholar]
- 13.Albert, R., and H. Othmer. 2003. The topology of the regulatory interactions predicts the expression pattern of the Drosophila segment polarity genes. J. Theor. Biol. 223:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaves, M., R. Albert, and E. Sontag. 2005. Robustness and fragility of Boolean models for genetic regulatory networks. J. Theor. Biol. 235:431–449. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, E. 1954. Lattice theoretic properties of frontal switching functions. J. Math. Phys. 33:57–67. [Google Scholar]
- 16.Minsky, M. 1967. Computation: Finite and Infinite Machines. Prentice-Hall, Englewood Cliffs, NJ.
- 17.Aracena, J., J. Demongeot, and E. Goles. 2004. On limit cycles of monotone functions with symmetric connection graph. Theor. Comput. Sci. 322:237–244. [Google Scholar]
- 18.Aracena, J., J. Demongeot, and E. Goles. 2004. Positive and negative circuits in discrete neural networks. IEEE Trans. Neural Netw. 15:77–83. [DOI] [PubMed] [Google Scholar]
- 19.Goles, E., and G. Hernández. 2000. Dynamical behavior of Kauffman networks with AND-OR gates. J. Biol. Syst. 8:151–175. [Google Scholar]
- 20.Smith, H. 1995. Monotone dynamical systems: an introduction to the theory of competitive and cooperative systems. In Mathematical Surveys and Monographs, Vol. 41. AMS, Providence, RI.
- 21.Anderson, I. 2002. Combinatorics of Finite Sets. Dover Publications, Mineola, NY.
- 22.Tosic, P. T., and G. A. Agha. 2004. Characterizing configuration spaces of simple threshold cellular automata. In Cellular Automata. Vol. 3305/2004, Lecture Notes in Computer Science. Springer, Heidelberg, Germany.
- 23.Greil, F., and B. Drossel. 2007. Kauffman networks with threshold functions. Eur. Phys. J. B. 57:109–113. [Google Scholar]
- 24.DasGupta, B., G. Enciso, E. Sontag, and Y. Zhang. 2007. Algorithmic and complexity aspects of decompositions of biological networks into monotone subsystems. Biosystems. 90:161–178. [DOI] [PubMed] [Google Scholar]
- 25.Sontag, E. 2007. Monotone and near-monotone biochemical networks. J. Sys. Synth. Biol. 1:59–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maayan, A., R. Iyengar, and E. D. Sontag. 2007. Intracellular regulatory networks are close to monotone systems. Technical report. Nature Proc. NPRE-2007-25-1.
- 27.Hadeler, K., and D. Glas. 1983. Quasimonotone systems and convergence to equilibrium in a population genetics model. J. Math. Anal. Appl. 95:297–303. [Google Scholar]
- 28.Hirsch, M. 1984. The dynamical systems approach to differential equations. Bull. AMS. 11:1–64. [Google Scholar]
- 29.Hirsch, M., and H. Smith. 2005. Monotone dynamical systems. In Handbook of Differential Equations, Ordinary Differential Equations, Vol. 2. Elsevier, Amsterdam, The Netherlands.
- 30.Dancer, E. 1998. Some remarks on a boundedness assumption for monotone dynamical systems. Proc. AMS. 126:801–807. [Google Scholar]
- 31.Hirsch, M. 1983. Differential equations and convergence almost everywhere in strongly monotone flows. Contemp. Math. 17:267–285. [Google Scholar]
- 32.Hirsch, M. 1985. Systems of differential equations that are competitive or cooperative. II: Convergence almost everywhere. SIAM J. Math. Anal. 16:423–439. [Google Scholar]
- 33.Angeli, D., and E. Sontag. 2008. Translation-invariant monotone systems, and a global convergence result for enzymatic futile cycles. Nonlinear Anal. B Real World Appl. 9:128–140. [Google Scholar]
- 34.Wang, L., and E. Sontag. 2008. Singularly perturbed monotone systems and an application to double phosphorylation cycles. J. Nonlinear Sci. In press.
- 35.Dasgupta, B., G. Enciso, E. Sontag, and Y. Zhang. 2006. Algorithmic and complexity results for decompositions of biological networks into monotone subsystems. In Lecture Notes in Computer Science: Experimental Algorithms. 5th International Workshop, WEA 2006. Springer-Verlag, Berlin, Germany.
- 36.Hüffner, F., N. Betzler, and R. Niedermeier. 2007. Optimal edge deletions for signed graph balancing. In Proceedings of the 6th Workshop on Experimental Algorithms (WEA07), June 6–8, 2007, Rome. Springer-Verlag, Berlin, Germany.
- 37.Zaslavsky, T. 1998. Bibliography of signed and gain graphs. Electr. J. Combinatorics. DS8.
- 38.Harary, F. 1953. On the notion of balance of a signed graph. Mich. Math. J. 2:143–146. [Google Scholar]
- 39.Shen-Orr, S., R. Milo, S. Mangan, and U. Alon. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31:64–68. [DOI] [PubMed] [Google Scholar]
- 40.Jarrah, A., R. Laubenbacher, and H. Vastani. 2004. DVD: discrete visual dynamics. www://http:dvd.vbi.vt.edu.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.