Abstract
Single-molecule force-clamp spectroscopy is a valuable tool to analyze unfolding kinetics of proteins. Previous force-clamp spectroscopy experiments have demonstrated that the mechanical unfolding of ubiquitin deviates from the generally assumed Markovian behavior and involves the features of glassy dynamics. Here we use single molecule force-clamp spectroscopy to study the unfolding kinetics of a computationally designed fast-folding mutant of the small protein GB1, which shares a similar β-grasp fold as ubiquitin. By treating the mechanical unfolding of polyproteins as the superposition of multiple identical Poisson processes, we developed a simple stochastic analysis approach to analyze the dwell time distribution of individual unfolding events in polyprotein unfolding trajectories. Our results unambiguously demonstrate that the mechanical unfolding of NuG2 fulfills all criteria of a memoryless Markovian process. This result, in contrast with the complex mechanical unfolding behaviors observed for ubiquitin, serves as a direct experimental demonstration of the Markovian behavior for the mechanical unfolding of a protein and reveals the complexity of the unfolding dynamics among structurally similar proteins. Furthermore, we extended our method into a robust and efficient pseudo-dwell-time analysis method, which allows one to make full use of all the unfolding events obtained in force-clamp experiments without categorizing the unfolding events. This method enabled us to measure the key parameters characterizing the mechanical unfolding energy landscape of NuG2 with improved precision. We anticipate that the methods demonstrated here will find broad applications in single-molecule force-clamp spectroscopy studies for a wide range of proteins.
INTRODUCTION
Folding and unfolding processes of biomacromolecules, including proteins and RNAs, are fundamental issues in life science. The folding and unfolding reactions of small RNAs and single-domain proteins are often described as Markovian processes (Poisson process) (1), which are memoryless and history-independent. The hallmark of Poisson processes is that their dwell times ti (interarrival time in the classical example of arrival process in stochastic analysis) are independent and follow identical single-exponential distribution with a rate constant of α: αe−αt, where t is time (2). This behavior has been directly demonstrated on small RNA systems using single-molecule fluorescence spectroscopy and optical tweezers (3,4). However, experimental verification of the Markovian behaviors of proteins folding/unfolding has been challenging (5). The recently developed single-molecule force-clamp spectroscopy offers tremendous promise to rigorously examine the Markovian nature of the mechanical unfolding of proteins as well as to determine how stretching force affects the mechanical unfolding kinetics of proteins. In single-molecule force-clamp spectroscopy experiments, polyproteins made of identical tandem repeats of the protein of interest are used. Under a constant stretching force, individual domains in the polyprotein unfold one by one, resulting in trajectories of stepwise elongation of the polyprotein. The currently used method to evaluate the Markovian behavior of protein unfolding is based on averaging normalized individual unfolding trajectories of polyprotein (6,7). However, important information about protein unfolding, such as the correlation between unfolding events as well as alternative pathways, will be lost during the averaging of individual trajectories. Hence, analyzing the dwell-time distribution of the individual unfolding events will be of critical importance to the examination of the Markovian nature of the unfolding processes.
Recently, an order-statistics-based formalism has been developed to analyze the unfolding kinetics of polyproteins with simple Markovian behavior as well as involving unfolding intermediate states (6–10). Using order-statistical analysis, Fernandez and colleagues (6–9) investigated the mechanical unfolding kinetics of ubiquitin and demonstrated that the mechanical unfolding of ubiquitin deviates from the generally assumed Markovian behavior and involves the features of glassy dynamics (7–9). However, it remains to be examined whether proteins belonging to the same protein fold family as ubiquitin will share similar unfolding characteristics.
Using single-molecule force-clamp spectroscopy, here we examine the mechanical unfolding kinetics of a small protein NuG2. NuG2, which has a similar β-grasp fold as ubiquitin, is a computationally designed fast-folding mutant of the small protein GB1 (11). It has been shown that the chemical unfolding of NuG2 follows a simple two-state kinetics behavior, e.g., Markovian process (11). By treating the mechanical unfolding of polyproteins as the superposition of multiple identical and independent Poisson processes, we developed a simple stochastic analysis approach to analyze the dwell-time distribution of individual unfolding events in polyprotein unfolding trajectories and test the validity of the Markovian assumption of mechanical unfolding of NuG2. Our results unambiguously demonstrate that the mechanical unfolding of NuG2 fulfills all criteria of a memoryless Markovian process. This result, in contrast with the complex mechanical unfolding behaviors observed for ubiquitin, serves as a direct experimental demonstration of the Markovian behavior for the mechanical unfolding of a protein and reveals the complexity of the unfolding dynamics among structurally similar proteins.
MATERIALS AND METHODS
Plasmid containing NuG2 gene was kindly provided by Prof. David Baker. NuG2 is a computationally designed mutant (11) of protein GB1. DNA encoding NuG2 was amplified via polymerase chain reaction using forward and reverse primers containing restriction sites BamHI and BglII followed by KpnI, respectively (12). The sequence of NuG2 gene was confirmed by direct DNA sequencing. Polyprotein (NuG2)8 was constructed using a well-established multistep cloning strategy based on the identity of the sticky ends generated by BamHI and BglII sites. The polyprotein was overexpressed in DH5α strain and purified using Co2+-NTA affinity chromatography.
Single-molecule force-clamp experiments were carried out on a custom-built atomic force microscope in the constant-force mode, which was constructed as described previously (13). The spring constant of each individual cantilever was calibrated in solution using the equipartition theorem before and after each experiment (14,15). All the force-extension measurements were carried out in phosphate buffered saline. A 1 μl polyprotein sample (∼500 ng) was added onto a clean glass coverslip covered by phosphate buffered saline and was allowed to absorb for 5 min before proceeding to atomic force microscopy measurements.
RESULTS AND DISCUSSION
Unfolding trajectories of polyprotein of (NuG2)8
To use single-molecule force-clamp spectroscopy (8,9,13,16) to directly probe the unfolding kinetics of NuG2, we constructed a polyprotein of (NuG2)8, which is made of eight identical tandem repeats of NuG2. In single-molecule force-clamp spectroscopy studies, polyproteins made of identical tandem repeats of the protein of interest are required to identify single-molecule stretching events unambiguously. Stretching polyprotein (NuG2)8 results in characteristic stepwise elongation of the polyprotein (Fig. 1 B), where the individual step corresponds to the mechanical unfolding of individual NuG2 domains. Fig. 1 B shows a typical extension-time recording of NuG2 in response to a constant stretching force of 70 pN. The upper trace in Fig. 1 B shows the time evolution of the force, in which spikes, transient relaxation of force to a lower value, were observed as a result of the finite response time of our analog electronic force feedback system in response to domain unfolding. The unfolding steps are spaced in time and are of equal sizes of ∼15 nm at 70 pN, indicating that NuG2 will elongate by 15 nm on unfolding at 70 pN, in good agreement with the force-extension measurements on the same protein. The unfolding of NuG2 occurs in sharp steps, indicating the stochastic nature of the unfolding reaction.
FIGURE 1.
Mechanical unfolding of (NuG2)8 under constant force. (A) Schematic trace of spontaneous miniature end-plate potentials (mEPPs). Dwell time ti is the time interval between successive mEPPs, which follow single-exponential distribution. The same schematic can also describe the Poisson arrival process in classical stochastic analysis. (B) Force-time and length-time curves of (NuG2)8. In the force-time curve, the force is clamped at a constant value of 70 pN. The force trace shows spikes caused by the finite response time of the force feedback system. The length-time curve is marked by ∼15-nm stepwise elongation of the end-to-end distance of the protein. Each step corresponds to the mechanical unfolding event of a protein domain in the polyprotein chain. ti defines the time over which any one of the i remaining folded domains unfolds. (C) Five representative length-time trajectories of the unfolding of the same (NuG2)8 polyprotein at 70 pN.
A simple stochastic analysis method to analyze dwell-time distribution
Dwell times ti are usually defined as the interarrival time in the classical arrival process in stochastic analysis (2). For example, in neurophysiology, the time interval between successive spontaneous miniature end-plate potentials is defined as the dwell time ti and has been shown to follow a single-exponential distribution (Fig. 1 A) (17). Although the use of polyproteins in force-clamp spectroscopy studies facilitates the identification of single-molecule stretching events, the dwell times ti (as defined in Fig. 1 B) are no longer independent and do not follow identical single-exponential decay. Thus, ti cannot be used to directly probe the Markovian nature of the mechanical unfolding process because of the finite number of NuG2 domains present in polyprotein.
To overcome such limitations, different stochastic analysis methods have to be used or developed. One approach is the order statistics analysis used by Fernandez and co-workers (9) in which a binomial distribution of dwell times was used to describe the stochastic dynamics of the unfolding of ubiquitin. Here, we develop a new stochastic analysis method to use dwell time, ti, to directly evaluate the Markov property of protein unfolding. This method will have the unique advantage of allowing one to pool all the available dwell times together to significantly improve the statistics and thus the precision of the measurements.
If the mechanical unfolding of NuG2 can be described as a Poisson process, we can then treat the unfolding trajectory shown in Fig. 1 B as superposition of independent and identical Poisson processes. As shown in Fig. 1 B, we define dwell time ti as the inter-unfolding time, representing the time from the last unfolding until the time at which any one of the i folded domains unfolds. If i domains remain folded in the polyprotein, the process of unfolding any one of these i domains can be considered as the superposition of i identical Poisson processes when unfoldings of each of the i domains are mutually independent (2). Therefore, the probability of observing the unfolding of any one of the i folded domains by time t is:
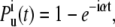 |
(1) |
where α is the unfolding rate constant of each domain. Hence, the probability density function for unfolding can be written as:
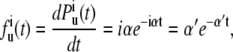 |
(2) |
where α′ = iα. It is evident that the probability distribution (Eq. 1) and probability density (Eq. 2) for the unfolding of a domain from the polyprotein are not stationary but dependent on the number of domains that remain folded (i). Thus, we can categorize the unfolding events with respect to the number of folded domains i in the polyprotein chain regardless of the total number of domains in a particular trajectory (Fig. 1, B and C). The dwell time of each category will follow Eq. 2. with the same apparent rate constant of α′. This simple relation allows us to take into account the nonstationary nature of polyprotein unfolding and accurately measure the unfolding rate constant, if the unfolding process of NuG2 is a Poisson process.
Accurately categorizing dwell time ti according to the number of domains that remain folded in the polyprotein chain
To use Eq. 2. to measure the unfolding rate constant accurately, it is critical to know the exact number of domains that remain folded (i) for each individual unfolding event in a given unfolding trajectory. However, because the polyprotein is picked up by the atomic force microscopy tip randomly along its contour, it is difficult to have prior knowledge of the number of domains in the chain being stretched. Because detachment of polyprotein from either the cantilever or the substrate can happen at any time, and an unfolding event could have occurred beyond the time window for a particular experiment, it is not reliable to determine i directly from the number of steps present in a given length-time curve (unfolding trajectory). Therefore, we used the following repetitive stretching-relaxation protocol to determine i accurately without any ambiguity.
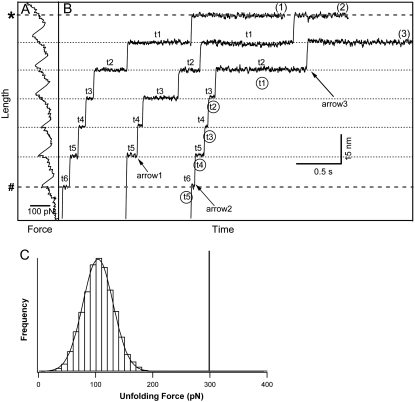
In our stretching-relaxation scheme, we first picked up a NuG2 polyprotein manually and stretched it at constant velocity until all the domains in the polyprotein chain unfolded (Fig. 2 A), and the force reached a high value of 300 pN (as the solid line in Fig. 2 C). The unfolding force histogram of NuG2 at a pulling speed of 400 nm/s is shown in Fig. 2 C. The average unfolding force of NuG2 is 105 ± 20 pN (average ± SD, n = 1773). It is clear that the probability for any NuG2 domain to remain folded at a force of 300 pN or higher is 0. Therefore, stretching the NuG2 polyprotein to 300 pN allowed us to ensure that all the NuG2 domains have unfolded and the exact number to be correctly counted. From this force-extension curve we also know the resting length (indicated by number sign (#) in Fig. 2) of the fully folded polyprotein at 70 pN at which we are going to carry out force-clamp experiment, and the length of the fully unfolded polyprotein (indicated by asterisk (*) in Fig. 2). We know as well the total number of domains N in this particular polyprotein (N = 6 in this example). We then relaxed the molecule to zero force and waited for a few seconds to allow all the domains to refold. We then carried out our force-clamp experiment, as shown in Fig. 2 B. The line indicated by # and the line indicated by * indicate the resting length for a fully folded polyprotein and the length of the fully unfolded polyprotein at the defined stretching force, respectively.
FIGURE 2.
Categorizing dwell time according to the number of folded domains in the chain. (A) Representative force-extension curve of polyprotein (NuG2)8. (B) Extension-time trajectories of the same (NuG2)8 during the force-clamp experiment using repetitive stretching protocols. (C) The comparison of the unfolding force histogram of NuG2 versus the high force (∼300 pN) used to ensure that all the NuG2 domains have been unfolded and the number of NuG2 domains in the chain is correctly counted.
From the resting length of each individual constant-force unfolding trajectory, we can readily assign ti to each unfolding event (see curve 1 in Fig. 2 B, for example). In curve 2, the resting length of the same polyprotein is ∼15 nm longer than the resting length for a fully folded polyprotein, indicating that one NuG2 domain did not fold, and curve B represents the unfolding trajectory of five folded NuG2 domains plus one unfolded NuG2 domain. Thus, the dwell time of the first event (indicated by arrow 1) will be assigned as t5. In curve 3, the resting length is the same as that of a fully folded polyprotein, but the final length is ∼15 nm shorter than that for a completely unfolded polyprotein, indicating that one NuG2 domain did not unfold within the given time window. Therefore, the dwell time for the first unfolding event (indicated by arrow 2) will be assigned as t6, and the last event (as indicated by arrow 3) will be assigned as t2. It is clear that if we did not have knowledge about the number of domains in the given polyprotein chain, the dwell times of the unfolding events in curve 3 would have been assigned incorrectly (as shown by circled ti). Therefore, it is critical to have knowledge of the number of domains that remain folded for each individual unfolding event, and in this article we only used the data obtained from stretching-relaxing experiments for which we have 100% certainty in ti assignment.
The mechanical unfolding of NuG2 is a Markovian process
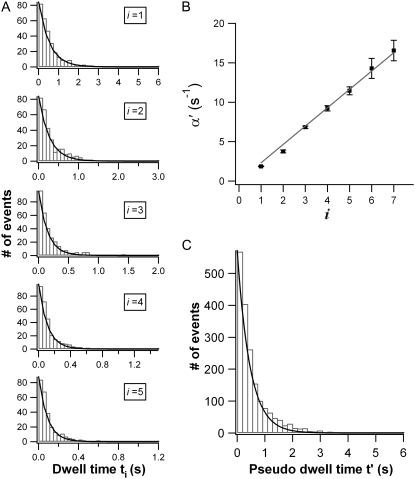
To test the Markovian nature of the mechanical unfolding of NuG2 experimentally, we separated dwell time into categories t1, t2 … tn as shown in Fig. 1 B. Using the method described above, we can determine i accurately without any ambiguity and assign i to each unfolding event with 100% certainty. Fig. 1 C shows five typical trajectories obtained from an experiment using such protocols. We categorized the dwell times of 1809 unfolding events. Histograms for ti (i = 1–5) are shown in Fig. 3 A, and those for ti (i = 6, 7) are shown in Fig. S1, Data S1, of the Supplementary Material. Each category of ti can be well described by a single exponential decay with the coefficient of determination R2 ranging from 0.984 to 0.995 (for 1 ≤ i ≤5, Table S1, Data S1, in the Supplementary Material), strongly indicating that Eq. 2, which is derived based on i mutually independent and identical Poisson processes, is an excellent description of the mechanical unfolding of NuG2 polyproteins. Hence, we can come to the conclusion that the mechanical unfolding of NuG2 is a history-independent Markovian process, and there is no correlation between the unfolding events of NuG2 domains.
FIGURE 3.
Unfolding kinetics of NuG2. (A) Dwell time ti follows exponential distribution. Solid lines are single exponential fits to the experimental data. (B) The relation of apparent unfolding rate constant α′(i) and the number i of domains that remain folded. α′(i) are proportional to i following the relation of α′(i) = iα. Linear fit (solid line) to the data measures an unfolding rate constant α of 2.33 s−1 for NuG2 at 70 pN. The error bars for all the data points correspond to the fitting error (Fig. 2 B), and some of the errors are so small that the error bars become invisible. The fitting error for each data point is (from left to right): 0.05, 0.18, 0.16, 0.34, 0.50, 1.27, and 1.30. (C) The distribution of pseudo-dwell time (t′) shows single exponential distribution. The pseudo-dwell times for all i are pooled together, which increases the statistics dramatically. Single-exponential fit (solid line) to the histogram measures an unfolding rate constant of 2.08 ± 0.08 s−1.
We also tested the appropriateness of other models (double exponential and power law) in describing the dwell-time ti distributions. As shown in Fig. S2 and Table S1 in the Supplementary Material, Data S1, it is evident that power law does not fit our data at all, excluding the possibility of glassy dynamics in the mechanical unfolding of NuG2. A double exponential function fits the experimental data with R2 comparable to those of single-exponential fits. However, the double-exponential fits are dominated by only one exponential term (amplitude is generally >90%), which has a rate constant very close to that of the single-exponential fits. These results strongly indicate that our measured dwell-time distribution can be genuinely described by a single-exponential distribution, corroborating the Markovian nature of the mechanical unfolding of NuG2.
Fitting each ti histogram to a single exponential function allows us to measure the apparent rate constant α′(i). Fig. 2 B plots α′(i) versus i. As predicted, α′(i) is linearly proportional to i following the equation of α′(i) = iα. A linear regression measures an α of 2.33 ± 0.04 s−1 (R2 = 0.995).
The confirmation that the stochastic model based on superposition of Poisson processes accurately describes the unfolding behavior of NuG2 polyprotein in force-clamp experiments presents direct proof that the mechanical unfolding of NuG2 is a Markovian process; that is, the unfolding of NuG2 is memoryless and does not depend on history. This result contrasts with the complex unfolding behaviors of ubiquitin, which has a similar β-grasp fold as NuG2 yet shows glassy dynamics in its unfolding behaviors (8), highlighting the complexity of folding/unfolding dynamics of structurally similar proteins. One of the possible reasons underlying such contrast could be the fact that NuG2 is a computationally designed protein (11). Because of the iterative energy minimization during the design, it is likely that frustration in the energy landscape of NuG2 has been largely removed, leading to a relative smooth energy landscape. Nevertheless, our finding provides an ideal model protein system for further investigation into the energy landscape underlying the unfolding and folding reactions of proteins. Moreover, these results also corroborate that the mechanical unfolding of individual NuG2 domains in the polyprotein (NuG2)8 are mutually independent, and therefore, there is no correlation between their unfolding events. Because this result is obtained at a stretching force of 70 pN, it remains to be demonstrated whether the same conclusion will hold for NuG2 at low forces.
Pseudo-dwell-time analysis allows accurate determination of key parameters characterizing the mechanical unfolding of NuG2
The mechanical unfolding of NuG2 is characterized by two key parameters: the spontaneous unfolding rate constant α0 at zero force and the distance to the transition state Δxu. The Bell-Evans model (18–20) has been widely used to extract these two parameters from force-spectroscopy measurements on polyproteins. It was shown that unfolding rate constant α(F) depends exponentially on the applied stretching force (18):
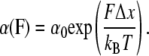 |
(3) |
However, the Bell-Evans model implicitly assumes two-state unfolding (Markovian) behavior, a characteristic that has been rarely tested rigorously especially for polyproteins made of identical tandem repeats. Having rigorously demonstrated that the mechanical unfolding of NuG2 domains in the polyprotein (NuG2)8 can be described as mutually independent Markovian processes, we can now extract these two key parameters using the dwell-time analysis we developed here. However, in this dwell-time analysis, categorizing dwell time ti according to i is a necessary step, which significantly reduced the sample size and hence reduced the precision of the measurement of unfolding rate constant dramatically (as the precision of the measurements is inversely proportional to the square root of the number of observations). For example, the sample size for ti is reduced from a pool of ∼1800 events to 301 for i = 1 and to 95 for i = 7. Because of the technical challenge of force-clamp experiments, it is not realistic to acquire sufficient data as in Fig. 3 at different stretching forces to confer a high-precision measurement of the unfolding rate constant at different forces. In extreme cases, it may become impossible to categorize dwell times ti according to i because of the limited number of unfolding events. It is thus of critical importance to further develop a more practical method that can make full use of all the available unfolding events by pooling all the dwell times ti together regardless of i.
Toward that goal, we now define a pseudo-dwell time t′ = i·t; hence, Eq. 1 becomes
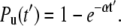 |
(4) |
It is of note that the new probability distribution (Eq. 4) is independent of i. Accordingly, the probability density function of unfolding with respect to pseudo-dwell time t′ can be written as
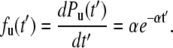 |
(5) |
The probability density function of unfolding with respect to t′ now has the form identical to that of a Poisson process and is independent of i. Thus, we can make full use of all the t′ from all the unfolding events. Fig. 3 C shows the histogram of t′ compiled from all the 1809 unfolding events obtained in our experiments. We can see that t′ shows a single exponential distribution with significantly improved statistics. Fitting the experimental data to Eq. 5 measures an average rate constant α of 2.08 ± 0.08 s−1 (R2 = 0.995).
Equation 5 provides a robust and efficient method for analyzing the force-clamp spectroscopy data. This method provides the possibility to use limited experimental observations to measure the unfolding rate constant with respectable precision for a Markovian-type unfolding reaction. We can now use this method to determine α0 and unfolding distance Δxu by carrying out force-clamp experiments on (NuG2)8 at different forces. We carried out force-clamp spectroscopy experiments on NuG2 at different stretching forces (53, 66, 70, 76, 89, 94, 107, 120 pN) and applied the pseudo-dwell time analysis to measure the unfolding rate constant at a given stretching force. Fig. 4 shows the force dependence of the unfolding rate constant of NuG2. As predicted by the Bell-Evans model, the logarithm of the unfolding rate constant α is linearly dependent on the stretching force: the higher the force is, the faster the unfolding rate is. Fitting the Bell-Evans model to the force dependence of the unfolding rate constant, we directly measured the spontaneous unfolding rate constant α0 of 0.031 ± 0.001 s−1 for NuG2 at zero force and the unfolding distance Δxu of 0.25 ± 0.01 nm for NuG2 (R2 = 0.988). From the spontaneous unfolding rate constant α0, one could then estimate the free energy barrier for unfolding. These two important parameters, free energy barrier and unfolding distance between the native state and transition state of NuG2, will provide quantitative information about the energy landscape for the mechanical unfolding of NuG2. It is of note that the measured Δxu for NuG2 is slightly bigger than that for ubiquitin (0.17 nm) (6) and similar to that for I27 (0.25 nm) (7). Moreover, the measured α0 and Δxu for NuG2 using force-clamp spectroscopy are similar to those measured for NuG2 using conventional force-extension measurements (α0 = 0.075 s−1 and Δxu = 0.25 nm, Y. Cao and H. Li, unpublished data). Despite the apparent close agreement between these two measurements for NuG2, the force-clamp-spectroscopy-based method demonstrated here has unique advantages. In conventional force-extension measurements, the relation between average unfolding force and pulling speed does not bear an analytical solution, and thus true analytical fitting is not feasible. Instead, Monte Carlo simulation or numerical solution is often used to estimate α0 and Δxu. In contrast, Eq. 3 makes it possible to fit the data as those in Fig. 4 to measure α0 and Δxu subjectively and accurately.
FIGURE 4.
Unfolding rate constant depends exponentially on the force. Using pseudo-dwell-time analysis, we measured the unfolding rate constants of NuG2 at different forces. The error bars for all the data points correspond to the fitting error. Some of the errors are so small that the error bars become invisible. The number of events and fitting errors for each individual data points, from left to right, are 298, 0.06; 569, 0.45; 1809, 0.07; 871, 0.06; 959, 0.15 148, 0.82; 278, 0.87; 178, and 1.57. Fitting Eq. 3 to the experimental data measured α0 of 0.031 ± 0.001 s−1 at zero force and unfolding distance Δxu of 0.25 ± 0.01 nm.
CONCLUSION
We have developed a simple stochastic analysis method allowing us to directly analyze the dwell-time distribution measured from the unfolding trajectories of polyproteins made of identical tandem repeats of the protein of interest. Using this method, we have directly demonstrated that the mechanical unfolding of NuG2 is a Markovian process, which is memoryless and independent of history. This observation contrasts with the glassy dynamics of the mechanical unfolding of ubiquitin, revealing the complexity of the unfolding kinetics among the proteins within the same protein fold. The robust pseudo-dwell-time analysis method makes it possible to use a limited amount of unfolding trajectories to measure the unfolding rate constant with respectable precision. These new approaches allow us to determine fundamental parameters characterizing the mechanical unfolding energy landscape of proteins. We anticipate that the methods demonstrated here will find a broad range of applications in single-molecule force-clamp spectroscopy studies for a wide range of proteins.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Prof. David Baker for providing the plasmid encoding NuG2.
This work is supported by the Natural Sciences and Engineering Research Council of Canada, the Canada Research Chairs program, and the Canada Foundation for Innovation.
Editor: Peter Hinterdorfer.
References
- 1.Fersht, A. R. 1992. Structure and Mechanism in Protein Science. Freeman, New York.
- 2.Cinlar, E. 1975. Introduction to Stochastic Processes. Prentice-Hall, Englewood Cliffs, NJ.
- 3.Zhuang, X., L. E. Bartley, H. P. Babcock, R. Russell, T. Ha, D. Herschlag, and S. Chu. 2000. A single-molecule study of RNA catalysis and folding. Science. 288:2048–2051. [DOI] [PubMed] [Google Scholar]
- 4.Liphardt, J., B. Onoa, S. B. Smith, I. J. Tinoco, and C. Bustamante. 2001. Reversible unfolding of single RNA molecules by mechanical force. Science. 292:733–737. [DOI] [PubMed] [Google Scholar]
- 5.Rhoades, E., M. Cohen, B. Schuler, and G. Haran. 2004. Two-state folding observed in individual protein molecules. J. Am. Chem. Soc. 126:14686–14687. [DOI] [PubMed] [Google Scholar]
- 6.Schlierf, M., H. Li, and J. M. Fernandez. 2004. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc. Natl. Acad. Sci. USA. 101:7299–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Manyes, S., J. Brujic, C. L. Badilla, and J. M. Fernandez. 2007. Force-clamp spectroscopy of single-protein monomers reveals the individual unfolding and folding pathways of I27 and ubiquitin. Biophys. J. 93:2436–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brujic, J., R. I. Hermans, K. A. Walther, and J. M. Fernandez. 2006. Single-molecule force spectroscopy reveals signatures of glassy dynamics in the energy landscape of ubiquitin. Nature Physics. 2:282–286. [Google Scholar]
- 9.Brujic, J., R. I. Z. Hermans, S. Garcia-Manyes, K. A. Walther, and J. M. Fernandez. 2007. Dwell-time distribution analysis of polyprotein unfolding using force-clamp spectroscopy. Biophys. J. 92:2896–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bura, E., D. K. Klimov, and V. Barsegov. 2007. Analyzing forced unfolding of protein tandems by ordered variates, 1: Independent unfolding times. Biophys. J. 93:1100–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nauli, S., B. Kuhlman, and D. Baker. 2001. Computer-based redesign of a protein folding pathway. Nat. Struct. Biol. 8:602–605. [DOI] [PubMed] [Google Scholar]
- 12.Carrion-Vazquez, M., A. F. Oberhauser, S. B. Fowler, P. E. Marszalek, S. E. Broedel, J. Clarke, and J. M. Fernandez. 1999. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA. 96:3694–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez, J. M., and H. Li. 2004. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 303:1674–1678. [DOI] [PubMed] [Google Scholar]
- 14.Florin, E. L., M. Rief, H. Lehmann, M. Ludwig, C. Dornmair, V. T. Moy, and H. E. Gaub. 1995. Sensing specific molecular-interactions with the atomic-force microscope. Biosens. Bioelectron. 10:895–901. [Google Scholar]
- 15.Hutter, J. L., and J. Bechhoefer. 1993. Erratum to Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64:1868–1873. [Google Scholar]
- 16.Oberhauser, A. F., P. K. Hansma, M. Carrion-Vazquez, and J. M. Fernandez. 2001. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc. Natl. Acad. Sci. USA. 98:468–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnston, D., and S. M.-S. Wu. 1995. Foundations of Cellular Neurophysiology. The MIT Press, Cambridge, MA.
- 18.Bell, G. I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. [DOI] [PubMed] [Google Scholar]
- 19.Evans, E. 2001. Probing the relation between force–lifetime–and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 30:105–128. [DOI] [PubMed] [Google Scholar]
- 20.Evans, E., and K. Ritchie. 1997. Dynamic strength of molecular adhesion bonds. Biophys. J. 72:1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.