Abstract
Solid-state NMR study shows that the 22-residue K3 peptide (Ser20-Lys41) from β2-microglobulin (β2m) adopts a β-strand-loop-β-strand conformation in its fibril state. Residue Pro32 has a trans conformation in the fibril state of the peptide, while it adopts a cis conformation in the native state of full-length β2m. To get insights into the structural properties of the K3 peptide, and determine whether the strand-loop-strand conformation is encoded at the monomeric level, we run all-atom explicit solvent replica exchange molecular dynamics on both the cis and trans variants. Our simulations show that the conformational space of the trans- and cis-K3 peptides is very different, with 1% of the sampled conformations in common at room temperature. In addition, both variants display only 0.3–0.5% of the conformations with β-strand-loop-β-strand character. This finding, compared to results on the Alzheimer's Aβ peptide, suggests that the biases toward aggregation leading to the β-strand-loop-β-strand conformation in fibrils are peptide-dependent.
INTRODUCTION
Protein aggregation plays a key role in many neurodegenerative diseases. For example, aggregates of the 40-residue Aβ-amyloid, the 210-residue prion, and the 99-residue β2-microgobulin are linked to Alzheimer's and Creutzfeldt-Jakob diseases, as well as dialysis-related amyloidosis, respectively. These proteins lack significant sequence identity and length similarity, yet they all form amyloid fibrils with a similar cross-β structure (1), suggesting the existence of a common assembly mechanism controlling the formation of these structures. Because the oligomeric species en route to fibril are transient, however, a detailed knowledge of their structures at the atomic level is still missing. Nevertheless, indirect evidence point to universal properties. For instance, it has been established that some unknown species are toxic and share similar morphological features based on antibody recognition experiments (2,3).
The human β2m(1–99) protein has five proline residues, with Pro32 adopting a cis conformation in the native state (4). In patients with chronic renal failure, as a result of long-term dialysis, the full-length protein aggregates into amyloid fibrils that often deposit on osteoarticular tissues, inducing severe bone/joint complications. It is well established that transition from the immunoglobulin fold in solution to the fibril state involves a cis-trans isomerization (5,6) and a nativelike, marginally populated intermediate with a trans-Pro32 appears to be a key precursor to fibril formation (7).
Recently, the 22-residue K3 peptide (Ser20-Lys41 fragment from human β2m(1–99) protein) was found to form amyloid fibrils in vitro (8,9). The 3D-structure reconstruction, based on solid-state NMR experiments (10), suggests that K3 fibrils consist of two-layered, parallel, and staggered β-sheets. In the fibrils, K3 folds into a β-strand (Asn21-Ser28)-loop (Gly29-Pro32)-β-strand (Ser33-Lys40) conformation as in the native full-length protein, and the residues Phe22, Asn24, Tyr26, and Ser28 are buried in the fibril while they are exposed to the surface in the native state.
Remarkably, the strand-loop-strand conformation is shared by many other peptides in their fibrillar states, for example the Alzheimer's Aβ(1–40) (11) and Aβ(1–42) peptides (12), the HET-s fragment (13), the second WW domain of human CA150 transcriptional activator (14), and a 19-residue fragment of the murine prion protein (15). This observation raises the possibility that the strand-loop-strand could be a marginally populated structure in solution, acting as an aggregation-prone building block for fibril assembly. Such a hypothesis has been proposed for Aβ(10–35) and is still a matter of debate. As we know, this peptide was described as random coil in solution from NMR. A transient hairpinlike strand-loop-strand, satisfying most of the NOE in solution (16), however, was recently observed in a 1.2 μs molecular dynamics (MD) trajectory at low pH (17). Because of the sampling limitations of MD, it is not clear whether this state is stable or only transient.
One of the main features of the K3 fibril is the presence of a trans His31-Pro32 peptide bond, while the corresponding bond in the native β2m(1–99) has a cis conformation. Although the relation between isomerization and amyloid structure is not clear, protein self-assembly with cis-trans proline isomerization has been discussed recently for the yeast prion protein Ure2 (18) and human cystatin C, the key protein in cerebral amyloid angiopathy (19). Increased interest in proline isomerization also comes from the recent finding that in Alzheimer's disease the prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-β production (20). Therefore, characterizing the free energy landscape of the trans-K3 and cis-K3 peptides in their monomeric states could provide insights into their structural differences in solution, and help develop a first qualitative picture of their differences in aggregation-prone properties.
In this study, we first examine the stability of a protofibril model using all-atom explicit solvent MD simulations with the Pro32 amino acid either in trans (as determined by the NMR chemical-shift data) or in cis conformations. Next, we use replica exchange molecular dynamics (REMD) simulations to explore the equilibrium structures of the trans-Pro32 and cis-Pro32 K3 peptides in solution. REMD is an enhanced sampling protocol and provides a better exploration of conformational space than long MD simulations (21–24).
MATERIALS AND METHODS
The amino-acid sequence of K3 peptide is SNFLNCYVSGFHPSDIEVDLLK. The residues involved in cis/trans isomerization are indicated in bold. To mimic the experimental acid conditions of pH <2.5, the side chains of Cys, Tyr, His, Glu, Asp, and Lys (Cys0, Tyr0, His+, Glu0, Asp0, and Lys+) and the N- and C-termini (NH3+, COOH) are protonated. The net charge of the peptide is +3. In all simulations, water is treated by the simple point charge model (25).
K3 protofibril
Two 40-ns MD simulations of the K3 protofibrils with the peptide in trans-Pro32 and cis-Pro32 conformations are performed using periodic boundary conditions in a rectangular water box. The protofibril model consists of eight peptides in a bilayer, parallel β-sheet, with each sheet being modeled by eight parallel β-strands. The initial intersheet Cα-Cα distance is set to 0.95 nm. Both the trans-K3 and cis-K3 protofibrils are solvated by 14,000 water molecules. The box dimensions are 7 nm × 8 nm × 8.2 nm.
K3 monomer
The structure of the cis-K3 peptide, taken from the full-length human protein (PDB code: 1JNJ (4)), is subject to a 10-ns MD simulation at 500 K. The final resulting structure shows no correlation with the initial state and is used as the starting point for the cis-K3 REMD simulation. The trans-K3 structure is taken from the fibril model of Goto et al. (PDB code: 2E8D (10)) and is also subject to a 10-ns MD run at 500 K. The final conformation is used as the initial state of the trans-K3 REMD simulation.
The peptides are solvated in a truncated octahedron box of 6000 water molecules (the minimum distance between the peptide and the box wall is 1.0 nm) and simulated using periodic boundary conditions. Before REMD production, a 1000-step steepest-descent minimization and MD equilibration of 1 ns are performed at each desired temperature.
MD and REMD simulations
MD simulations are performed in the NPT ensemble using GROMACS software (26) and OPLS-AA force field (27). The SETTLE algorithm is used to constrain the bond lengths and bond angles of water molecules. The bond lengths of the peptide are constrained by the LINCS algorithm. This allows an MD integration time step of 2 fs. A twin-range cutoff 1.0/1.4 nm is used for the nonbonded interactions, and a reaction-field correction with dielectric permittivity ε = 80 is used to calculate long-range electrostatics interactions. The temperature is controlled using the Berendsen method (28) with a coupling constant of 0.1 ps. The solute and solvent are separately coupled to external temperature and pressure baths. The pressure is kept constant at 1 bar using a coupling time of 1 ps (28).
REMD simulations are carried out in the NVT ensemble using 64 replicas, each of 82 ns, at temperatures exponentially spaced between 275 K and 475 K. The swap time between neighboring replicas is 1 ps and the acceptance ratio varies between 16% and 28%. In the REMD analysis, the first 22 ns of each replica are discarded. Clustering of the structures is carried out using the GROMOS method with a RMSD cutoff of 0.3 nm for residues 22–39. Secondary structure composition is calculated on the full sequence using the PROSS program (29).
RESULTS
OPLS force field recognizes native from nonnative protofibrils on short timescales
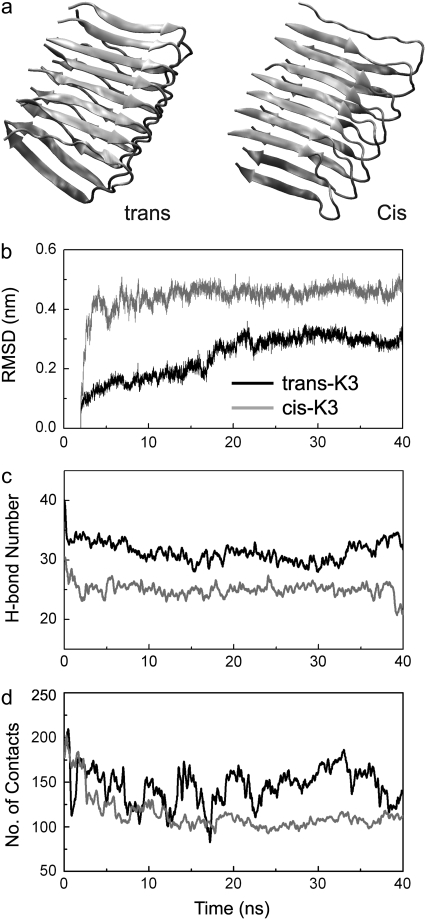
Based on the protofibril designed by Goto et al. (10) for the trans-K3 peptide, we construct a protofibril model for the cis-K3 peptide. Because of finite-size effects, we monitor the time evolution of several parameters involving the four central units of both layers. In Fig. 1 a, we see that for trans-K3 the Cα RMSD increases progressively in the first 20 ns and then stabilizes at ∼0.3 nm within the present 40-ns timescale. The RMSD profile for the cis-K3 protofibril is significantly different: a rapid increase to 0.38 nm in the first 3 ns followed by fluctuations ∼0.45 nm. Higher stability in the trans peptide is also seen in the time evolution of the number of intermolecular main chain H-bonds (Fig. 1 b) and intermolecular side-chain atomic contacts (Fig. 1 c). Here, a hydrogen bond (H-bond) is taken as formed if the donor-acceptor distance is <0.35 nm and the donor-hydrogen-acceptor angle is >150°, and two heavy atoms are in contact if their distances come within 0.54 nm. Time evolution of all parameters used indicates higher stability of the K3 fibril with the trans His31-Pro32 peptide bond on a 40-ns timescale. We recognize that force fields may lead to different results on long timescales, but μs simulations of fibrils are still out-of-reach.
FIGURE 1.
MD simulations of the trans-K3 and cis-K3 protofibrils. Both models are shown in panel a. The parameters used for comparison are calculated using the four central units of both sheets. (b) Cα-RMSD of the four central chains with respect to the MD-generated trans- and cis-K3 protofibril at 2 ns. Note that the trans structure at 2 ns deviates by 0.14 nm from the solid-state NMR-derived model. (c) Time evolution of the total number of intermolecular main-chain hydrogen bonds. (d) Time evolution of the number of intermolecular side-chain-side-chain atomic contacts between the two groups of residues: Ile35, Val37, Leu39 and Phe22, Asn24, Tyr26, and Phe30.
Free energy landscapes of the trans- and cis-K3 peptides in solution
To probe the conformational space of the trans and cis isomers, each peptide is subject to a REMD run starting from a distinct point, shown in Fig. 2 a. A sufficient sampling requires the trajectories to visit most/all of the available conformational space.
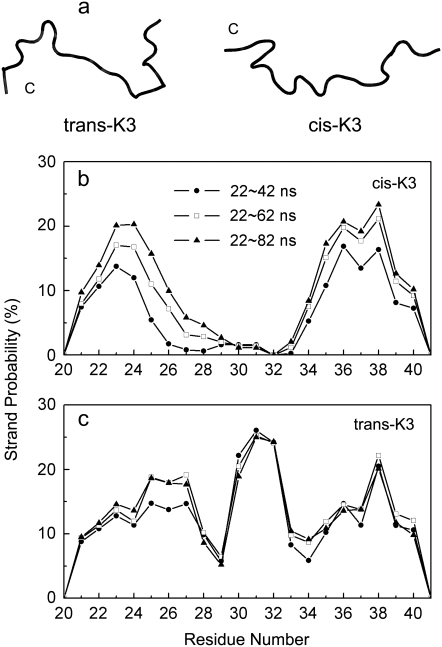
FIGURE 2.
Initial structures and first convergence test on the REMD runs. (a) Initial structures of the trans- and cis-K3 peptides used for REMD simulations. The position of the C-terminus is indicated. (b and c) The REMD-averaged β-strand probability of each residue at 298 K using the time intervals: 22–42 ns, 22–62 ns, and 22–82 ns.
Convergence of REMD runs is first examined by comparing the β-strand probability of each residue at 298 K for both peptides using four independent time intervals 2–22, 22–42, 42–62, and 62–82 ns. As seen in the Supplementary Material Data S1, Fig. S1, while the four distributions superpose well for the amino acids 21–23 and 29–40 in trans-K3 (Fig. 2 a), these profiles evolve considerably in cis-K3 (Fig. 2 a), in particular between the later intervals 42–62 ns and 62–82 ns and the interval 2–22 ns. The very low β-strand probability profile using 2–22 ns likely results from the strong structural correlation between the starting structures used by all replicas that remains in the first 22 ns. To decrease the bias associated with the initial state, data from 2 to 22 ns are therefore excluded for analysis.
The convergence of the REMD runs is next verified by comparing the β-stand probability of each residue at 298 K using now the cumulative time intervals: 22–42, 22–62, and 22–82 ns. Fig. 2 shows that the proportion of β-strands in the cis- and trans-K3 peptides reaches a relatively stable distribution after ∼62 ns, with deviations being within 4% from 22–62 ns to 22–82 ns for cis-K3 (Fig. 2 b), and 2% for trans-K3 (Fig. 2 c), indicating a convergence of the REMD simulations.
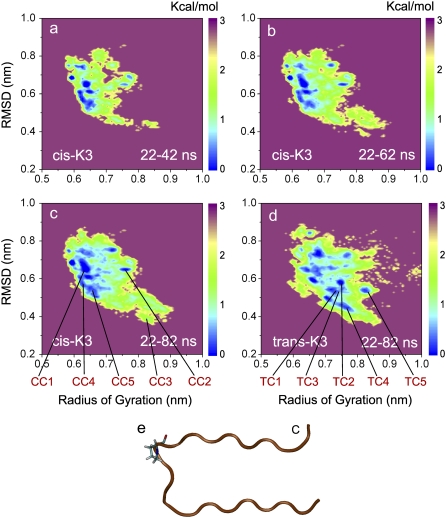
We can also see that the simulations have converged by comparing the free energy landscape of the cis-K3 peptide for different durations of the simulation: 22–42 ns (Fig. 3 a), 22–62 ns (Fig. 3 b), and 22–82 ns (Fig. 3 c). Here, the free energy landscape is projected on the Cα radius of gyration and the Cα RMSD with respect to the conformation of residues Phe22-Leu39 in the fibril state (Fig. 3 e, corresponding to the last frame and Chain C in the PDB code: 2E8D). As seen in Fig. 3, there is little change in the location of the minima and the size of the basins after 62 ns, demonstrating again reasonable convergence of the simulation. Such a long convergence time for the free energy landscape is not surprising, since metastable states can exist that slow down the sampling, as was recently discussed for the monomer of Aβ(1–42) (30).
FIGURE 3.
Free energy surfaces (in kcal/mol) of the trans- and cis-K3 peptides. Evolution of the cis-K3 free energy surface using the 22–42 (a), 22–62 (b), and 22–82 (c) ns intervals; free energy surface of the trans-K3 peptide using the 22–82 ns interval (d). The two reaction coordinates used are the Cα radius of gyration and the Cα-RMSD with respect to the strand-loop-strand structure in K3 fibril shown in panel e. Pro32 is shown in all-atom representation.
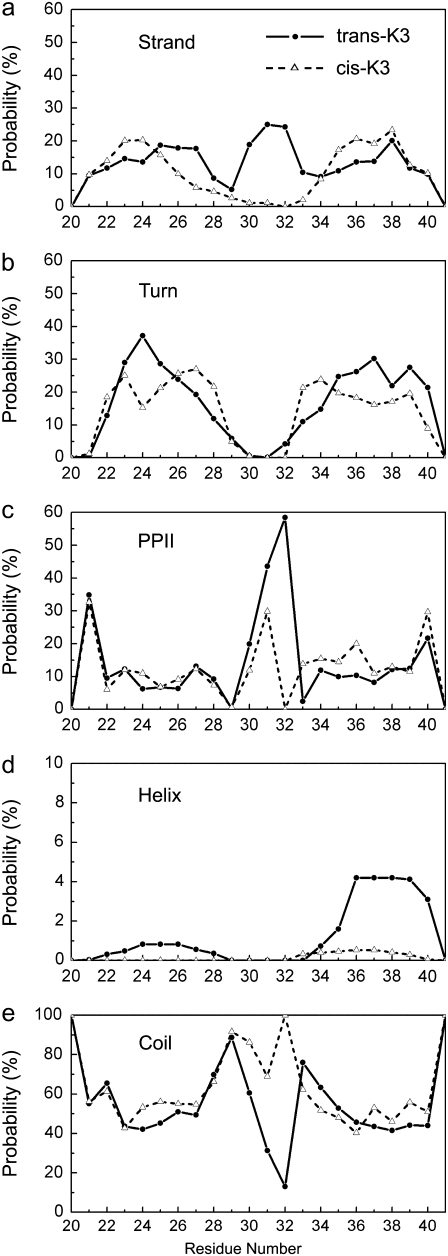
The secondary structure probability of each residue at 298 K, as assigned by the PROSS program, is shown in Fig. 4. PROSS uses a five-letter code based solely on backbone dihedral angles: β-strand, β-turn, PPII, helix, and coil (29). We recall that the coil character of an amino acid and the random coil character of an equilibrium structure have different meanings, and a protein structure with random coil character contains a low percentage of α- and β-secondary structures.
FIGURE 4.
Secondary structure probabilities of each residue in the trans- and cis-K3 peptides: (a) β-strand, (b) β-turn, (c) PPII, (d) helix, and (e) coil using the PROSS program.
We see that the trans-K3 and cis-K3 peptides share very similar secondary structure probabilities or profiles in the regions A (Asn21-Val27) and B (Asp34-Leu39). The β-strand content is 10–20% (Fig. 4 a) and the helix signal is very small, <5% (Fig. 4 d). The β-turn probability of residues 24–26 is slightly higher in trans (Fig. 4 b) than in cis. Overall, the regions A and B are, however, mostly assigned as coil (Fig. 4 e).
Not surprisingly, the greatest impact of trans/cis-Pro32 is observed in the region Ser28-Ser33, where the difference between the two peptides is striking. Cis-K3 displays a negligible β-strand signal and a small PPII content, but a coil content between 70 and 100%. By contrast, trans-K3 displays β-strand, PPII, and coil signals. Note that the PPII probability of residue Pro32 is zero in cis-K3 by definition in PROSS. The secondary structure probabilities at 298 K of the amino acids defined at least twice in the sequence are detailed in Data S1, Table S1. While we see small fluctuations between the time probabilities of many amino acids for a secondary structure in trans- and cis-peptides—5% for β-strand and PPII, and 9% for β-turn averaged over 13 amino acids—amino acids of the same type such Asn21 and Asn24 or Leu39 and Leu40 behave differently, indicating that the REMD-derived conformational preferences are not systematically individual propensities and thus are context-dependent. For instance, Asn21 is 1% β-turn, but Asn24 is 37% β-turn in trans.
Overall, both peptides are essentially random in solution, but still display a significant tendency to visit conformations compatible with the native β2m(1–99) protein and K3 fibril structures. In particular, the β-strand observed with a high probability in regions A and B match exactly the location of the β-strands found in both the native and fibril states.
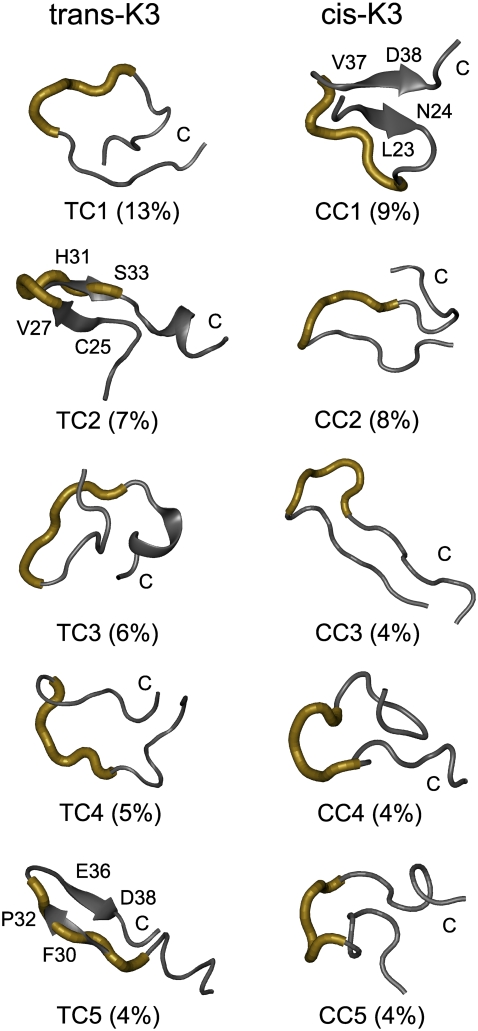
The free energy landscapes of the trans-K3 and cis-K3 peptides at 298 K are shown in Fig. 3, c and d, projected on the two reaction coordinates described above. The locations of the low free energy basins corresponding to the first five most populated clusters are shown, where TCi and CCi (i = 1…5) represent the central structures of the ith cluster for the trans-K3 and cis-K3 peptides, respectively.
Comparing the free energy surfaces of cis-K3 (Fig. 3 c) and trans-K3 (Fig. 3 d), we observe that the area spanned by the trans-K3 peptide is slightly larger than that visited by the cis-K3 peptide. The trans-K3 appears therefore more flexible and able to visit a wider range of conformations than the cis-K3 peptide. This observation is reflected by the result of cluster analysis: the total number of clusters for the trans-K3 and cis-K3 peptides is 140 and 110 using a Cα RMSD cutoff of 0.3 nm for residues 22–39, respectively.
The TCi and CCi structures along with their populations are shown in Fig. 5. These five TCi and CCi states represent 35% and 29% of the total conformations available to the trans- and cis-K3 peptides, respectively. Residues Ser28-Ser33 are shown by tube representation. Comparing the two sets of centers, we see that 11% and 9% of conformations display β-sheet structure for the trans-K3 (TC2 and TC5) and cis-K3 (CC1) peptides, respectively. The β-sheet involves Cys25-Tyr26-Val27 and His31-Pro32-Ser33 in TC2, and Phe30-His31-Pro32 and Glu36-Val37-Asp38 in TC5. In contrast, the β-sheet spans Leu23-Asn24 and Val37-Asp38 in CC1. Overall, the impact of Pro32 isomerization on the visited conformations is significant and the trans-K3 and cis-K3 peptides only share two common clusters, representing a population of 1%, among all those identified in these simulations. Thus, the equilibrated trans-K3 and cis-K3 peptides visit a very different set of structures at 298 K.
FIGURE 5.
Centers of the first five most-populated structures of the trans- and cis-K3 peptides. Boltzmann population at 298 K is given in parentheses.
Probability analysis on the structures with β-strand-loop-β-strand character shows that the cis-K3 peptide populates only 0.3% of the conformations with a Cα22-Cα39 RMSD <0.4 nm from the solid-state NMR-derived structure, and the trans-K3 peptide populates 0.5% of the conformations. In this analysis, a strand is considered formed if at least four consecutive residues are assigned in β-strand conformation by PROSS. All these structures display a variety of β-hairpins with various loop lengths and registers of H-bonds. Because the strand-loop-strand topology matters more than the exact conformation (17), we also calculate the populations of β-strand-loop-β-strand conformations with high RMSD deviations. We find that a population shift to 0% using a Cα22-Cα39 RMSD >0.4 nm, with respect to the solid-state NMR structure.
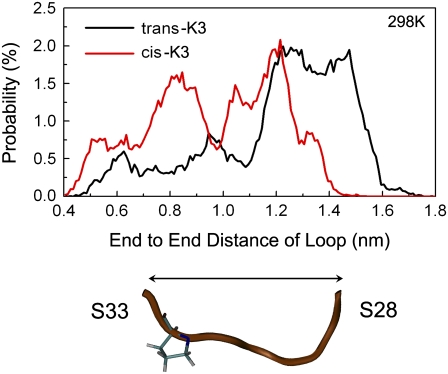
To probe the effect of the cis-trans isomerization on the loop region, the distance distribution between the Cα atom of Ser28 and that of Ser33 is calculated and shown in Fig. 6. In line with the structure analysis, the trans- and cis-K3 peptides exhibit a distinct average Cα-Cα distance between these two residues. The Cα28-Cα33 distance is ∼1 nm in both the native β2m(1–99) and K3 fibrils. Our calculation shows that 27% of trans-K3 conformations and 52% of cis-K3 conformations display a Cα28-Cα33 distance between 0.8 and 1.2 nm, and the percentage shifts to 12% for trans-K3 and 22% for cis-K3 conformations within 0.9 ∼ 1.1 nm. Most of these conformations deviate within 0.1 ∼ 0.2 nm RMSD from the loop conformation in the fibril state.
FIGURE 6.
Probability distribution of the Cα28-Cα33 distance for the trans- and cis-K3 peptides.
The impact of proline isomerization on the K3 peptide structures can also be estimated by computing the formation time probabilities of the side-chain-side-chain contacts present in the fibril and native structures. Table 1 gives the list of interactions with native character (Leu23-Leu39, Cys25-Val37, Val27-Ile35, Cys25-Leu39, and Val27-Val37) and fibril character (Phe22-Leu39, Asn24-Val37, Tyr26-Ile35, Phe22-Val37, Asn24-Ile35, and Tyr26-Phe30). A contact is considered formed when aliphatic carbon atoms of two side chains come to within 0.54 nm of each other. It can be seen from Table 1 that all native and fibril contacts are populated in both the trans and cis predicted equilibrium structures, with formation time probabilities varying between 4% and 21%. This result, along with previous analysis, indicates that neither peptide displays a strong preference for the fibril or the native state. Interestingly, there is very little variation in the surface-accessible area of each amino acid in both cis and trans equilibrium structures (see Data S1, Fig. S2), indicating that neither species is more aggregation-prone than the other by exposing more hydrophobic groups and main-chain amide and carbonyl groups. We cannot therefore explain the preference for the trans conformation in the K3 fibril at the monomeric level, and simulations of higher order species such as dimers and trimers are required.
TABLE 1.
The formation time probabilities of the side-chain-side-chain contacts present in the trans- and cis-K3 peptides with respect to those in the native structure of the full-length β2m protein and the K3 fibril
| trans-K3 | cis-K3 | ||
|---|---|---|---|
| Native contact | |||
| 1 | L23-L39 | 21% | 17% |
| 2 | C25-V37 | 5% | 10% |
| 3 | V27-I35 | 4% | 7% |
| 4 | C25-L39 | 12% | 12% |
| 5 | V27-V37 | 4% | 5% |
| Fibril contact | |||
| 1 | F22-L39 | 18% | 12% |
| 2 | N24-V37 | 6% | 9% |
| 3 | Y26-I35 | 8% | 10% |
| 4 | F22-V37 | 19% | 17% |
| 5 | N24-I35 | 7% | 6% |
| 6 | Y26-F30 | 21% | 24% |
DISCUSSION AND CONCLUSIONS
Replica exchange molecular dynamics simulations based on 64 replicas, each of 82 ns, reveal that the K3 peptide exists as an ensemble of heterogeneous conformations at low pH, independently of the cis/trans character of the His31-Pro32 peptide bond. This conformational variability is in qualitative agreement with our current structural knowledge of most amyloid-forming peptides in solution, such as Aβ(10–35) and Aβ(1–40). Our simulations clearly indicate that the isomerization state of the His31-Pro32 peptide bond leads to two distinct ensemble averages, with only 1% of the conformations being in common. Both peptides display a 10–20% β-strand content at positions Asn21-Val27 and Asp34-Leu40, where the β-strands forms in both the K3 fibril and the native full-length β2m protein, but overall both peptides are essentially random coil in character.
The trans-K3 peptide is found to be much more flexible than its cis-counterpart, which is reflected by a larger number of clusters and a larger free energy landscape. The landscapes differ because the cis His31-Pro32 peptide bond affects the dimensions of the chain locally, leading to a decreased average distance between Ser28 and Ser33. This effect of cis isomerization on the probability distribution of the Cα28-Cα33 distance is fully consistent with recent all-atom simulations of the proline-containing (Ser)3-Pro-(Ser)3 peptide, which show, using a hard-sphere model, that cis prolyl isomers has a largely restricted conformational space and shorter end-to-end distances compared to trans isomers (31). The probability distribution of the number of side-chain-side-chain atomic contacts within the Ser28-Ser33 region (Data S1, Fig. S3) indicates stronger steric effects in the loop region of the cis isomer than that of the trans isomer, reducing the conformational space of the cis-K3 peptide.
Although the trans- and cis-K3 isomers adopt distinct conformations in solution, neither isomer is found to be more aggregation-prone than the other, indicating that the preference for trans conformation in the K3 fibril results from intermolecular interactions. While our simulations on the fibrils are rather short, they clearly show a better packing in the trans model than in the cis model. Note that the proline's cis conformation in the native state of full-length β2m is explained by the NMR solution study of the mutant P32G β2m, which shows that trans Pro32 impacts the native edge β-strands A and D, favoring aggregation (7).
We also find that although the trans and cis isomers display rather different energy landscapes, they both visit a minority of structures with a β-strand-loop-β-strand topology as observed in the native full-length β2m protein and in the K3 fibril, albeit with a very small probability, only 0.3 ∼ 0.5% at 298 K. While small, this value cannot be considered as totally negligible. For example, Radford et al. (7) reported that the population of the amyloidogenic full-length β2m precursor is ∼3%. Similar studies on the prion protein report an intermediate monomeric species with a population of 1% (32). We emphasize that the β-strand-loop-β-strand conformation does not need to be strictly identical to that observed in the fibril structure to be a related precursor. In addition, the strand-loop-strand topology may not be required to accelerate fibrillization. It was shown experimentally by Meredith et al. (33) that enforcing loop formation by a lactam bridge suffices to increase Aβ40 aggregation rate by three orders of magnitude.
MD simulations of the Aβ(10–35) peptide in explicit solvent under acid pH condition show that the probability of a β-strand-loop-β-strand structure is 21% at 300 K (17). This runs in contrast with the present results of the K3 peptide at low pH, which indicates that the β-strand-loop-β-strand conformation is not encoded at the monomeric level (population of 0.5%). The high probability of Aβ(10–35) for β-strand-loop-β-strand might be due to an intrinsic propensity of the region Asp22-Lys28 to form a loop (34). The different probabilities for this conformation in the monomers of Aβ(10–35) and K3 peptides might therefore result from the existence/absence of favorable electrostatic interactions to stabilize the turn. In Aβ(10–35), there are three charged residues within the loop: Asp22−, Glu23−, and Lys28+. In the K3 peptide, however, there is only one charged residue, His31+, within the loop. This suggests that the biases toward aggregation leading to the β-strand-loop-β-strand conformation in fibrils are sequence-dependent.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We are grateful to the Réseau Québécois de Calcul de Haute Performance, the Shanghai Supercomputing Center, and the National High Performance Computing Center of Fudan University for their generous allocation of computational resources.
P.D. and N.M. thank Fudan University for its hospitality and financial support through a Senior Visiting Scholar Grant. P.D. thanks Centre National de la Recherche Scientifique and University of Paris 7 Denis Diderot. N.M. acknowledges partial support from the Natural Sciences and Engineering Research Council (Canada), Fonds de Recherche sur la Nature et les Technologies Québec (Québec), and the Canada Research Chair program. G.W. thanks the National Natural Science Foundation of China (grant No. 10674029), Program for Changjiang Scholars and Innovative Research Team in University (i.e., PCSIRT), and the Young Foundation of Fudan University.
Editor: Kathleen B. Hall.
References
- 1.Lashuel, H. A., and P. T. J. Lansbury. 2006. Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q. Rev. Biophys. 36:167–201. [DOI] [PubMed] [Google Scholar]
- 2.Kayed, R., E. Head, J. L. Thompson, T. M. McIntire, S. C. Milton, C. W. Cotman, and C. G. Glabe. 2003. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 300:486–489. [DOI] [PubMed] [Google Scholar]
- 3.Kayed, R., and C. G. Glabe. 2006. Conformation-dependent anti-amyloid oligomer antibodies. Methods Enzymol. 413:326–344. [DOI] [PubMed] [Google Scholar]
- 4.Verdone, G., A. Corazza, P. Viglino, F. Pettirossi, S. Giorgetti, P. Mangione, A. Andreola, M. Stoppini, V. Bellotti, and G. Esposito. 2002. The solution structure of human β2-microglobulin reveals the prodromes of its amyloid transition. Protein Sci. 11:487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiba, T., T. Hagihara, Y. Higurashi, K. Hasegawa, H. Naiki, and Y. Goto. 2003. Amyloid fibril formation in the context of full-length protein: effects of proline mutations on the amyloid fibril formation of β2-microglobulin. J. Biol. Chem. 278:47016–47024. [DOI] [PubMed] [Google Scholar]
- 6.Corazza, A., F. Pettirossi, P. Viglino, G. Verdone, J. Garcia, P. Dumy, S. Giorgetti, P. Mangione, S. Raimondi, M. Stoppini, V. Bellotti, and G. Esposito. 2004. Properties of some variants of human β2-microglobulin and amyloidogenesis. J. Biol. Chem. 279:9176–9189. [DOI] [PubMed] [Google Scholar]
- 7.Jahn, T. R., M. J. Parker, S. W. Homans, and S. E. Radford. 2006. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat. Struct. Mol. Biol. 13:195–201. [DOI] [PubMed] [Google Scholar]
- 8.Kozhukh, G. V., Y. Hagihara, T. Kawakami, H. Hasegawa, K. Naiki, and Y. Goto. 2002. Investigation of a peptide responsible for amyloid fibril formation of β2-microglobulin by Achromobacter protease I. J. Biol. Chem. 277:1310–1315. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi, K., S. Takahashi, T. Kawai, H. Naiki, and Y. Goto. 2005. Seeding-dependent propagation and maturation of amyloid fibril conformation. J. Mol. Biol. 352:952–960. [DOI] [PubMed] [Google Scholar]
- 10.Iwata, K., T. Fujiwara, Y. Matsuki, H. Akutsu, S. Takahashi, H. Naiki, and Y. Goto. 2006. 3D structure of amyloid protofilaments of β2-microglobulin fragment probed by solid-state NMR. Proc. Natl. Acad. Sci. USA. 103:18119–18124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petkova, A. T., Y. Ishii, J. J. Balbach, O. N. Antzutkin, R. D. Leapman, F. Delaglio, and R. Tycko. 2002. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA. 99:16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luhrs, T., C. Ritter, M. Adrian, D. Riek-Loher, B. Bohrmann, H. Dobeli, D. Schubert, and R. Riek. 2005. 3D structure of Alzheimer's amyloid-β(1–42) fibrils. Proc. Natl. Acad. Sci. USA. 102:17342–17347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritter, C., M. L. Maddelein, A. B. Siemer, T. Luhrs, M. Ernst, B. H. Meier, S. J. Saupe, and R. Riek. 2005. Correlation of structural elements and infectivity of the HET-s prion. Nature. 435:844–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson, N., J. Becker, H. Tidow, S. Tremmel, T. D. Sharpe, G. Krause, J. Flinders, M. Petrovich, J. Berriman, H. Oschkinat, and A. R. Fersht. 2006. General structural motifs of amyloid protofilaments. Proc. Natl. Acad. Sci. USA. 103:16248–16253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croixmarie, V., F. Briki, G. David, Y. M. Coic, L. Ovtracht, J. Doucet, N. Jamin, and A. Sanson. 2005. A cylinder-shaped double ribbon structure formed by an amyloid hairpin peptide derived from the β-sheet of murine PrP: an x-ray and molecular dynamics simulation study. J. Struct. Biol. 150:284–299. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, S., K. Iwata, M. J. Lachenmann, J. W. Peng, S. Li, E. R. Stimson, Y. Lu, A. M. Felix, J. E. Maggio, and J. P. Lee. 2000. The Alzheimer's peptide aβ adopts a collapsed coil structure in water. J. Struct. Biol. 130:130–141. [DOI] [PubMed] [Google Scholar]
- 17.Han, W., and Y. D. Wu. 2005. A strand-loop-strand structure is a possible intermediate in fibril elongation: long time simulations of amyloid-β peptide (10–35). J. Am. Chem. Soc. 127:15408–15416. [DOI] [PubMed] [Google Scholar]
- 18.Galani, D., A. R. Fersht, and S. Perrett. 2002. Folding of the yeast prion protein Ure2: kinetic evidence for folding and unfolding intermediates. J. Mol. Biol. 315:213–227. [DOI] [PubMed] [Google Scholar]
- 19.Jenko Kokalj, S., G. Guncar, I. Stern, G. Morgan, S. Rabzelj, R. A. Kenig, M. F. Staniforth, J. P. Waltho, E. Zerovnik, and D. Turk. 2007. Essential role of proline isomerization in stefin B tetramer formation. J. Mol. Biol. 366:1569–1579. [DOI] [PubMed] [Google Scholar]
- 20.Pastorino, L., A. Sun, P. J. Lu, X. Z. Zhou, M. Balastik, G. Finn, G. Wulf, J. Lim, S. H. Li, X. Li, W. Xia, L. K. Nicholson, and K. P. Lu. 2006. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-β production. Nature. 440:528–534. [DOI] [PubMed] [Google Scholar]
- 21.Sugita, Y., and Y. Okamoto. 1999. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 314:141–151. [Google Scholar]
- 22.Okazaki, K., N. Koga, S. Takada, J. N. Onuchic, and P. G. Wolynes. 2006. Multiple-basin energy landscapes for large-amplitude conformational motions of proteins: structure-based molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 103:11844–11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Simone, A., A. Zagari, and P. Derreumaux. 2007. Structural and hydration properties of the partially unfolded states of the prion protein. Biophys. J. 93:1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang, C., P. Derreumaux, and G. Wei. 2007. Structure and aggregation mechanism of β2-microglobulin(83–99) peptides studied by molecular dynamics simulations. Biophys. J. 93:3353–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berendsen, H. J. C. 1981. Interaction Models for Water in Relation to Protein Hydration, Intermolecular Forces. Reidel, Dordrecht, The Netherlands.
- 26.Lindahl, E., B. Hess, and D. Van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306–317. [Google Scholar]
- 27.Jorgensen, W. L., and J. Tirado-Rives. 1988. The OPLS potential functions for proteins: energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 110:1657–1666. [DOI] [PubMed] [Google Scholar]
- 28.Berendsen, H. J. C., J. P. M. Postma, A. Di Nola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 29.Fleming, P. J., H. Gong, and G. D. Rose. 2006. Secondary structure determines protein topology. Protein Sci. 15:1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sgourakis, N. G., Y. Yan, S. A. McCallum, C. Wang, and A. E. Garcia. 2007. The Alzheimer's peptides aβ40 and 42 adopt distinct conformations in water: a combined MD/NMR study. J. Mol. Biol. 368:1448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krieger, F., A. Moglich, and T. Kiefhaber. 2005. Effect of proline and glycine residues on dynamics and barriers of loop formation in polypeptide chains. J. Am. Chem. Soc. 127:3346–3352. [DOI] [PubMed] [Google Scholar]
- 32.Kuwata, K., H. Li, H. Yamada, G. Legname, S. B. Prusiner, K. Akasaka, and T. L. James. 2002. Locally disordered conformer of the hamster prion protein: a crucial intermediate to PrPSc? Biochemistry. 41:12277–12283. [DOI] [PubMed] [Google Scholar]
- 33.Sciarretta, K. L., D. J. Gordon, A. T. Petkova, R. Tycko, and S. C. Meredith. 2005. Aβ40-Lactam(D23/K28) models a conformation highly favorable for nucleation of amyloid. Biochemistry. 44:6003–6014. [DOI] [PubMed] [Google Scholar]
- 34.Lazo, N., M. Grant, M. Condron, A. Rigby, and D. B. Teplow. 2005. On the nucleation of amyloid β-protein monomer folding. Protein Sci. 14:1581–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.