Abstract
Presbycusis – age-related hearing loss – is the number one communicative disorder and one of the top three chronic medical condition of our aged population. High-throughput technologies potentially can be used to identify differentially expressed genes that may be better diagnostic and therapeutic targets for sensory and neural disorders. Here we analyzed gene expression for a set of GABA receptors in the cochlea of aging CBA mice using the Affymetrix GeneChip MOE430A. Functional phenotypic hearing measures were made, including auditory brainstem response (ABR) thresholds and distortion-product otoacoustic emission (DPOAE) amplitudes (four age groups). Four specific criteria were used to assess gene expression changes from RMA normalized microarray data (40 replicates). Linear regression models were used to fit the neurophysiological hearing measurements to probe-set expression profiles. These data were first subjected to one-way ANOVA, and then linear regression was performed. In addition, the log signal ratio was converted to fold change, and selected gene expression changes were confirmed by relative real-time PCR. Major findings: expression of GABA-A receptor subunit α6 was upregulated with age and hearing loss, whereas subunit α1 was repressed. In addition, GABA-A receptor associated protein like-1 and GABA-A receptor associated protein like-2 were strongly downregulated with age and hearing impairment. Lastly, gene expression measures were correlated with pathway/network relationships relevant to the inner ear using Pathway Architect, to identify key pathways consistent with the gene expression changes observed.
Keywords: Presbycusis, Aging, GABA receptors, Microarray, Relative real-time PCR, Auditory brainstem response, Distortion-product otoacoustic emission
1. Introduction
A landmark epidemiological study by Gates et al. (1999) revealed that age-related hearing loss – presbycusis – has a genetic component. But in contrast to congenital deafness, where over 100 genes have been discovered to be responsible in humans, no genes have yet been identified to be causative for presbycusis. One approach to uncovering genes involved in presbycusis is to perform high-throughput experiments in mammals of different ages to identify genes whose expression changes with age and hearing loss. Then gene and protein expression studies can be done in humans, using the animal studies as specific guides (e.g., Xiao et al., 2004).
The present study uses a unique approach for selecting biologically relevant genes from large set of RMA normalized microarray data using statistical tools in stages, such as one-way ANOVA and linear regression of auditory physiological measures and gene expression fold change values. The probe-sets selected with these criteria for further study were then confirmed quantitatively with relative real-time PCR.
Gamma aminobutyric acid (GABA) is a primary inhibitory neurotransmitter of the mammalian brain, and the key inhibitory neurotransmitter of the auditory system. Previous immunocytochemical and biochemical investigations of the aging auditory system have revealed a selective decline in the GABA inhibitory neurotransmitter systems in the auditory brainstem, particularly at the level of the auditory midbrain (inferior colliculus; Caspary et al., 1999; Frisina and Rajan, 2005). In contrast to this convincing demonstration of an age-linked decline in GABA in the central auditory system, there have been no previous reports on age changes in the cochlear GABA systems, despite the prominent role that this inhibitory neurotransmitter plays in the auditory efferent feedback system from the brainstem back to the inner ear to modulate sound processing on an ongoing basis.
The goal of the present study was to investigate GABA receptor gene expression trends using 31 probe-sets of RMA normalized microarray data for aging mice, and to utilize physiological hearing measures from each mouse, employing a novel statistical approach. The data obtained from relative real-time PCR of a set of GABA receptor probe-sets confirmed key expression trends from the microarray data. The outcome resulted in one probe-set, the GABA-A receptor subunit α6, displaying upregulation in aging mouse cochlea and GABA-A receptor subunit α1 which was repressed with age and hearing loss. In addition, GABA-A receptor associated protein like-1 and GABA-A receptor associated protein like-2 were strongly downregulated with age and hearing loss. To critically evaluate our statistical approach, we have also included random selection of GABA receptor probe-sets. In the discussion we make specific comparisons of the microarray data with relative real-time PCR and the biological interaction pathways of GABA receptors derived from Pathway Architect.
2. Results
A total of 40 microarrays, one for each pair of cochleae of each mouse, were analyzed with various statistical approaches to maximize the statistical power of the microarray-derived results, including functional correlations employing individual mouse hearing test results (no grouping of cochlear tissue or hearing measures from different mice prior to array processing). As reported earlier (Tadros et al., 2007a,b) the subject group size was large, ranging from 6 to 17 mice in each age group. Amplified cochlear RNA was utilized in the microarrays and non-amplified RNA with amplified cDNA for the relative real-time PCR runs. As reported in our previous studies (Tadros et al., 2007a,b) four subject groups were classified on the basis of their functional hearing abilities: auditory brainstems response (ABR) thresholds and distortion-product otoacoustic emission (DPOAE) amplitudes. The four subject groups included: young adults with good hearing; middle aged with good hearing; old mice with mild hearing loss, and old mice with severe presbycusis. A striking elevation in ABR thresholds was seen comparing the middle aged to the old age groups. The old mice were divided into mild presbycusic, who still displayed significant DPOAE responses; and severe presbycusic, who had poor ABR thresholds, and virtually no DPOAE responses (Table 1). There were numerous genes that showed apparently continuous changes in expression associated with age and hearing loss, as now explained in detail.
Table 1.
Four age groups derived from functional hearing abilities based on DPOAE amplitudes and ABR thresholds in CBA mice
| Group | No. of mice | No. of chips (1 chip/mouse) | Age-months ± range | Gender |
|---|---|---|---|---|
| Young, good hearing | 8 | 8 | 3.5 ± 0.4 | Male = 5, female = 4 |
| Middle aged, good hearing | 17 | 17 | 12.3 ± 1.5 | Male = 8, female = 9 |
| Mild presbycusis | 9 | 9 | 27.7 ± 3.4 | Male = 4, female = 5 |
| Severe presbycusis | 6 | 6 | 30.6 ± 1.9 | Male = 2, female = 4 |
cDNA was made from amplified cochlear RNA from each mouse, fluorescent labeled cDNA was hybridized to an Affymetrix MOE430A GeneChip. The intensity of the gene expression measurements of the 40 GeneChips (in four age-groups) were normalized by RMA (Irizarry et al., 2003). MOE430A GeneChip (RMA normalized) consisted of 22,600 probe-sets; there were 31 GABA receptor probe-sets in the 22,600 probe on each GeneChip. Data for this group of probe-sets were imported into an Excel database used for our present study. This group of 31 GABA receptor probe-sets, whose gene expression changed significantly with age or hearing loss: 1 probe-set (Gabra6, 1451706_at) was upregulated in all age-groups and 3 probe-sets (Gabarapl1, Gabarapl2 transporter Slc6a13) were downregulated consistently in all age-groups, 5 probe-sets (Gabrq, Gabrr2, Gabra3, Gabrd and Gabrb2) showed no change in gene expression, while all others (mixed) were either up and down in different age-groups. For example, Gabra6 (1451706_at) was included in the upregulation category, where it was upregulated in all age groups: middle age, old mild and severe presbycusis. Because we were interested in the overall influence that GABA receptors might play in the cochleae of aging mice, we focused our attention on other GABA receptor-related genes as well, with more subtle fold changes with age, relative to the expression of the young adult controls with good hearing.
Four criteria were utilized to select genes for further study from the GABA receptor-related genes on the microarray: (1) Microarray RMA normalized log signal ratio (LSR) data were subjected to one-way ANOVA (compares group means) with post-hoc Bonferroni tests (pairwise tests corrected for multiple comparisons), and Kruskal-Wallis tests (compares the medians of the groups); (2) Genes with a significant p-value from regression analysis for microarray LSR data and the physiological hearing data (ABR and DPOAE); (3) High fold changes for the RMA normalized gene expression data (Fold change cut-off above 2 or below −2) when compared to young adult, good hearing control mice; (4) A final selection of GABA receptor probe-sets with low fold changes (1.0–1.4) was done randomly from the microarray gene expression data of 31 GABA receptor probe-sets. In all cases, selected probe-sets were furthermore validated by relative real-time PCR studies.
2.1. Criterion 1
31 GABA receptor probe-sets were subjected to one-way ANOVA with the post-hoc tests (Bonferroni analysis: pair-wise test of means corrected for multiple comparisons), which yielded 1 probe-set Gabrg3 (p < 0.04, F = 3.4, d.f. = 3, 32; Fig. 1A). Second, Kruskal–Wallis post hoc tests (compares the medians of the groups) yielded five probe-sets showing statistically significant changes with age and hearing loss: Gabra1 (1421280_at), Gabbr1 (1422051_at), Gabrg3 (1422187_at), Gabarapl2 (1423187_at) and Gabrg1 (1427227_at). Gene expression changes of two of these probe-sets: GABA-A receptor subunit γ1 (Gabrg1, Kruskal–Wallis statistic = 6.27, p < 0.04, d.f. = 3, 32), MP vs. SP: p = 0.05, and GABA-B receptor 1 (Gabbr1, Kruskal–Wallis statistic = 6.19, p < 0.04, d.f. = 3, 32), MP vs. SP = 0.05 were confirmed by relative real-time PCR (Fig. 1B). Utilizing this first criterion, the three probe-sets Gabrg3, Gabrg1 and Gabbr1 were confirmed by relative real-time PCR. Gabrg3 (Bonferroni post hoc test), indicates, only the middle aged group fold change was in agreement between the GeneChip and the relative real-time PCR data, while the old mild and severe presbycusis groups did not show agreement (Fig. 1A). The probe-set Gabrg1 and Gabbr1 (Criterion 1, Kruskal–Wallis post hoc test) confirmed by relative real-time PCR, validated the GeneChip findings for all three age and hearing loss subject groups (Fig. 1B).
Fig. 1.
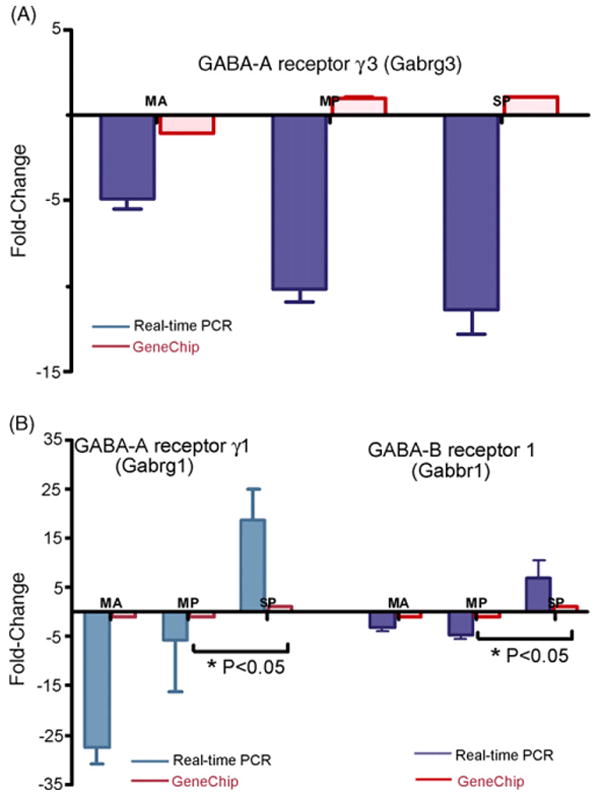
Validation of microarray gene expression patterns by relative real-time PCR according to the first selection criterion. One gene was found to be statistically significant for the group main effect by one-way ANOVA (Bonferroni multiple comparison). (A) GABA-A receptor subunit γ3 (p < 0.04, F = 3.4, d.f. = 3, 32). Five genes were found to be statistically significant for the ANOVA (Kruskal–Wallis, compares the medians of the groups), two probe-sets confirmed are: (B) GABA-B receptor γ1, Gabrg1, (Kruskal–Wallis statistic = 6.27, p < 0.04, d.f. = 3, 32), MP vs. SP: p < 0.05; and GABA-B receptor 1 (Gabbr1, Kruskal–Wallis statistic = 6.19, p < 0.04, d.f. = 3, 32), MP vs. SP: p < 0.05. The results are expressed as fold change in middle aged, old mild presbycusis and old severe presbycusis subject groups relative to the mean gene expression levels of the young adult group with good hearing. GAPDH was used as an endogenous control to derive the ΔCt values, while ΔΔCt was derived from the mean of the young adults with good hearing ΔCt of all the replicates for relative real-time PCR. SEM is indicated, MA = middle age, MP = old mild presbycusis, SP = old severe presbycusis.
A closer examination of the gene annotations and functions of the 31 GABA receptor genes revealed functional relationships among most of the probe-sets. For example, GABA-B receptor 1, which is a G-coupled protein receptor, is associated with metabotropic glutamate GABA-B-like receptor activity. Four probe-sets from different regions of the GABA-B receptor 1 gene (1422051_a_at, 1455021_at, 1425595_at and 1450300_at) were represented on the Affymetrix GeneChip MOE430A. This has particular functional significance for hearing because GABA-B receptors have been reported previously to be prominently expressed in cochlear spiral ganglion neurons (Lin et al., 2000), most likely as receptors for GABA released by the auditory efferent feedback system from the brainstem to the cochlea. This system is important for regulating sound transmission through the cochlea to the central auditory system. GABA-B receptors are also located in pre- and post-synaptic membranes of vertebrate neurons to modulate neurotransmission through G-protein coupled pathways as shown by Kaupmann et al. (1997). Two of the three GABA-B receptor1 probe-set data in microarray and relative real-time PCR of the present study were upregulated in old severe presbycusis when compared to young controls, which may be in response to age-related excessive glutamatergic neurotransmission or excitotoxicity in the aging mouse cochlea.
2.2. Criterion 2
GABA-A receptor associated protein like-1 (Gabarapl1), GABA-A receptor subunit π, α3, γ2 and GABA-B receptor1 (Gabbr1, 1425595_at), were the genes identified according to the second criterion. Linear regression analysis of Gabarapl1 exhibited p < 0.01 and p < 0.04 and r2 = 0.18 and 0.12 (Fig. 2A) respectively, for low- and mid-frequency DPOAE amplitudes in relation to microarray data. While Gabbr1 showed p < 0.01, p < 0.03, p < 0.01, and, r2 = 0.20, 0.14, 0.19 (Fig. 3A) respectively for 12, 24, and 32 kHz ABR thresholds and microarray data (Table 2). Age-related hearing loss can be monitored by elevation in ABR thresholds and declines in DPOAE amplitudes. A marked elevation of ABR thresholds at 24, 32, and 48 kHz (p < 0.0001), and declines in DPOAE amplitudes that occurred in CBA mice by 24 months of age, strongly indicate the presence of presbycusis in the two old age groups. Furthermore, the validation of Gabarapl1 (Fig. 2B) by relative real-time PCR confirmed the microarray gene expression changes, in terms of magnitude and fold change. The relative real-time PCR differed for Gabbr1 (Fig. 3B), in the old mild presbycusic group, Gabbr1 was found in decreased levels in old mild presbycusis, whereas elevated levels were observed in the middle aged and old severe presbycusis mice relative to the young adult, good hearing controls. The PCR gene expression displayed greater fold changes than in the microarray data. Specifically, Gabarapl1 is a cytoskeletal binding protein, which was found in decreased levels in the middle age and old groups.
Fig. 2.
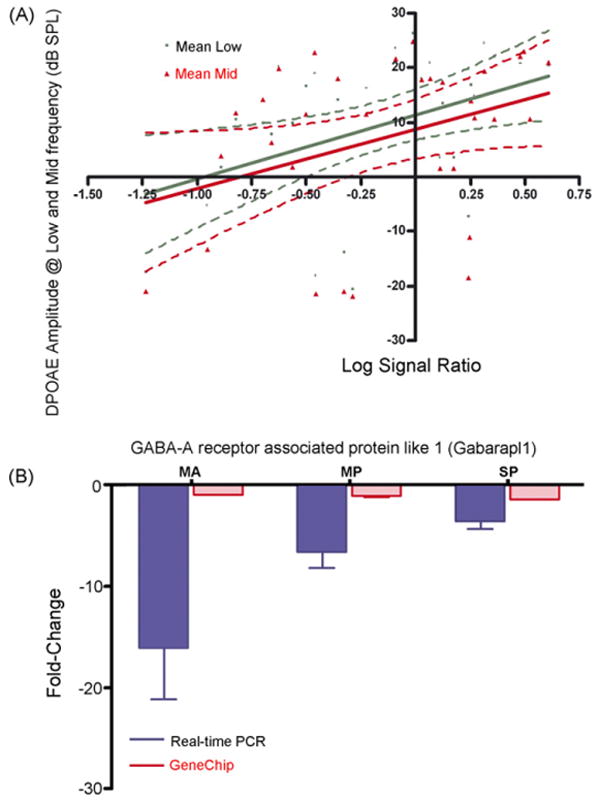
Confirmation of microarray gene expression trends by relative real-time PCR, according to the second criterion, correlation to DPOAE test: (A) correlation of hearing test to the gene expression of GABA-B receptor associated protein like 1 by linear regression. A linear regression between the microarray LSR and DPOAE amplitudes showed that Gabarapl1 p < 0.01 for low frequency DPOAEs, and p < 0.04 for middle frequency DPOAE's. The linear regression between LSR of Gabrapl1 gene expression and DPOAE amplitudes of low and middle frequency range reveal significant correlation with hearing test. (B) GABA receptor associated protein like 1. Significant downregulation in the expression of Gabarapl1 were observed in aging mice in the microarrays, and similar trend was confirmed by relative real-time PCR in all age-groups. S.E.M. is indicated. MA = middle age, MP = old mild presbycusis, SP = old severe presbycusis.
Fig. 3.
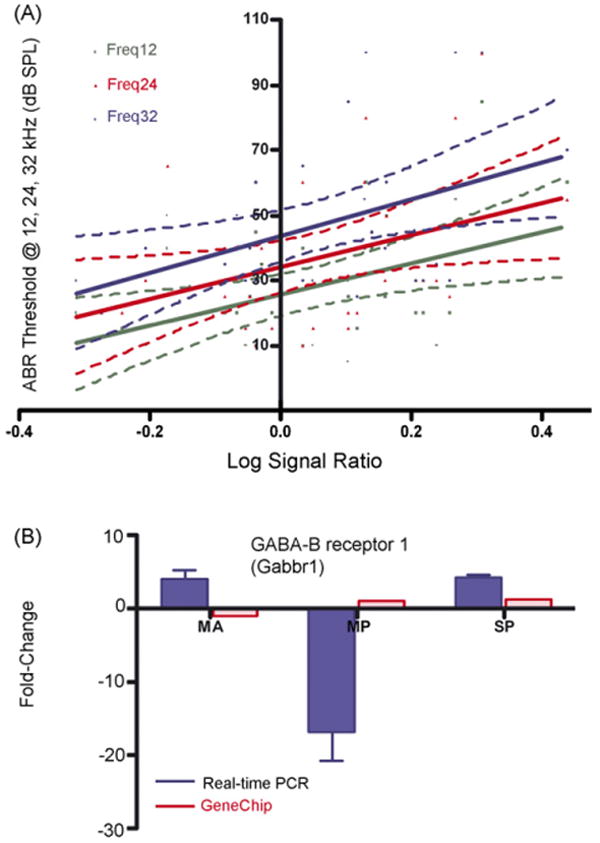
Confirmation of microarray gene expression trends by relative real-time PCR, according to the second criterion, correlation to ABR: (A) linear regression of GABA-B receptor 1 gene expression to the ABR threshold: A linear regression between the microarray LSR and ABR thresholds showed that Gabbr1 p < 0.01, 0.03 and 0.01 for ABR frequency 12, 24 and 32 kHz, respectively. (B) GABA-B receptor 1, the relative real-time PCR differed for Gabbr1, in the old mild presbycusis group Gabbr1 was found at decreased levels, whereas elevated levels were observed in the middle aged and old severe presbycusis mice relative to the young adult, good hearing controls. S.E.M. is indicated. MA = middle age, MP = old mild presbycusis, SP = old severe presbycusis.
Table 2.
Linear regression analyses of microarray RMA normalized LSR (expression profiles) and ABR thresholds or DPOAE amplitudes (functional hearing measures)
| Affymetrix probe-set ID–gene name–gene abbreviation | Data type | Regression values | |
|---|---|---|---|
| p-Value | r2 | ||
| 1416418_at –GABA-A receptor associated protein like-1–Gabarapl1 | GeneChip vs. low frequency DPOAE | 0.01 | 0.18 |
| 1416418_at–GAB A-A receptor associated protein like-1–Gabarapl1 | GeneChip vs. mid-frequency DPOAE | 0.04 | 0.12 |
| 1421263_at–GABA-A receptor α3–Gabra3 | GeneChip vs. 3 kHz data of ABR | 0.01 | 0.18 |
| 1421536_at–GABA-A receptor π–Gabrp | 0.04 | 0.12 | |
| 1418177_at–GABA-A receptor γ2–Gabrg2 | GeneChip vs. 6 kHz data of ABR | 0.04 | 0.12 |
| 1421536_at–GABA-A receptor π–Gabrp | 0.001 | 0.28 | |
| 1418177_at–GABA-A receptor γ2–Gabrg2 | GeneChip vs. 12 kHz data of ABR | 0.02 | 0.14 |
| 1421536_at–GABA-A receptor π–Gabrp | 0.002 | 0.27 | |
| 1425595_at–GABA-B receptor 1–Gabbr1 | 0.01 | 0.20 | |
| 1418177_at–GABA-A receptor γ2–Gabrg2 | GeneChip vs. 24 kHz data of ABR | 0.01 | 0.16 |
| 1425595_at–GABA-B receptor 1–Gabbr1 | 0.03 | 0.14 | |
| 1416418_at–GABA-A receptor associated protein like-1–Gabarapl1 | GeneChip vs. 32 kHz data of ABR | 0.01 | 0.18 |
| 1425595_at–GABA-B receptor 1–Gabbr1 | 0.01 | 0.19 | |
| 1416418_at–GABA-A receptor associated protein like-1–Gabarapl1 | GeneChip vs. 48 kHz data of ABR | 0.03 | 0.13 |
The last two columns show statistically significant values.
2.3. Criterion 3
Genes with high fold changes (greater than 2 or less than −2) for the microarray gene expression data relative to the young adults were selected for further study. GABA-A receptor subunit α6 (Gabra6) was a particularly unique probe-set, displaying high fold changes with age, and displaying statistical significance p < 0.05 value in both the 22,600 probe-set (complete probe-set of GeneChip) and in the 31 GABA receptor probe-set of microarray data. Specifically, Gabra6 revealed a greater than twofold change in expression levels in the old severe presbycusic mice when compared to young adults, as shown in Fig. 4A. Here, the magnitude of the age-dependent GABA receptor upregulation observed in the microarray experiments was in good agreement with the relative real-time PCR analyses in middle aged and old severe presbycusis. PCR results exhibited even greater fold changes. In addition, Gabarapl2 was downregulated in MA, MP and SP when compared to young controls for both the microarrays and for the relative real-time PCR data (Fig. 4B). Both the microarray and relative real-time PCR techniques revealed the same trends for this gene, with fairly strong overall concordance of gene expression changes for all of the subject groups.
Fig. 4.
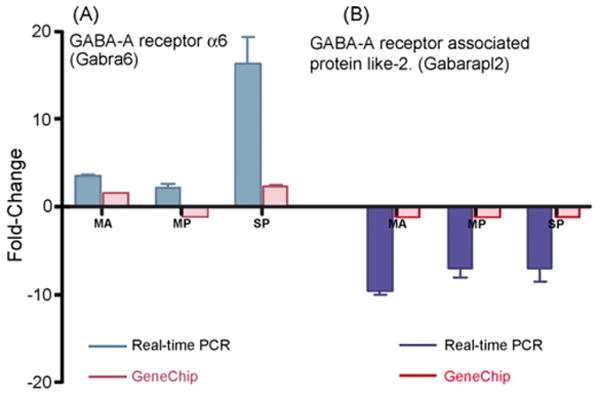
Validation of microarray RMA normalized expression data according to the third criterion. (A) GABA-A receptor subunit α6 expression trends showed greater than twofold upregulation in the microarray data. Cochlea Gabra6 expression was upregulated in all three age-groups in relative real-time PCR, whereas in GeneChip the elevated levels were seen in middle aged and old severe presbycusis and decreased levels in old mild presbycusis mice. (B) GABA associated protein like two was downregulated and the trend was confirmed in both GeneChip and relative real-time PCR in all age-groups. Relative real-time PCR exhibited high levels of fold changes and confirmed the microarray expression trends with age and hearing loss. S.E.M. is indicated. MA = middle age, MP = old mild presbycusis, SP = old severe presbycusis.
2.4. Criterion 4
The final criterion consisted of examining statistically significant probe-sets with low fold change differences (1.0–1.4) between the young adult and older subject groups. Groups of genes in this category were of the GABA-A and B receptor family, namely Gabra1 (1421280_at) and Gabbr1 (1455021_at). They were tested by relative real-time PCR by this array-guided approach for selection of interesting genes which are biologically relevant for auditory processing and neurotransmission. The results of relative real-time PCR data utilizing this selection criterion are shown in Table 3. In addition, 50% of the primer/probes meeting this criterion produced PCR validated results, while the other 50% did not result in confirmation of the magnitude of the microarray findings, as shown in Table 3. The mean fold change of microarray data was typically small: the average difference was less than 1.5. Differences of such small magnitude usually displayed a much higher fold change in the relative real-time PCR runs.
Table 3.
Relative real-time PCR confirmation according to Criterion 4: random selection of two GABA receptor probe-sets, Gabrr1 and Gabra1, from 31 GABA receptor probe-sets: the microarray expression trends were validated by relative real-time PCR using ABI Taqman primer/probes
| Gene name–gene abbreviation | Fold change compared to young controls (good hearing) | |||
|---|---|---|---|---|
| Age group | MA | MP | SP | |
| GABA-B receptor 1–Gabbr1 | GeneChip | 1.09 | 1.08 | 1.05 |
| Real-time PCR | 3.95 | −3.53 | −3.38 | |
| GABA-A receptor αl–Gabra1 | GeneChip | −1.0 | −1.0 | 1.09 |
| Real-time PCR | −1.36 | −10.55 | −8.0 | |
Microarray and relative real-time PCR data represent the mean fold changes in the middle age (MA, 17 replicates), mild presbycusis (MP, 9 replicates), and severe presbycusis (SP, 6 replicates) groups, relative to the young adult controls (8 replicates). The relative real-time PCR and GeneChip gene expression fold changes for the Gabbr1 are consistent for the MA mice but not for the MP and SP subject groups. Gabra1 is consistent for MA and MP groups, but not for SP.
Applying the fourth criterion helped elucidate some of the extensive but subtle differences in inner ear GABA receptor-related gene expression changes between the different age-groups in terms of biological significance, as reflected in inner ear gene expression profiles of individual mice, but not necessarily in terms of strong group tendencies. For example, reduced gene expression levels of Gabra1 were seen in the cochlea of old mild and severe presbycusic mice in GeneChip, whereas the relative real-time PCR gene expression of the same gene was much higher (Table 3). Studies from Gabra1 knock-outs by Milbrandt et al. (1997) reveal an interesting murine phenotype, which approximates certain human clinical deafness conditions. The fourth and final analysis criterion for probe-sets with subtle fold changes also provides evidence for the usefulness of targeting biologically important genes when analyzing microarray data.
As mentioned earlier, the present GeneChip study employed amplified RNA from each cochlea which yields approximately 200–300 ng total RNA for GeneChip and non-amplified RNA, but amplified cDNA was used for relative real-time PCR studies. The results of validation of relative real-time PCR of GeneChip data confirm the usefulness of the amplified RNA in microarray studies and non-amplified RNA in the relative real-time PCR validation experiments. Comparisons of the results of amplified mRNA microarray and non-amplified RNA relative real-time PCR showed strong overall concordance of differential expression trends, regardless of the RNA template. But the dynamic fold change range of differential expression seen with relative real-time PCR analysis was often much greater than that seen with the microarray analysis.
In an attempt to correlate the functional associations of differentially expressed GABA receptor genes in aging presbycusic mice, all of 31 GABA receptor genes were used to generate regulatory pathways by using the Pathway Architect software package. This software creates biological interaction networks using a natural processing algorithm to extract the binding interactions between the input gene sets, based upon known molecular interactions as shown by Nikitin et al. (2003). Of the 31 GABA receptor genes analyzed, 16 were retained as shown in Fig. 5. Out of these 16 probe-sets (pink oval with blue outline), which are linked to each other through nodes, only 8 probe-sets (8 of the 9 probe-sets of Table 4) indicated with arrows and the boxes listing the criterion selection and function of the gene, showed statistically significant changes with age or hearing loss in the present study. The genes (Fig. 5) linked to each other by binding or regulatory interactions are depicted as connecting lines between nodes. These gene interactions, and possibly novel pathways, have not been previously reported in the context of aging and hearing loss. It offers an approach to study gene interaction networks based upon differentially expressed GABA receptor types that differ in their expression based upon age and hearing status.
Fig. 5.
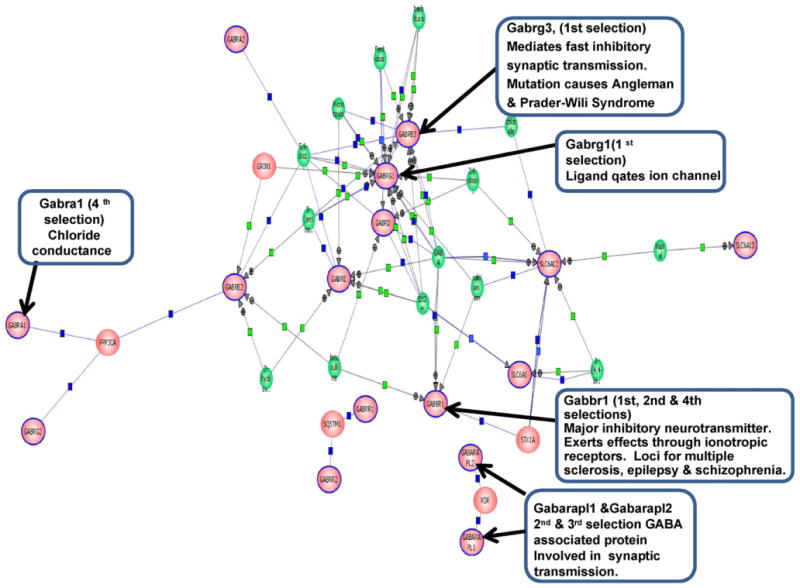
Biological interaction of GABA receptor network regulatory pathways generated by Pathway Architect using 31 GABA receptor-related genes from the microarray data of 40 CBA mice grouped according to their physiological hearing data, including auditory brainstem response thresholds and distortion-product otoacoustic emission amplitudes. Sixteen genes (pink oval with blue outline) out of 31 probe-sets are connected with the central nodes of the biological network of GABA receptor pathways, as generated by a curated database of gene ontology for the most prominently over-represented genes. The algorithm then builds the regulatory network, including genes (pink oval with blue outline are 16 genes from the 31 GABA receptor probe-sets of the present investigation), proteins (pink oval), small molecules (green circles), binding molecules (blue squares), regulatory molecules (green squares). For eight out of the nine genes from Table 4, indicated with arrows and the boxes describing their respective criterion selection and functions, were found to be significantly changing their expression patterns with age and hearing loss in the present investigation. (For interpretation of the references to color in this figure legend, the reader is referred to the web site and manual for Pathway Architect.)
Table 4.
Detailed information for the nine GABA receptor probe-sets, selected according to the four criteria described in Section 2 and Section 5
| Probe-set ID | Accession number | Target gene name (symbol) | Assay-on-demand Assay ID |
Gene-sequence region (mice) bp |
Best human homology gene name and (chromosome location) |
Amplicon length (bp) | Probe-set selection criteria and figure/table |
|---|---|---|---|---|---|---|---|
| 1422187_at | NM_008074 | GABA-A receptor, subunit γ3 (Gabrg3) | Mm00433494_m1 | Gabrg3 1403–1940 | GABRG3 (15q11-q13) | 92 | 1st one-way ANOVA Bonferroni Fig. 1A |
| 1427227_at | NM_010252 | GABA-A receptor, subunit γ1 (Gabrg1) | Mm00439047_m1 | Gabrg1 2772–3301 | GABRG1 (4p12) | 125 | 1st one-way ANOVA Kruskal–Wallis Fig. 1B |
| 1422051_at | NM_019439.2 | GABA-B receptor 1 (Gabbr1) | Mm00444578_m1 | Gabbr1 1938–2290 | GABBR1 (6q21-31) | 63 | 1st one-way ANOVA Kruskal–Wallis Fig. 1B |
| 1416418_at | NM_020590.3 | GABA-A receptor associated protein like-1 (Gabarapl1) | Mm00457882_m1 | Gabarapl1 1244–1316 | GABARAPL1 (12p13.2) | 90 | 2nd linear regression DPOAE Fig. 2B |
| 1425595_at | NM_019439.2 | GABA-B receptor 1 (Gabbr1) | Mm00444579_m1 | Gabbr1 3976–4257 | GABBR1 (p21-31) | 112 | 2nd linear regression ABR Fig. 3B |
| 1417121_at | NM_008068.1 | GABA-A receptor, subunit α6 (Gabra6) | Mm01227755_m1 | Gabra6 1956–2380 | GABRA6 (5q34) | 68 | 3rd high fold change Fig. 4A |
| 1423187_at | NM_022693 | GABA-A receptor associated protein-like2 (Gabarapl2) | Mm00786429_s1 | Gabarapl2 124–544 | GABARAPL2 (16q22.3-q24.1) | 125 | 3rd high fold change Fig. 4B |
| 1421280_at | NM_010250 | GABA-A receptor, subunit α1 (Gabra1) | Mm00439040_m1 | Gabra1 1563–2011 | GABRA1 (5q34-q35) | 109 | 4th random Table 3 |
| 1455021_at | NM_019439.3 | GABA-B receptor 1 (Gabbr1) | Mm01273514_m1 | Gabbr1 72–436 | GABBR1 (p21.31) | 65 | 4th random Table 3 |
3. Discussion
Using Affymetrix M430A microarrays and cochlear RNA of CBA mice of different age-groups to identify possible roles of GABA receptors in age-related hearing loss, a total of 31 GABA receptor probe-sets were identified to have differential expression. Before making any definitive conclusions about these gene expression changes, we designed four unique criteria to select members of this family of GABA receptors for further validation, including relative real-time PCR. Four criteria, which were associated with different statistical procedures, including ANOVA and linear regression of LSR, and with functional hearing measures were employed. For example, multiple microarray replicates within each group allowed us to examine the reproducibility of data trends for subjects, including the biological variance of individual CBA/CaJ mice.
Another feature of the present investigation was that since an individual mouse's cochlear RNA yield is minute; we employed two-independent amplification techniques for the Affymetrix arrays and relative real-time PCR. Namely RNA was first amplified and then transcribed to cDNA for Affymetrix arrays (Invitrogen Kit), while for relative real-time PCR, we used RNA (5 ng) to transcribe to cDNA first, which was than amplified to (10 μg) using the NuGen Kit. The consistency of results obtained between GeneChip and relative real-time PCR data indicated that differential gene expression changes were reproducible, in the face of amplification of either RNA or cDNA. Furthermore the higher sensitivity of fold changes for relative real-time PCR data helped define the fold change most precisely. Specifically here, relative real-time PCR validated the GABA receptor subtype gene expression changes observed in RMA normalized GeneChip data. It remains to be seen whether these receptors participate in downstream pathways of transcription factors like Math-1, Pax-3, CREB or SOX10, which are important in the mammalian cochlea.
In an attempt to identify GABA receptors that play roles in age-related hearing loss, we used linear regression models to fit the functional physiological hearing data to the gene expression data. The highest correlations were for Gabarapl1, Gabra3, and Gabbr1. Gabarapl1 was downregulated with age, while Gabbr1 exhibited decreased levels in mild presbycusis, and an increase in the severe presbycusic mice.
GABA-A receptor subtypes γ1, γ3, α1, and Gabrapl1 and Gabrapl2 show overall reduction in gene expression and in relative real-time PCR in middle aged, old mild and severe presbycusic mice when compared to young adult mice. Mice with a Gabra1 deletion are normal in most regards, including hearing, but display reduced effects of the actions of ethanol as described by Lin et al. (2000). Previous studies of age-related changes in the inferior colliculus—auditory midbrain-showed decreases in subunits α1 and increases in Gabarg1 protein levels in F1 hybrid rats as described by Sieghart and Sperk (2002) and Kralic et al. (2005).
Another interesting gene, Gabra6, showed a gradual up-upregulation in middle-aged good hearing subjects, and mild presbycusic mice. Severe presbycusic mice exhibit very high levels of gene expression when compared to young adults with good hearing. It has been reported that ectopic overexpression of Gabra6 in hippocampal pyramidal neurons of mice results in an increased tonic conductance of these neurons as shown by Wisden et al. (2002). It is possible that the upregulation of the Gabra6 subunit may provide a mechanism to compensate for age-related cochlear hearing loss physiological changes.
Of the 31 probe-sets in the RMA normalized microarray data 26 are functionally annotated as having neurotransmitter receptor ligand ion channel gated activity; 5 are cytoskeletal binding proteins; one shows G-coupled protein activity, and one displays metabotropic glutamate activity. These functional categories can be plausibly related to presbycusis, for example the Gabbr1 gene showed-induced gene expression level changes with age and hearing loss. This gene displays G protein receptor and metabotropic glutamate activity. This could lead to glutamate excitotoxicity and contribute to hair cell death or impaired hair cells in old age, thus leading to reduce hearing sensitivity levels.
Several molecular or biochemical pathways may help regulate GABA receptors in cochlea as a function of hearing loss and age. Biological GABA receptor regulatory network pathways are depicted in Fig. 5, where eight out of the nine (Table 4) statistically significant GABA receptor probe-sets examined in our current study are indicated with arrows. The concordance in GABA receptor gene expression patterns observed in presbycusic CBA mice and the pathways generated by Pathway Architect provide a convenient and practical avenue to explore the gene interaction pathways models that may play important roles in the genetic etiologies of age-related sensory dysfunction. These models could be pursued furthermore at two levels: first, to look at expression levels of gene product, and secondly to look into the influence of these gene products as they interact in complex ways with metabolic and physiological processes during aging in the inner ear of mammals, which subserves both auditory and vestibular functions.
4. Conclusion
A focus on individual mice in different age groups for this GeneChip study provides key insights into possible molecular genetic bases of presbycusis. The unique approaches to microarray probe-set selection criteria along with the linear regression modeling of functional data of hearing abilities, i.e., ABR thresholds and DPOAE amplitude's, were utilized for selection of biologically important genes in the aging inner ear to reveal biologically relevant changes in the GABA neurotransmitter system. The present investigation complements previous discoveries of age-related changes in gene expression for neurotransmitter systems of the auditory brainstem, and provides new conceptual and statistical approaches for analyzing gene microarray data for studies of the aging nervous system and sensory function.
5. Material and methods
Animal groups and lab procedural details were the same as our previous microarray investigations as in Tadros et al. (2007a,b).
5.1. Mice
CBA/CaJ mice were bred in-house according to the University of Rochester Vivarium and Animal Use Committee protocols. Original breeding pairs were obtained from Jackson Laboratories. All animals had similar environmental and non-ototoxic histories, being raised together in a relatively quiet vivarium room.
5.2. Functional hearing assessment
DPOAE amplitudes and ABR thresholds were measured, and four groups of mice were employed as given in Table 1, and as described in detail by Tadros et al. (2007a,b).
5.3. Cochlea isolation
Tissue isolation, RNA preparation, cDNA synthesis, and GeneChip target hybridization, washing, staining and scanning is as described in Tadros et al. (2007a,b).
5.4. Microarrays MOE430A
Microarrays were purchased from Affymetrix, MOE430A, as described in Affymetrix technical notes (2003), and the chip contained 22,690 unique probe-sets. Oligonucleotide identifiers were 25 nt in length, centrally positioned with a 9 probe-set of PM (perfect match) and MM (mismatch) rows designed for Mus musculus (mouse) RefSeq sequences. Each array had 14,484 full-length transcripts, 9450 transcripts of non-ESTs (excluding full-length) and 21,103 transcripts from ESTs with a strong evidence for polyadenylation. The sequence selection region was a 600 bp area selected from the consensus with regions based on strong evidence for polyadenylation, a full-length 3′ end, and consensus sequence ends. Locuslink database: http://www.affymetrix.com.
5.5. Normalization of microarray data
The commercially available mouse chip MOE430A was used for all microarray experiments and probe-set information is available at http://www.affymetrix.com.
Hybridization intensities for each chip were scaled in comparison to young controls (8 chips). Intensity values were normalized using robust multi-array analysis (RMA) as described in Irizarry et al. (2003) and Iobion Informatics Gene Traffic 3.1 (2005). This strategy tends to include low intensity signals off PM relative to MM. The software used for normalization was GeneTraffic 3.1, for more information available at Iobion, http://www.iobion.com.
A two-class unpaired analysis of the data was performed to identify genes that differ by ≥1.5 fold from the young controls at a false discovery rate (FDR) ≤1% in middle aged, old mild and old severe presbycusic subject groups. RMA analysis using LSR probe-set expression values identified 31 GABA receptor-related probe-sets, including GABA-A, -B, and -C receptors, transporters and associated-like proteins.
5.6. Taqman primer/probes for relative real-time PCR
Taqman primer/probes (ABI, Applied Biosystems Incorporation) were used to confirm the microarray data. The probes were tagged with a fluorescence dye (FAM) for passive fluorescence (ROX). Relative real-time PCR analysis of samples from the GeneChip experiments was done in the four subject groups. Relative real-time PCR analysis as described by Livak ABI Bulletin #2 (1997) and Livak and Schmittgen (2001) was done on triplicates of each sample on an ABI 7500 thermocycler (PE Applied Biosystems) with a TaqMan universal master mix (Applied Biosystems). See Table 4 for Taqman primer/probe details. The following PCR profile was applied: incubation for 2 min at 50 °C, then initial denaturation at 10 min at 95 °C, followed by 50 cycles of 15 s denaturation at 95 °C, and 1 min annealing at 60 °C.
5.7. cDNA synthesis for relative real-time PCR
Total RNA was utilized from samples that were saved from the original microarray experiments. Five nanograms total RNA was reverse transcribed to cDNA using the NuGen kit Protocol according to the manufacturer's instructions (NuGen, http://www.NuGen.com).
5.8. Relative real-time PCR
A unique strategy was developed for the selection of probe-sets for further analysis, from our RMA normalized microarray data set. The probe-set selection criterion was based upon the following steps:
Microarray RMA normalized log signal ratio data of 31 GABA receptor probe-sets were subjected to one-way ANOVA (compares group means) with post hoc Bonferroni tests (pair-wise tests corrected for multiple comparisons), and Kruskal–Wallis tests (compares the medians of the groups).
Genes with a significant p-value from regression analysis for microarray LSR data and the physiological hearing data ABR and DPOAE.
High fold changes for the RMA normalized gene expression data (Fold change cut-off above 2 or below −2) when compared to young adult, good hearing control mice.
A final selection of GABA receptor probe-sets with low fold changes (1.0–1.4) was done randomly from the 31 GABA receptor probe-set of microarray gene expression data.
Nine probe-sets (genes, Table 4) were purchased from ABI (Applied Biosystems Inc.), to perform relative real-time PCR. RNA (saved from the microarray samples) was independently reverse transcribed to cDNA. Using the NuGen Kit protocol, cDNA was amplified and was used as the template source for the real-time PCR. Nine microliters (90 ng) of cDNA was included in a 20-μl PCR reaction. For a quantitative analysis, relative real-time Taqman PCR technology was used (Applied Biosystems). The RNA expression fold Change of relative real-time PCR was calculated according to 2−ΔΔCt as described by Livak ABI Bulletin #2 (1997) and Livak and Schmittgen (2001).
5.9. Statistical analysis
Data were analyzed by using Prism 4.0 (GraphPad). All data were presented as LSR or fold change ±S.D. and subjected to one-way ANOVA with an appropriate post hoc test corrected for multiple comparisons of pair-wise differences in groups.
5.10. Pathway Architect
Biological interacting GABA receptor regulatory network pathways were derived from microarray data of the 31 GABA receptor probe-sets from four subject groups, young adults with good hearing; middle aged with good hearing; old mice with mild hearing loss, and old mice with severe presbycusis. Eight of Table 4 probe-sets are included in Fig. 5 of Pathway Architect. The Pathway Architect software details are as in http://www.stratagene.com.
Acknowledgments
Supported by NIH Grants P01 AG09524 from the Nat. Inst. on Aging, P30 DC05409 from the Nat. Inst. on Deafness & Communication Disorders, and the International Center for Hearing & Speech Research, Rochester, NY.
Abbreviations
- GABA
gamma aminobutyric acid
- RMA
robust multi-chip average
- LSR
log signal ratio
- ABR
auditory brainstem response
- DPOAE
distortion-product otoacoustic emission
- PCR
polymerase chain reaction
- IC
inferior colliculus
- S.E.M.
standard error mean
References
- Affymetrix technical notes, 2003. Array Design and Performance of the GeneChip Mouse Expression Set 430.
- Caspary DM, Holder TM, Hughes LF, Milbrandt JC, McKernan RM, Naritoku DK. Age-related changes in GABAA receptor subunit composition and function in rat auditory system. Neuroscience. 1999;93:307–12. doi: 10.1016/s0306-4522(99)00121-9. [DOI] [PubMed] [Google Scholar]
- Frisina RD, Rajan R. Inferior colliculus: aging and plasticity. In: Winer J, Schreiner C, editors. The inferior colliculus. New York: Springer; 2005. pp. 559–84. Ch. 18. [Google Scholar]
- Gates GA, Couropmitree N, Myers RH. Genetic associations in age-related hearing loss. Arch Otolaryngol Head Neck Surg. 1999;125:645–59. doi: 10.1001/archotol.125.6.654. [DOI] [PubMed] [Google Scholar]
- Iobion Informatics Gene Traffic 3.1, 2005. Analysis, visualization of Microarray expression data. Compliant with MIAME and MAML standard.
- Irizarry RA, Bolstad BM, Collin F, Cope L, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupmann K, Huggel K, Heid J, Flor PJ, Bishoff S, Mickel SJ, McMaster G, Angst C, Bittiger H, Froestl W, Bettler B. Expression cloning of GABAB receptors uncovers similarity to metabotropic glutamate receptors. Nature. 1997;386:239–46. doi: 10.1038/386239a0. [DOI] [PubMed] [Google Scholar]
- Kralic JE, Criswell HE, Osterman JL, O'Buckley TK, Wilkie ME, Matthews DB, Hamre K, Breese GR, Homanics GE, Morrow AL. Genetic essential tremor in γ-aminobutyric acid A receptor α1 subunit knockout mice. J Clin Invest. 2005;115:774–9. doi: 10.1172/JCI23625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Shanping C, Ping C. Activation of metabotropic GABAB receptors inhibited glutamate responses in spiral ganglion neurons of mice. Neuroreport. 2000;11:957–61. doi: 10.1097/00001756-200004070-00012. [DOI] [PubMed] [Google Scholar]
- Livak KJ. ABI Pims 7700 Sequencer detection system. User Bulletin # 2. PE Appl Biosyst. 1997 [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2-ΔΔCT. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Milbrandt JC, Hunter C, Caspary DM. Alterations of GABAA receptor subunit mRNA levels in the aging Fischer 344 rat inferior colliculus. J Comp Neurol. 1997;379:455–65. doi: 10.1002/(sici)1096-9861(19970317)379:3<455::aid-cne10>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazzo I. Pathway studio—the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–7. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- Sieghart W, Sperk G. Subunit composition, distribution and function of GABAA receptor subtypes. Curr Top Med Chem. 2002;2:795–816. doi: 10.2174/1568026023393507. [DOI] [PubMed] [Google Scholar]
- Tadros S, D'Souza M, Zettel ML, Zhu X, Waxmonsky NC, Frisina RD. Glutamate related gene expression changes with age in the mouse auditory midbrain. Brain Res. 2007a;1127:1–9. doi: 10.1016/j.brainres.2006.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros S, D'Souza M, Zettel ML, Zhu X, Erhardt ML, Frisina RD. Serotonin 2B receptor: upregulated with age and hearing loss in mouse auditory system. Neurobiol Aging. 2007b;28:1112–23. doi: 10.1016/j.neurobiolaging.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Wisden W, Cope D, Klausberger T, Hauer B, Sinkkonen ST, Tretter V, Lujan R, Jones A, Korpi ER, Mody I, Sieghart W, Somogyi P. Ectopic expression of the GABAA receptor α6 subunit in hippocampal pyramidal neurons produces extrasynaptic receptors and an increased tonic inhibition. Neuropharmacology. 2002;43:530–49. doi: 10.1016/s0028-3908(02)00151-x. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Frisina RD, Gordon A, Klebanov L, Yakovlev A. Multivariate search for differentially expressed gene combinations. BMC Bioinform. 2004;5:164–84. doi: 10.1186/1471-2105-5-164. [DOI] [PMC free article] [PubMed] [Google Scholar]


