Abstract
The myristoylated alanine-rich C kinase substrate (MARCKS) is a prominent protein kinase C (PKC) substrate in brain that is expressed highly in hippocampal granule cells and their axons, the mossy fibers. Here, we examined hippocampal infrapyramidal mossy fiber (IP-MF) limb length and spatial learning in heterozygous Macs mutant mice that exhibit an ≈50% reduction in MARCKS expression relative to wild-type controls. On a 129B6(N3) background, the Macs mutation produced IP-MF hyperplasia, a significant increase in hippocampal PKCɛ expression, and proficient spatial learning relative to wild-type controls. However, wild-type 129B6(N3) mice exhibited phenotypic characteristics resembling inbred 129Sv mice, including IP-MF hypoplasia relative to inbred C57BL/6J mice and impaired spatial-reversal learning, suggesting a significant contribution of 129Sv background genes to wild-type and possibly mutant phenotypes. Indeed, when these mice were backcrossed with inbred C57BL/6J mice for nine generations to reduce 129Sv background genes, the Macs mutation did not effect IP-MF length or hippocampal PKCɛ expression and impaired spatial learning relative to wild-type controls, which now showed proficient spatial learning. Moreover, in a different strain (B6SJL(N1), the Macs mutation also produced a significant impairment in spatial learning that was reversed by transgenic expression of MARCKS. Collectively, these data indicate that the heterozygous Macs mutation modifies the expression of linked 129Sv gene(s), affecting hippocampal mossy fiber development and spatial learning performance, and that MARCKS plays a significant role in spatial learning processes.
Keywords: protein kinase C/hippocampus/Macs
Protein kinase C (PKC) is a family of serine/threonine kinases implicated in synaptic plasticity and information storage processes (24). The myristoylated alanine-rich C kinase substrate (MARCKS) is a primary PKC substrate that binds plasma membrane via N-terminal myristoylation and electrostatic interactions with the phosphorylation site domain, binds calmodulin in a calcium-dependent manner, and cross-links filamentous (F) actin, all in a PKC phosphorylation-reversible manner (1, 3). Accordingly, MARCKS is proposed to regulate membrane–cytoskeleton plasticity in response to PKC- and calcium-mediated signaling (1, 3). Moreover, MARCKS is developmentally regulated (13), is enriched in neuronal growth cones (R.K.M., E. A. Wees, P. J. Meberg, T. B. Kuhn, and R.H.L., unpublished data), and is highly expressed in select regions of the developing and adult rat brain (12, 13). In particular, the granule cell layer of the hippocampus exhibits a high level of MARCKS gene expression (12, 13). In addition, the granule cell axons (mossy fibers), which have been implicated in the spatial learning ability of inbred mice (reviewed in ref. 17), express MARCKS protein at high levels in the adult brain (14). Because granule cell neogenesis occurs well into adulthood in a manner regulated by environmental demands (9), MARCKS may contribute to mossy fiber maturation and synaptic plasticity throughout the life of the animal.
The creation of a mutant mouse strain that exhibits reductions in MARCKS expression has permitted the assessment of the contribution of MARCKS to hippocampal mossy fiber development and associated spatial learning processes. Homozygous mutant mice that do not express MARCKS exhibit abnormal brain development characterized by decreased brain size, collosal and commissural agenesis, cortical lamination abnormalities, particularly in the neocortex and hippocampus, exencephaly, and perinatal lethality (20). This phenotype is “rescued” (complemented) by transgenic expression of epitope-tagged human MARCKS driven by 3.4 kilobases of human MACS promoter (21), indicating that the phenotype is caused by the MARCKS deficiency alone. In the present study, we used heterozygous Macs mutant mice, which do not exhibit any gross neuroanatomical abnormalities, and MACS transgenic mice to assess the role of MARCKS in hippocampal mossy fiber development and spatial learning in the Morris water maze.
METHODS
Animals.
The generation of mutant and transgenic mice is described in detail elsewhere (20, 21). In brief, heterozygous Macs mutant mice were created by homologous recombination using 129Sv (E14TG2a) embryonic stem cells; chimeric mice were mated with C57BL/6 (B6) mice. These heterozygous mutant 129B6(N2) mice were crossed with B6SJL(N1) mice bearing the epitope-tagged MACS transgene (Tg+), and resulting progeny possessing both the heterozygous Macs mutation and the MACS Tg+ were used in the rescue experiment. Adult (4–5 months) male and female, mutant and wild-type litter mates from three different gene backgrounds were used: (i) 129Sv/C57BL/6(N3) (129B6(N3)), (ii) 129Sv/C57BL/6(N9) (129B6(N9)), and (iii) C57BL/6/SJL(N1) (B6SJL(N1)). For purposes of baseline comparisons in the assessment of hippocampal mossy fiber morphology, adult (2 months) male C57BL/6 mice were purchased from The Jackson Laboratory. Experimental animals were housed with same-sex siblings with 4–6 animals per cage in specific pathogen-free Murine Facilities at the University of Florida maintained on a 12:12 light:dark cycle. All behavioral testing was conducted during the light portion of the cycle. Food and water were available ad libitum. Genotyping by Southern blot analysis was conducted after data collection by using DNA extracted from tail clips, as described (20, 21).
Quantitative Timm’s Stain Assay.
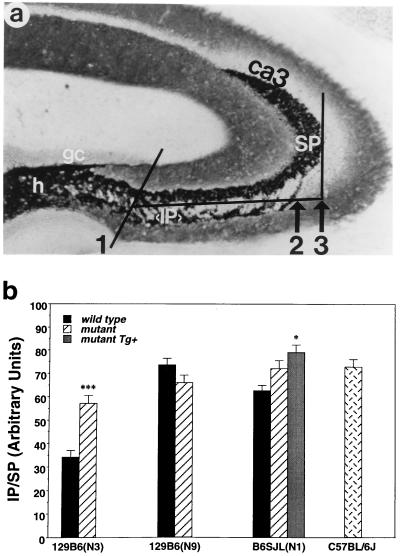
Brains were fixed, and coronal sections (35 μm) were subjected to Timm’s staining as described (18). Mossy fiber (MF) projections were visualized with a light microscope (10× lens) interfaced with a video-imaging system and quantitated with National Institutes of Health image 1.6 software. The length of the infrapyramidal (IP) and suprapyramidal (SP) limbs were measured from a common point on a line intersecting the tips of the dorsal and ventral blades of the granule cell layer (see Fig. 2a). The length of the IP limb was normalized against the length of the SP limb to control for potential differences in hippocampal size. Multiple sections (n = 12) taken from the dorsoseptal aspect of the hippocampus were used for each mouse. The investigator performing the quantitation was blind to genotype. After quantitation, mean IP/SP ratios for individual mice were grouped according to genotype, and differences were assessed with a one-way ANOVA.
Figure 2.
(a) Photomicrographs illustrating a Timm’s stained coronal hippocampal section from the midseptotemporal region of a representative inbred C57BL/6J mouse brain (gc, granule cell layer; h, hilus; IP, infrapyramidal mossy fiber limb, SP, suprapyramidal limb, ca3, ca3 pyramidal cell field), as well as the quantitative parameters: (1) A line intersecting the tips of the dorsal and ventral blades of the granule cell layer providing a common point from which IP and SP lengths can be measured. (2) Demarcation of the extent of the IP limb length. (3) Demarcation of the extent of the SP limb length. (b) Quantitation of the IP limb length relative to the SP limb length (IP/SP) in wild-type and mutant 129B6(N3), 129B6(N9), B6SJL(N1) mice, in mutant B6SJL(N1) Tg+ mice, and in inbred C57BL6/J mice. Data expressed as mean ± SEM. ∗, P < 0.05; ∗∗∗, P < 0.001 compared with wild-type control means.
RNase Protection Assay.
Total RNA was extracted from the hippocampi or cortex of naive mice as described (5). The biotinylated antisense MARCKS riboprobe was 466 nucleotides long and protected a 386-bp fragment, and the cyclophilin riboprobe (Ambion, Austin, TX) protected a 103-bp fragment. The assay was conducted according to Ambion protocol. Autoradiographs were quantitated by scanning densitometry using National Institutes of Health image 1.47 software, and MARCKS values were normalized to cyclophilin to control for loading/transfer variability. Data (MARCKS:cyclophilin ratios) then were grouped according to genotype, and differences were assessed with a one-way ANOVA.
Western Blot Assay.
Total protein (5–20 μg) extracted from the hippocampi and cortex of naive mice was analyzed by quantitative Western blot as described (25). Primary antibody titers and suppliers were as follows: MARCKS (1:36,000, R.H.L.), RC3 (1:1000, D. Gerendasy, Scripps Institute), GAP-43 (1:3000, Sigma), PKCβII (1:5000) and PKCδ (1:750, Santa Cruz Biotechnology), PKCα (1:2500) and PKCɛ (1:8000, Transduction Laboratories, Lexington, KY), PKCγ (1:800; Seikaguku Kogyo, Tokyo), and hemagglutinin monoclonal 12CA5 antibody (1:800; Babco, Richmond, VA). The dilutions for the secondary antibody were 1:24,000 (MARCKS), 1:20,000 (PKCβII, PKCδ, PKCγ, PKCɛ, RC3, and GAP-43), 1:10,000 (PKCα), and 1:1000 (12CA5). Anti-mouse and anti-rabbit IgG F(ab′)2 conjugated to horseradish peroxidase were purchased from Bio-Rad, chemiluminescent substrate was purchased from Pierce, and Hyperfilm ECL was purchased from Amersham. Equal loading was determined from Coomassie-stained gels. On all gels, standard curves of known protein concentrations were run to guide subsequent film exposure times to be within the linear range. Autoradiographs were quantified by scanning densitometry with an image analysis system using National Institutes of Health image 1.47 software. Density values were compared with a one-way ANOVA.
In Situ Hybridization.
An antisense riboprobe corresponding to bases 354–1,505 of the murine MARCKS cDNA (19) was used. The specificity of this probe and in situ hybridization methodology used are described in detail elsewhere (12, 13).
Spatial Learning.
Spatial learning was assessed in the Morris water maze as described (11). The water maze consisted of a circular pool (diameter, 120 cm; height, 45 cm) filled with cool (25 ± 1°C), opaque water into which a circular escape platform (12 cm diameter), either visible (2 cm above the water surface exposing black sides) or submerged (2 cm below the water surface), was placed. Mice, run in groups of 5–6, first were trained (8 trials/day for 2 days) to escape onto a visible platform that was relocated to the center of each of the four pool quadrants between trials in a random manner (to obviate the accurate use of spatial cues), then to a submerged platform (8 trials/day for 5 days) maintained in the center of the northwest quadrant, followed by training (8 trials/day for 2 days and 4 trials for 1 day) to the submerged platform placed in the diagonally opposite (southeast) quadrant (reversal). Swim-path length (distance in centimeters), escape latency (seconds), and swim speed (centimeters per second) were recorded. The following groups of mice were tested in the Morris water maze: (i) 129B6(N3) (wild-type: n = 20, mutant: n = 20), (ii) 129B6(N9), (wild-type: n = 9, mutant: n = 17), and (iii) B6SJL(N1) (wild-type: n = 11, mutant: n = 10, mutant Tg+: n = 14). Each group was composed of ≈50:50 male:female, and statistical analyses indicated that there were no significant sex by strain interactions for any strain (data not shown). Distance, latency, and swim speed data were analyzed with an ANOVA with repeated measures, and the number of target platform crossings relative to nontarget crossings during the probe trials were analyzed with a one-way ANOVA. In case of multiple group analyses (rescue experiment), post hoc comparisons were conducted by using ANOVA with repeated measures.
RESULTS
Demonstration of Reduced MARCKS Expression in Adult Mice.
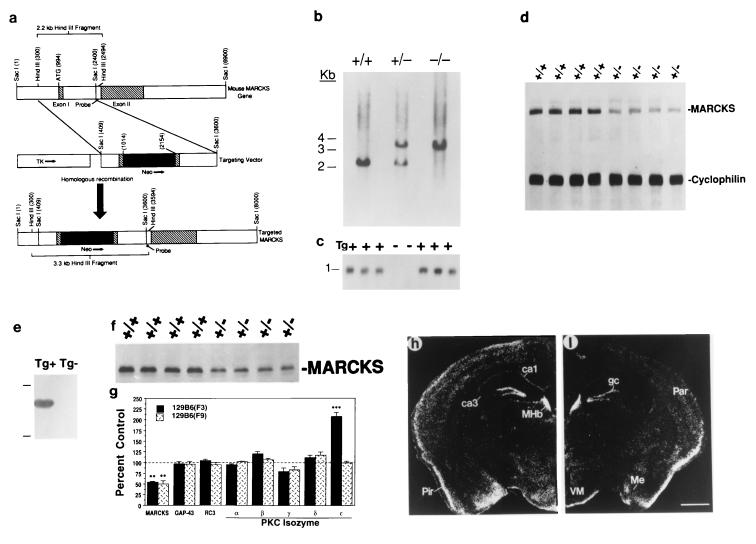
The endogenous MARCKS gene, the MARCKS sequence targeting vector, and the predicted homologously recombined MARCKS gene is illustrated in Fig. 1a. Resultant wild-type (+/+), heterozygous (+/−), and homozygous (−/−) offspring were identified by genomic Southern blot analysis (Fig. 1b). Presence of the MACS transgene (Tg) also was determined by genomic Southern blot analysis (Fig. 1c). MARCKS mRNA was reduced by 51.2 ± 0.01% (mean ± SEM) in hippocampal homogenates of adult heterozygous mutant 129B6(N3) mice relative to wild-type controls (Fig. 1d). MARCKS protein also was reduced by ≈50 percent in each of the three strains: (i) 129B6(N3) (54.3 ± 2.3%; Fig. 1g), (ii) 129B6(N9) (50.5 ± 6.6; Fig. 1 f and g), and (iii) B6SJL(N1) (56 ± 2.5%). These data confirm that MARCKS is expressed in an allele dose-dependent fashion in adult mice. In the 129B6(N3) mouse hippocampus, MARCKS was reduced in both the cytosolic (47.5 ± 7.8%) and membrane (52 ± 3.8%) fractions, as well as in cortical homogenates (48 ± 5.3%) relative to wild-type controls. Expression of the epitope-tagged human MACS transgene (Tg+; Fig. 1e) elevated MARCKS protein expression in brain by ≈50% relative to endogenous expression in wild-type mice (21). Examination of MARCKS gene expression in the adult mouse forebrain by using in situ hybridization revealed high expression in hippocampal granule cells, medial habenula, amygdala, piriform cortex, and periventricular hypothalamic nuclei (Fig. 1h), identical to that described for the adult rat brain (12). Comparison of hippocampal MARCKS mRNA expression in 129B6(N3) wild-type and mutant mice revealed reductions in CA1-CA3 and the granule cell layer (Fig. 1h vs. Fig. 1i).
Figure 1.
(a) Schematic diagram showing the endogenous MARCKS gene, the MARCKS sequence targeting vector, and the predicted homologously recombined MARCKS gene. The MARCKS intron is depicted as an open box, exons I and II are depicted as hatched boxes, and untranslated sequences are depicted as open boxes. The dark box under the gene represents the DNA fragment used as a hybridization probe for performing genomic Southern blots. The bracketed sections indicate the expected HindIII restriction fragments in Southern blots: 2.2 kilobases for the endogenous gene and 3.3 kilobases for the homologously recombined gene. (b) Identification of wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mutant mice by genomic Southern blot analysis. (c) Identification of the MACS transgene (Tg) by genomic Southern blot analysis. (d) Autoradiograph illustrating MARCKS and cyclophilin mRNA expression in wild-type (+/+) and heterozygous mutant (+/−) 129B6(N3) mouse brain. (e) Autoradiograph from Western blot analyses illustrating transgenic human MARCKS expression in Tg+ and Tg− B6SJL(N1) mice [(Top) Mr 97.4 × 109. (Bottom) 68 × 109]. (f) Autoradiograph from Western blot analyses illustrating MARCKS immunoreactive bands in hippocampal homogenates derived from wild-type (+/+) and heterozygous mutant (+/−) 129B6(N3) mice. (g) Quantitative analyses of MARCKS, GAP-43, RC3, and PKC isozyme expression in hippocampal homogenates from 129B6(N3) and 129B6(N9) mutant mice (expressed as percent of wild-type control; ∗∗, P < 0.01; ∗∗∗, P < 0.0001 relative to wild-type control). Shown is localization of MARCKS gene expression in wild-type (h) and heterozygous mutant (i) 129B6(N3) mouse forebrain by using in situ hybridization histochemistry. gc, granule cell layer; MHb, medial habenula; Par, parietal cortex; Pir, piriform cortex; Me, medial amygdaloid nuclei; VM, ventromedial thalamic nuclei. (Bar = 1 mm.)
Elevation in PKCɛ Expression Resulting from Macs Mutation–Gene Background Interactions.
To determine whether the heterozygous Macs mutation elicited compensatory alterations in the expression of related molecules, we examined the expression of PKC isozymes α, βII, γ, δ, and ɛ, for which MARCKS is a key substrate (23), the presynaptic PKC substrate GAP-43, and the postsynaptic PKC substrate RC3 in hippocampal homogenates. We observed a dramatic, 100% elevation in PKCɛ, but none of the other PKC isozymes or PKC substrates (GAP-43/RC3) in mutant 129B6(N3) mice relative to wild-type controls (P < 0.0002; Fig. 1g). However, mutant 129B6(N9) mice did not exhibit any significant alterations in PKC isozyme or substrate (GAP-43/RC3) expression relative to wild-type controls (Fig. 1g), suggesting that the Macs mutation interacted with the 129B6(N3) background to up-regulate PKCɛ expression. Hippocampal PKCɛ expression did not differ between wild-type and mutant B6SJL(N1) mice (P = 0.72) or between mutant Tg- and mutant Tg+ B6SJL(N1) mice (P = 0.58).
Role of MARCKS and Background Genes in Mossy Fiber Development.
The IP-MF limb (examined as an IP/SP ratio) of mutant 129B6(N3) mice (n = 10) was significantly longer than the wild-type IP-MF limb (n = 7; P = 0.004; Fig. 2) whereas the length of the SP-MF limb did not differ significantly (P = 0.07; data not shown). However, the IP-MF limb of wild-type 129B6(N3) mice was significantly shorter relative to inbred C57BL/6J mice (P = 0.0001; Fig. 2c). The IP-MF limb of mutant 129B6(N3) mice was also significantly shorter relative to inbred C57BL/6J mice (P = 0.01). Conversely, the length of the IP-MF limb of mutant 129B6(N9) mice (n = 17) did not differ significantly from wild-type controls (n = 13; P = 0.11; Fig. 2), and neither did the length of the SP-MF limb (P = 0.90; data not shown). Furthermore, the lengths of IP-MF limb of neither wild-type (P = 0.87) nor mutant (P = 0.30) 129B6(N9) mice differed significantly from the IP-MF limb length of inbred C57BL/6J mice. However, the IP-MF limb length of wild-type 129B6(N9) mice differed significantly from the IP-MF limb of wild-type 129B6(N3) mice (P = 0.0001) whereas the IP-MF limb length of mutant 129B6(N9) mice did not differ significantly from mutant 129B6(N3) mice (P = 0.08). Together, these data indicate that the IP-MF hypoplasia exhibited by wild-type 129B6(N3) mice is caused by residual 129Sv background genes and that the Macs mutation interacts with this background to enhance IP-MF outgrowth. The IP-MF limb of mutant B6SJL(N1) mice (n = 9) did not differ significantly from either wild-type controls (n = 11; P = 0.08) or inbred C57BL/6J mice (P = 0.62), and neither did the length of the SP-MF limb relative to wild-type controls (P = 0.58; data not shown). However, the IP-MF limb of wild-type B6SJL(N1) mice was significantly shorter relative to inbred C57BL/6J mice (P = 0.03). The IP-MF limb of mutant B6SJL(N1) Tg+ mice (n = 14) was marginally (≈17%) longer relative to the IP-MF limb of wild-type B6SJL(N1) mice (P = 0.01) but did not differ significantly from mutant B6SJL(N1) mice (P = 0.17). In none of the strains examined was a significant sex by strain interaction observed for the IP/SP length measure.
Role of MARCKS and Background Genes in Spatial Learning.
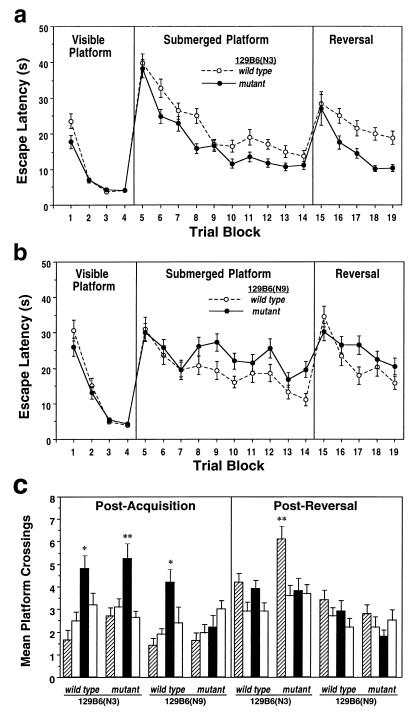
For each strain tested on the tasks described below, distance data (not shown) exhibited the same pattern of performance and levels of significance as escape latency data, and swim speed did not differ significantly between wild-type and mutant mice (data not shown). On the visible platform task, wild-type and mutant 129B6(N3) mice exhibited comparable escape latencies [Group (G) F(1, 58) =2.88, P = 0.09; Trial (T) F(3, 474) = 114, P = 0.001; G×T F(3, 474) = 4.02, P = 0.05] (Fig. 3a), as did wild-type and mutant 129B6(N9) mice [G, F(1, 114) = 1.07, P = 0.3; T, F(3, 342) = 93.3, P < 0.001; G×T, F(3, 342) = 1.1, P = 0.36] (Fig. 3b). Of interest, mutant B6SJL(N1) mice exhibited significantly shorter escape latencies relative to wild-type mice on this task [G, F(1, 82) = 6.5, P = 0.01; T, F(3, 246) = 46.9, P = 0.001; G×T, F(3, 246) = 0.65, P = 0.58] (Fig. 4a). Together, these data indicate the Macs mutation did not impair gross sensorimotor function, escape motivation, or simple associative learning processes in any of these three strains.
Figure 3.
The latency (in seconds) taken to locate the visible platform (Visible Platform), the submerged platform (Submerged Platform), and the submerged platform moved to the diagonally opposite quadrant (Reversal). A trial block is composed of four trials, each beginning from one of four different start locations. (a) Performance of wild-type and mutant 129B6(N3) mice. (b) Performance of wild-type and mutant 129B6(N9) mice. (c) Performance of wild-type and mutant 129B6(N3) and 129B6(N9) mice during the postacquisition and post-reversal probe trials. Presented are the number of crossings over the location where the platform was located during initial acquisition training (solid bar) and during reversal training (hatched bar) as well as the equivalent location in the other two quadrants (open bars). Data are presented as group mean ± SEM. ∗, P < 0.05; ∗∗, P < 0.01 compared with the next highest, nontarget, platform crossing.
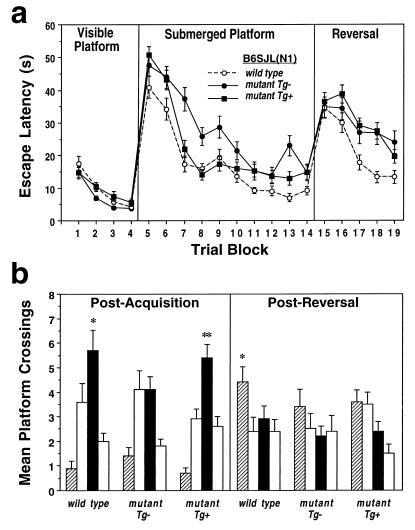
Figure 4.
(a) Escape latencies (in seconds) of wild-type, mutant Tg−, and mutant Tg+ B6SJL(N1) mice during the visible, submerged, and reversal platform tasks. (b) Performance of wild-type, mutant, and mutant Tg+ B6SJL(N1) mice during the postacquisition and post-reversal probe trials. Presented are the number of crossings over the location where the platform was located during initial acquisition training (solid bar) and during reversal training (hatched bar) as well as the equivalent location in the other two quadrants (open bars). Data are presented as group mean ± SEM. ∗, P < 0.05; ∗∗, P < 0.01 compared with the next highest, nontarget, platform crossing.
When the visible platform was replaced with the stationary submerged platform (Fig. 3a), mutant 129B6(N3) mice exhibited faster escape latencies relative to wild-type mice [G, F(1, 150) = 19.2, P = 0.001; T, F(9, 1350) =48.9, P = 0.001; G×T, F(9, 1350) = 1.86, P = 0.29]. In contrast, mutant 129B6(N9) mice exhibited significantly longer escape latencies relative to wild-type controls [G, F(1, 110) = 9.06, P = 0.003; T, F(9, 990) = 6.98, P = 0.0001; G×T, F(9, 1350) = 0.9, P = 0.54] (Fig. 3b). Mutant B6SJL(N1) mice also exhibited significantly longer escape latencies relative to wild-type controls [G, F(1, 82) = 29.7, P = 0.0001; T, F(9, 738) = 45.8, P = 0.0001; G×T, F(9, 738) = 2.03, P = 0.03] (Fig. 4a). During the postacquisition probe trial, both mutant and wild-type 129B6(N3) mice exhibited a similar bias for the target quadrant, and their number of target platform crossings did not differ significantly (P = 0.64; Fig. 3c). However, wild-type 129B6(N9) mice exhibited a greater number of target platform crossings relative to the next-preferred platform location (P = 0.05) and to mutant target platform crossings (P = 0.01), and mutant mice failed to exhibit a spatial bias for the platform location (Fig. 3c). Similarly, wild-type B6SJL(N1) mice exhibited a greater number of crossings over the target platform location relative to the next preferred platform location (P = 0.05) as well as to mutant target platform crossings (P = 0.01; Fig. 4b).
When the submerged platform was moved to the diagonally opposite quadrant, mutant 129B6(N3) mice exhibited significantly shorter escape latencies relative to wild-type controls [G, F(1, 150) = 15.1, P = 0.001; T, F(4, 600) = 13.3, P = 0.001; G×T, F(4, 600) = 1.15, P = 0.33] (Fig. 3a). In contrast, mutant 129B6(N9) mice exhibited a trend toward longer escape latencies relative to wild-type mice, though this did not reach statistical significance [G, F(1, 110) = 1.5, P = 0.22; T, F(4, 440) = 10.9, P = 0.001; G×T, F(4, 440) = 2.3, P = 0.06] (Fig. 3b). However, mutant B6SJL(N1) mice exhibited significantly longer escape latencies relative to wild-type controls [G, F(1, 82) = 8.54, P = 0.004; T, F(9, 328) = 17.2, P = 0.0001; G×T, F(4, 328) = 2.1, P = 0.07] (Fig. 4a). During the postreversal probe trial, wild-type 129B6(N3) mice failed to exhibit a significant bias for the reversed platform location relative to the next-preferred platform location (P = 0.65) whereas mutant 129B6(N3) mice did exhibit a bias relative to the next-preferred platform location (P = 0.01) and to wild-type crossings (P = 0.05; Fig. 3c). However, neither wild-type nor mutant 129B6(N9) mice exhibited a significant bias for the new correct platform location (Fig. 3c). In contrast, wild-type B6SJL(N1) mice exhibited a significant bias for the target platform location relative to the next preferred platform location (P = 0.05) whereas mutant mice did not exhibit such a bias (P = 0.65; Fig. 4b).
Transgenic Rescue of Impaired Spatial Learning.
In this experiment, wild-type mice, mutant mice not bearing the MACS transgene (Tg−), and mutant mice bearing the MACS transgene (Tg+) were compared (Fig. 4). On the visible platform task, there was no significant main effect of Group or Group-by-Trial interaction [G, F(2, 137) = 2.21, P = 0.11; T, F(3, 411) = 49.2, P = 0.0001; G×T, F(6, 411) = 1.1, P = 0.33] for escape latency (Fig. 4a) or distance (data not shown). During the submerged platform task, there was a significant Group, Trial, and Group-by-Trial interaction [G, F(2, 137) = 16.5, P = 0.0001; T, F(9, 1233) = 81.6, P = 0.0001; G×T, F(18, 1233) = 2.43, P = 0.001] for both escape latency (Fig. 4a) and distance (data not shown). The swim speeds of mutant Tg+ mice were significantly slower relative to wild-type controls (P = 0.01). Post hoc analysis of distance data, which are not confounded by differences in swim speed, revealed that mutant Tg+ and wild-type controls did not differ significantly [G, F(1, 98) = 2.5, P = 0.12; T, (F(9, 882) = 59.3, P = 0.001; G×T, F(9, 882) = 1.8, P = 0.07] whereas mutant Tg+ mice did differ significantly from mutant Tg− mice [G, F(1, 94) = 9.2, P = 0.003; T, F(9, 846) = 53.6, P = 0.0001; G×T, F(9, 846) = 3.4, P = 0.0004]. During the postacquisition probe trial, mutant Tg+ mice, unlike mutant Tg− mice, exhibited a significant bias for the target platform location relative to the next-preferred platform location (P = 0.01; Fig. 4b). During reversal training, there was a significant Group and Trial effect but not a significant Group-by-Trial interaction [G, F(2, 137) = 7.5, P = 0.0008; T, F(4, 548) = 26.1; P = 0.0001; G×T, F(8, 548) = 1.6, P = 0.13] for both escape latency (Fig. 4a) and distance data (data not shown). Mutant Tg+ mice exhibited significantly longer distances (data not shown) and escape latencies relative to wild-type controls [G, F(1, 98) = 17.1, P = 0.0001; T, F(4, 392) = 24.5, P = 0.0001; G×T, F(4, 392) = 1.9, P = 0.11] but not relative to mutant Tg− mice [G, F(1, 94) = 0.11, P = 0.73; T, F(4, 376) = 12.6, P = 0.0001; G×T, F(4, 376) = 0.79, P = 0.53] (Fig. 4a). During the post-reversal probe trial, mutant Tg+, like mutant Tg− mice, failed to exhibit a significant bias for the new target quadrant relative to the next-preferred platform location (P = 0.54; Fig. 4b).
DISCUSSION
The contribution of linked background genes to the phenotype of mutant mice has been documented (7) as have the significant behavioral differences between inbred mouse strains (6). The 129Sv strain used in the generation of our mutant mice exhibit IP-MF hypoplasia (2) and impaired spatial learning in the Morris water maze (6). In our study, comparison of 129B6(N3) mice, which posses on average 12.5% residual 129Sv-linked genes; 129B6(N9) mice, which posses on average 0.2% residual 129Sv-linked genes; and inbred C57BL/6J mice, which possess no 129Sv-linked genes, revealed the significant contribution of 129Sv background genes to the phenotype. First, mutant 129B6(N3) mice, but not mutant 129B6(N9) mice, exhibited a significant elevation in hippocampal PKCɛ expression relative to wild-type controls. Second, wild-type 129B6(N3) mice exhibited significant IP-MF hypoplasia relative to both inbred C57BL/6J and wild-type 129B6(N9) mice, which is consistent with the 129Sv phenotype (2), and likely contributed to their spatial-reversal learning impairment (16). Conversely, mutant 129B6(N3) mice exhibited IP-MF hyperplasia relative to wild-type controls, which may be secondary to reductions in MARCKS expression during development and may not have been observable in mutant 129B6(N9) mice because of the already near maximal IP-MF limb length. Alternatively, this IP-MF hyperplasia may be secondary to the observed elevation in hippocampal PKCɛ in mutant 129B6(N3) mice. Indeed, PKCɛ is highly expressed in hippocampal mossy fibers (15), and over-expression of PKCɛ significantly enhances NGF-mediated neurite outgrowth in PC12 cells (8). Collectively, these data highlight the significant contribution of background genes to both the wild-type and mutant phenotypes (see also ref. 22) and emphasize the necessity of using backcrossing and rescue strategies.
The effect of the Macs mutation was assessed additionally in two separate mouse strains, backcrossed 129B6(N9) mice as well as a different B6SJL(N1) strain. In both of these strains, the Macs mutation produced a significant spatial learning impairment, as evidenced by longer escape latencies and distances, and a failure to exhibit a bias for the target quadrant during the probe trial. This impairment was most apparent during initial training to the submerged platform and was exhibited more dramatically in B6SJL(N1) mice during reversal training. This impairment cannot be attributed to gross sensorimotor impairments or to deficits in escape motivation or simple associative learning processes because these mutant mice were not impaired on the visible platform task. Furthermore, the swim speeds of mutant and wild-type mice did not differ significantly, indicating motorical competence. Together, these data indicate that a heterozygous Macs mutation produces a selective impairment of associative spatial learning processes. To assess whether the spatial learning impairment resulting from the Macs mutation could be attributed to the reduction in MARCKS expression, we examined whether the Macs mutation produced compensatory alterations in the expression of related genes and whether the impairment could be rescued by transgenic expression of epitope-tagged human MARCKS. Mutant 129B6(N9), unlike 129B6(N3) mice, did not exhibit any alterations in hippocampal PKC isozyme or PKC substrate (GAP-43 and RC3) expression relative to wild-type controls. Additionally, during initial acquisition of the submerged platform, mutant mice expressing the MARCKS transgene exhibited significantly shorter distances relative to mutant mice not expressing the transgene and did not differ significantly from wild-type controls, indicating that MARCKS replacement did reverse the impairment. Although the Macs mutation also impaired spatial-reversal learning in B6SJL(N1) mice, it is of interest that the transgene did not attenuate this impairment, implying that reversal learning, which additionally requires the extinction of the former escape response, is not mediated by reductions in MARCKS expression alone.
To assess whether the impaired spatial learning produced by the Macs mutation was secondary to neurodevelopmental alterations, we assessed potential morphological alterations in hippocampal mossy fibers because they express MARCKS at high levels during development and in the adult (Fig. 1h and refs. 12–14) and because the length of the IP-MF limb is positively correlated with spatial learning performance in inbred mice (17). Neither the IP-MF limb nor the SP-MF limb lengths of mutant 129B6(N9) or B6SJL(N1) mice differed significantly from either wild-type controls or inbred C57BL/6J mice. These data indicate that the observed spatial learning impairment cannot be attributed to gross developmental alterations in IP- or SP-MF morphology. Moreover, expression of the MACS transgene in mutant Tg* mice did not alter IP-MF limb length relative to mutant Tg− mice, suggesting that the spatial learning impairment rescue cannot be attributed to differences in IP-MF limb length. Additionally, our findings provide only limited support for previous data indicating a positive correlation between IP-MF limb length and spatial learning ability (17): (i) Wild-type 129B6(N3) mice exhibited both IP-MF hypoplasia and impaired spatial–reversal learning, and (ii) a significant (df = 26, r = +0.43, P = 0.05, two-tailed test) correlation was observed between IP-MF limb length and platform crossings during the reversal probe trial in 129B6(N9). However, a significant correlation was not observed in B6SJL(N1) mice during the reversal probe trial.
In the Morris water maze, the differences in spatial learning exhibited by different mouse strains have been correlated with differing levels of hippocampal PKC activity (26) and PKC isozyme expression (4), and PKC has been demonstrated to play an integral role in synaptic plasticity (long-term potentiation/depression) and associative information storage processes (24). Furthermore, we recently have observed a differential expression of MARCKS in the hippocampus of adult DBA/2J and C57BL/6J mice (R.K.M., L.M.M., E.K.W., and R.H.L., unpublished data). In the present study, we demonstrate that MARCKS, a primary PKC substrate expressed at high levels in the adult hippocampus, is important for hippocampally mediated spatial learning processes, suggesting that MARCKS may be an important mediator of the neuroplastic actions of PKC. Indeed, PKC phosphorylation of MARCKS results in its translocation from the plasma membrane to the cytosol and prevents it from crosslinking F-actin and binding calmodulin whereas dephosphorylation reassociates MARCKS with the membrane and permits F-actin crosslinking and calmodulin binding (1, 3). This bi-directional modification of actin-membrane cytoskeletal tensile strength mediated by MARCKS phosphorylation, as well as MARCKS aggregation at synaptic sites (10), makes it well suited to mediate local intracellular calcium homeostasis and long term alterations in synaptic structural morphology in response to PKC activation. Therefore, 50% reductions in MARCKS expression may impair long term information storage through a disruption of pre-/postsynaptic intracellular calcium homeostasis, secondary to elevations in free-calmodulin, and/or cytoskeletal stabilization required for synaptic modification.
Acknowledgments
The authors gratefully acknowledge Ms. S. Sweeney, Ms. E. Wees, and Mr. K. Little for performing the quantitative RNase protection assay and Western blot analyses, Ms. N. Tian for her help in preparing the RNase protection assay probes, and Dr. D. Gerendasy for providing the RC3 antibody. This work was supported in part by National Institute of Mental Health Grant RO1 MH59959-01 to R.H.L. and a grant from the Medical Research Council/CIBA-Geigy to R.K.M.
ABBREVIATIONS
- PKC
protein kinase C
- MARCKS
myristoylated alanine-rich C kinase substrate
- MF
mossy fiber
- IP
infrapyramidal
- SP
suprapyramidal
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Aderem A. Cell. 1992;71:713–716. doi: 10.1016/0092-8674(92)90546-o. [DOI] [PubMed] [Google Scholar]
- 2.Barber R P, Vaughn J E, Wimer R E, Wimer C C. J Comp Neurol. 1974;156:417–434. doi: 10.1002/cne.901560404. [DOI] [PubMed] [Google Scholar]
- 3.Blackshear P J. J Biol Chem. 1993;268:1501–1504. [PubMed] [Google Scholar]
- 4.Bowers B J, Christensen S C, Pauley J R, Paylor R, Yuva L, Dunbar S E, Wehner J M. J Neurochem. 1995;64:2737–2746. doi: 10.1046/j.1471-4159.1995.64062737.x. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Crawley J N, Belknap J J, Collins A, Crabbe J C, Frankel W, Henderson N, Hitzemann R J, Maxson S C, Miner L L, Silva A J, et al. Psychopharmacology. 1997;132:107–124. doi: 10.1007/s002130050327. [DOI] [PubMed] [Google Scholar]
- 7.Gerlai R. Trends Neurosci. 1996;19:177–181. doi: 10.1016/s0166-2236(96)20020-7. [DOI] [PubMed] [Google Scholar]
- 8.Hundle B, McMahon T, Dadgar J, Messing R O. J Biol Chem. 1995;270:30134–30140. doi: 10.1074/jbc.270.50.30134. [DOI] [PubMed] [Google Scholar]
- 9.Kempermann G, Kuhn H G, Gage F H. Nature (London) 1997;386:493–495. doi: 10.1038/386493a0. [DOI] [PubMed] [Google Scholar]
- 10.Lu D, Lenox R H, Raizada M K. J Cell Biol. 1998;13:217–227. doi: 10.1083/jcb.142.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNamara R K, Kirkby R D, dePape G E, Corcoran M E. Behav Brain Res. 1992;50:65–175. doi: 10.1016/s0166-4328(05)80298-0. [DOI] [PubMed] [Google Scholar]
- 12.McNamara R K, Lenox R H. J Comp Neurol. 1997;379:2–27. [PubMed] [Google Scholar]
- 13.McNamara R K, Lenox R H. J Comp Neurol. 1998;397:337–356. [PubMed] [Google Scholar]
- 14.Ouimet C C, Wang J K T, Walaas S I, Albert K A, Greengard P. J Neurosci. 1990;10:1683–1698. doi: 10.1523/JNEUROSCI.10-05-01683.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito N, Itouji A, Totani Y, Osawa I, Koide H, Fujisawa N, Ogita K, Tanaka C. Brain Res. 1993;607:241–248. doi: 10.1016/0006-8993(93)91512-q. [DOI] [PubMed] [Google Scholar]
- 16.Schopke R, Wolfer D P, Lipp H-P, Leisinger-Trigona M-C. Hippocampus. 1991;1:315–328. doi: 10.1002/hipo.450010322. [DOI] [PubMed] [Google Scholar]
- 17.Schwegler H, Crusio W E. Behav Brain Res. 1995;67:29–41. doi: 10.1016/0166-4328(95)91998-3. [DOI] [PubMed] [Google Scholar]
- 18.Schwegler H, Lipp H-P. Behav Brain Res. 1983;7:1–38. doi: 10.1016/0166-4328(83)90002-5. [DOI] [PubMed] [Google Scholar]
- 19.Seykora J T, Ravetch J V, Aderem A. Proc Natl Acad Sci USA. 1991;88:2505–2509. doi: 10.1073/pnas.88.6.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stumpo D J, Bock C B, Tuttle J S, Blackshear P J. Proc Natl Acad Sci USA. 1995;92:944–948. doi: 10.1073/pnas.92.4.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swierczynski S L, Siddhanti S R, Tuttle J S, Blackshear P J. Dev Biol. 1996;179:135–147. doi: 10.1006/dbio.1996.0246. [DOI] [PubMed] [Google Scholar]
- 22.Threadgill D W, Dlugosz A A, Hansen L A, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris R C, et al. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- 23.Umberall F, Giselbrecht S, Hellbert K, Fresser F, Bauer B, Gschwendt M, Grunicke H H, Baier G. J Biol Chem. 1997;272:4072–4078. doi: 10.1074/jbc.272.7.4072. [DOI] [PubMed] [Google Scholar]
- 24.Van der Zee E A, Douma B R. Prog Neuropsychopharmacol Biol Psychiatry. 1997;21:379–406. doi: 10.1016/s0278-5846(97)00010-9. [DOI] [PubMed] [Google Scholar]
- 25.Watson D G, Watterson J M, Lenox R H. J Pharmacol Exp Ther. 1998;285:307–316. [PubMed] [Google Scholar]
- 26.Wehner J M, Sleight S, Upchurch M. Brain Res. 1990;523:181–187. doi: 10.1016/0006-8993(90)91485-y. [DOI] [PubMed] [Google Scholar]