Abstract
A large number of metabolites are found in each plant, most of which have not yet been identified. Development of a methodology is required to deal systematically with unknown metabolites, and to elucidate their biological roles in an integrated ‘omics’ framework. Here we report the development of a ‘metabolite annotation’ procedure. The metabolite annotation is a process by which structures and functions are inferred for metabolites. Tomato (Solanum lycopersicum cv. Micro-Tom) was used as a model for this study using LC-FTICR-MS. Collected mass spectral features, together with predicted molecular formulae and putative structures, were provided as metabolite annotations for 869 metabolites. Comparison with public databases suggests that 494 metabolites are novel. A grading system was introduced to describe the evidence supporting the annotations. Based on the comprehensive characterization of tomato fruit metabolites, we identified chemical building blocks that are frequently found in tomato fruit tissues, and predicted novel metabolic pathways for flavonoids and glycoalkaloids. These results demonstrate that metabolite annotation facilitates the systematic analysis of unknown metabolites and biological interpretation of their relationships, which provide a basis for integrating metabolite information into the system-level study of plant biology.
Keywords: LC-FTICR-MS, metabolite annotations, secondary metabolites, Solanum lycopersicum, tomato
Introduction
Large-scale biology studies supported by high-throughput data acquisition technologies require a method to bridge the gap between the data obtained and their biological interpretation. In genomics, without an analytical method to define genes, the nucleotide sequence of a whole genome is merely a series of letters (Ashburner, 2000). Using the process of annotation, by which information about the location and the number of genes and the functions of encoded proteins is inferred, researchers obtain biological meaning from the genome sequence (Stein, 2001). Metabolomics researchers are currently experiencing a similar situation to that which faced early genomics researchers. Recent progress in data acquisition technologies such as chromatography-coupled mass spectrometry has facilitated simultaneous detection and quantification of a large number of metabolite-derived peaks (Hall, 2006). However, the data obtained by high-throughput MS are merely a series of peaks without metabolite assignment. At this stage in metabolomics research, most of the peaks detected using MS cannot be assigned to identified metabolites. Such peaks are labeled as ‘unknown’ and usually are not characterized further. Thus the limited capability for metabolite identification has been one of the major obstacles in metabolomics (Kind and Fiehn, 2006; Wagner et al., 2003).
One approach to overcoming this obstacle is to quantify all detected peaks and compile them as un-annotated variables (Bino et al., 2005; Roessner et al., 2001; Schauer et al., 2005). This approach, non-targeted metabolic profiling, is frequently combined with statistical correlation analysis to hypothesize biological roles for the detected metabolites (Carrari et al., 2006; Schauer et al., 2006).
Another approach to overcoming the obstacle is to create a comprehensive dataset of plant metabolites by compiling various pieces of chemical information as has been done for human metabolites (Smith et al., 2005), and to provide annotations for the metabolites. FTICR-MS is a promising candidate technology to achieve this goal. FTICR-MS measurement provides mass values with very high accuracy and resolution. This technology has been employed for non-targeted analyses of metabolites, and has demonstrated its advantage in detecting differentially expressed metabolites (Aharoni et al., 2002; Murch et al., 2004; Oikawa et al., 2006). However, despite many technical advantages, FTICR-MS has a drawback in that it is incapable of separating isomers that have the same elemental compositions. It has been demonstrated recently that coupling of liquid chromatography to FTICR-MS facilitates the effective separation of isomers (Suzuki et al., 2008). However, a comprehensive metabolite dataset using chromatography-coupled FTICR-MS has not yet been produced.
In the present study, we propose a procedure for metabolite annotation using the data obtained by high-performance LC-FTICR-MS. Tomato (Solanum lycopersicum cv. Micro-Tom) fruit was analyzed as a model plant for two reasons. First, tomato contains a number of secondary metabolites that are not present in other model plants such as Arabidopsis and rice. Second, a tomato genome sequencing project is currently underway (Mueller et al., 2005) that will allow interpretation of metabolite data in conjunction with annotated gene functions.
Tomato metabolite data were collected in a non-targeted manner. We then compiled a dataset comprised of mass spectral features including retention time, UV/visible absorption spectrum, m/z value, m/z value of the MS/MS fragment, and relative intensity of the MS/MS fragment. These mass spectral features were attached as annotations to individual metabolites. This information allowed us to provide annotations of predicted molecular formulae for 869 metabolites. Comparison with public databases suggests that 494 of the metabolites are novel. Additionally, MS/MS fragmentation profile data allowed provision of annotations for a number of secondary metabolites with known chemical structures. We constructed a web-based database compiling the metabolite annotations (http://webs2.kazusa.or.jp/komics/). Based on comprehensive characterization of tomato fruit metabolites, we identified chemical building blocks that appear frequently in the tomato fruit tissues. We also assigned several unknown flavonoids and glycoalkaloids to novel metabolic pathways based on the annotations of putative structures. These results demonstrate that metabolite annotation allows us to systematically analyze unknown metabolites and facilitates biological interpretation of their roles in metabolic processes.
Results
Procedure of metabolite annotation
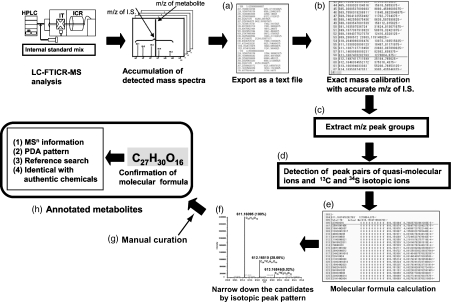
We developed a procedure to organize MS data in a metabolite-oriented manner, which hereafter is referred to as a metabolite annotation procedure. The procedure comprises eight sequential steps. First, the whole raw data set comprising data from successive mass scans were exported as a text file (Figure 1a). Second, the observed m/z values of mass signals were calibrated with those of internal standards detected in the same scan (Oikawa et al., 2006) (Figure 1b). After internal standard calibration, errors in m/z values decreased to less than 1 ppm (Table S1). Third, we grouped mass signals if the same m/z value was detected in consecutive scans, hereafter referred to as a ‘peak group’ (Figure 1c). An accurate m/z value for each peak group was calculated as the mean of the m/z values for the mass signals with the highest intensities (for details, see Experimental procedures). Fourth, we searched for pairs of peak groups that had m/z intervals (Δ) of 1.0033 and 1.9958 to identify 12C/13C1 isotopic peak pairs and 32S/34S1 isotopic peak pairs, respectively (Figure 1d). A peak group for the quasi-molecular ion accompanied by isotopic peaks was regarded as an individual ‘metabolite’. Fifth, molecular formulae were predicted from the accurate m/z values of the metabolites (Figure 1e). To avoid obtaining obviously unnatural formulae, we surveyed elemental compositions in the DNP database (Dictionary of Natural Products). Although the results for such a survey have been reported previously (Kind and Fiehn, 2007), we checked the maximum element numbers within our mass scan range (50–1500 Da). Our survey demonstrated that 95.65% of the DNP compounds (186 788 compounds in a range 50–1500 Da) consist of C, H, N, O, P and S within the ranges C 1–95, H 1–182, N 0–10, O 1–45, P 0–6 and S 0–5. Thus, we set these as upper limits for elemental compositions in the molecular formula calculations. Sixth, we narrowed down the number of candidate formulae using the relative intensity of the 13C1 and 34S1 isotopic ions (Figure 1f). A particular advantage of LC-FTICR-MS is that the resolution is high enough to separate the 34S1 isotopic ion from the 13C2 isotopic ion. Thus, we could use the relative intensity of the 34S1 isotopic ion as a constraint for the number of sulfur atoms. Seventh, we manually performed the isotopic peak group assignment and in-source fragment peak group assignment (Figure 1g). Assignment of the peak groups composed of adduct ions was also performed manually in this step. After these manual curation processes, metabolites were finally designated as ‘annotated metabolites’. In the eighth step, the mass spectral features (including retention time, m/z value, m/z value of the MS/MS fragment, relative intensity of the MS/MS fragment and UV/visible absorption spectrum) and database search results were attached to each metabolite as annotations (Figure 1h). All of the steps, except the manual curation process, are computerized. The annotated metabolites were classified using an annotation grading system (Figure 2, see Experimental procedures).
Figure 1.
Schematic flow of the metabolite annotation procedure. (a) Raw data acquisition. (b) m/z calibration with internal standards. (c) Extraction of peak groups. (d) Isotopic ion assignment. (e) Molecular formula calculation. (f) Molecular formula screening using the relative intensity of isotopic ions. (g) Manual curation of isotopic, fragment and adduct peak assignment. (h) Provision of metabolite annotations. This procedure aims to identify a putative ‘metabolite’, which is defined as a group of mass signals that are detected in consecutive scans to form a peak group, accompanied by isotopic ions.
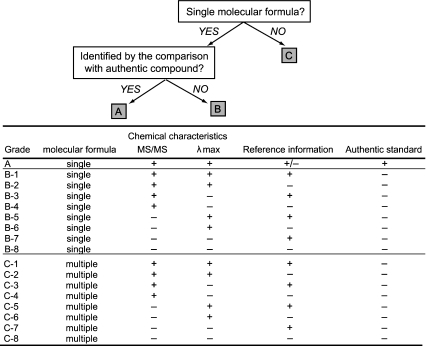
Figure 2.
Annotation grading system. Metabolite annotations were classified according to the evidence that supports the annotations. Grade A consists of metabolites with annotations supported by comparison with authentic compounds. Grade B consists of metabolites with a single molecular formula. Grade C consists of metabolites with multiple molecular formulae. Grades B and C were divided into eight sub-grades according to the availability of MS/MS, λmax and reference information.
Number of annotated metabolites in tomato fruit
We applied the metabolite annotation procedure to the MS data obtained from eight different tomato fruit tissues, comprising peel and flesh at the mature green, breaker, turning and the red stages. The number of detected mass signals ranged from 12 498 to 70 278 (Table 1). On average, 14.0 ± 3.6 mass signals were combined into one peak group. In both positive- and negative-ionization modes, 21 ± 1.7% of the peak groups were consistently assigned with the isotopic ions and recognized as metabolites. After manual curation, 57 ± 7.9% of the metabolites were provided with molecular formula annotations and designated as annotated metabolites. After removing the redundancy across samples, the total number of annotated metabolites was 869 (Table S2).
Table 1.
The numbers of mass signals, peak groups, metabolites and annotated metabolites in tomato fruits
| Annotation grade | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tissues | Ionization mode | Number of mass signalsa | Number of peak groupsa | Number of metabolitesa | Number of annotated metabolites | Total number of annotated metabolites in each tissueb | A | B | C |
| Mature green | |||||||||
| Flesh | Positive | 30 412 ± 3069 | 1470 ± 155 | 306 ± 35 | 154 | 267 | 13 | 146 | 108 |
| Negative | 17 292 ± 1483 | 1673 ± 102 | 305 ± 22 | 167 | |||||
| Peel | Positive | 42 734 ± 5067 | 2311 ± 260 | 479 ± 69 | 228 | 368 | 18 | 184 | 166 |
| Negative | 20 769 ± 2938 | 1925 ± 226 | 397 ± 51 | 228 | |||||
| Breaker | |||||||||
| Flesh | Positive | 28 782 ± 8835 | 1729 ± 271 | 357 ± 96 | 182 | 291 | 15 | 166 | 110 |
| Negative | 15 853 ± 4078 | 1604 ± 311 | 308 ± 66 | 168 | |||||
| Peel | Positive | 43 462 ± 9540 | 2621 ± 379 | 636 ± 119 | 250 | 440 | 23 | 236 | 181 |
| Negative | 32 675 ± 4440 | 2733 ± 376 | 602 ± 85 | 295 | |||||
| Turning | |||||||||
| Flesh | Positive | 24 353 ± 6111 | 1680 ± 58 | 352 ± 26 | 188 | 284 | 15 | 158 | 111 |
| Negative | 12 498 ± 4924 | 1239 ± 460 | 251 ± 134 | 156 | |||||
| Peel | Positive | 63 258 ± 6645 | 3495 ± 348 | 784 ± 112 | 358 | 611 | 26 | 329 | 256 |
| Negative | 39 274 ± 3449 | 3187 ± 364 | 676 ± 79 | 402 | |||||
| Red | |||||||||
| Flesh | Positive | 28 109 ± 1791 | 1700 ± 132 | 353 ± 42 | 179 | 263 | 18 | 147 | 98 |
| Negative | 13 808 ± 4403 | 1444 ± 414 | 266 ± 64 | 144 | |||||
| Peel | Positive | 70 278 ± 3619 | 4305 ± 288 | 1039 ± 77 | 445 | 696 | 29 | 372 | 295 |
| Negative | 55 429 ± 2452 | 4723 ± 301 | 1026 ± 68 | 428 | |||||
Numbers indicate means ± SD of three measurements.
Total numbers of non-redundant annotated metabolites detected in positive- and negative-ionization modes.
Only 3.6% of the metabolites were identified by comparison with authentic compounds (grade A, Table 1). Database searches in the DNP, KNApSAcK (Oikawa et al., 2006), Kyoto Encyclopedia of Genes and Genomes (KEGG) (Goto et al., 2002) and MotoDB (Moco et al., 2006) revealed that 494 of the annotated metabolites were not present in the databases, suggesting that they are novel metabolites.
The complete set of LC-FTICR-MS data and metabolite annotations is accessible at http://webs2.kazusa.or.jp/komics/.
Qualitative analysis of metabolite composition
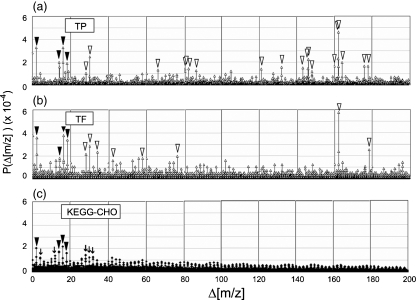
Based on the metabolite annotations (Table S2), we investigated the distribution of mass differences between metabolites. Given that a metabolite is generated from a pre-existing metabolite by substitution of chemical building blocks, mass differences may provide insights into the types of reactions that have occurred between two metabolites. The distribution of Δ[m/z ] values showed ‘spikes’, demonstrating that certain Δ[m/z ] values occurred more frequently than others (Figure 3; the threshold probability to identify Δ[m/z ] spikes was determined as described in Figure S1). The Δ[m/z ] spike profiles seen in tomato fruit samples were different from those of 10 743 compounds containing C, H and O listed in KEGG (Goto et al., 2002) (Figure 3c; for a complete list of the compounds, see Table S3). This demonstrates that the Δ[m/z ] spikes have a sample-specific profile. The Δ[m/z ] spikes that occurred in the tomato samples are listed in Table S4.
Figure 3.
Examples of the distribution of Δ[m/z ] values in the 0–200 Da range at 0.001 Da intervals. Actual calculation of Δ[m/z ] values was performed in the 500 Da range. Δ[m/z ] values were calculated to obtain insights into the chemical building blocks that occur frequently in a set of metabolites. Δ[m/z ] values calculated from m/z values detected in positive-ionization mode from (a) peel at the turning stage (TP) and (b) flesh at the turning stage (TF), and (c) from the theoretical molecular weight of KEGG CHO compounds (KEGG-CHO). Closed arrowheads indicate Δ[m/z ] spikes that were detected in all three sample types (TP, TF and KEGG-CHO). Open arrowheads indicate Δ[m/z ] spikes that were observed specifically in tomato samples TP and TF. Arrows indicate Δ[m/z ] spikes that were observed specifically in KEGG-CHO. P(Δ[m/z ]) indicates the probability of the occurrence of Δ[m/z ] values.
We then checked whether Δ[m/z ] spikes were generated from biologically relevant metabolite pairs, i.e. that Δ[m/z ] values were produced in combinations that reflect reaction relationships. This was achieved by inspecting the MS/MS fragmentation data (available at http://webs2.kazusa.or.jp/komics/). Biologically relevant metabolite pairs were screened according to two criteria. First, the Δ[m/z ] value observed between the metabolites must be observed in more than one pair of MS/MS fragments. Second, metabolite pairs must have more than one identical MS/MS fragment. The relative intensity of the MS/MS fragment ions was not taken into account. For example, Figure 4 shows the MS/MS spectra of a pair of metabolites with m/z values of 1372.5 (Figure 4a) and 1210.5 (Figure 4b), with a Δ[m/z ] value of 162.053 between the fragments. In addition, several common fragments were detected in the MS/MS spectra of these two metabolites. Thus, the pair is regarded as biologically relevant. We manually inspected the MS/MS spectra of all 2722 metabolite pairs that contributed to the formation of Δ[m/z ] spikes, and found that approximately 37% was biologically relevant (Table S4). Further screening for biologically relevant metabolite pairs was performed by inspecting annotations of putative structures and database hits to determine whether occurrence of a Δ[m/z ] value was possible based on knowledge of the biochemical reactions. The Δ[m/z ] values with the highest percentages of relevant metabolite pairs include those corresponding to chemical building blocks C3H7NO2S (121.020), caffeic acid (162.032), hexose (162.053 and 162.054), malonic acid (86.001) and the amino group (17.027) (Table 2). The Δ[m/z ] spike profiles show tissue- and ripening stage-dependent differences (Figure S2). To confirm the ripening stage-dependent changes, Δ[m/z ] values between metabolites in two consecutive stages were analyzed (for details, see Experimental procedures). The analysis indicated that addition of chemical building blocks such as an amino group, caffeic acid, a C3H7NO2S moiety or hexose occurred frequently during ripening. According to the annotations of putative structure and database hits, these chemical building blocks are frequently associated with secondary metabolism.
Figure 4.
An example of the MS/MS spectra comparison to confirm biological relevance of Δ[m/z ] values. MS/MS spectra of metabolite ID 275 (a) and metabolite ID 379 (b). The MS/MS spectral data for metabolite ID 275 and metabolite ID 379 are provided at http://webs2.kazusa.or.jp/komics/. Comparison of (a) and (b) demonstrates that an Δ[m/z ] value between the two metabolites was observed in a pair of MS/MS fragments (m/z 1372.5 and m/z 1210.5), and that there are several MS/MS fragments with identical m/z values suggesting that Δ[m/z ] observed between metabolite ID 275 and ID 379 is biologically relevant.
Table 2.
Biologically relevant Δ[m/z ] spikes estimated by inspection of MS/MS spectra, putative structures and database hits
| MS/MS inspection results | Elemental composition differencec | Putative chemical building blocks | |||
|---|---|---|---|---|---|
| Δ[m/z ] value | Relevant (%) | Not relevant (%) | No MS/MS (%) | Description | Description |
| 121.020 | 97.3 | 0.0 | 2.7 | C3H7NO2S | Addition of C3H7NO2S |
| 456.149a | 93.8 | 0.0 | 6.2 | C17H28O14 | NSd |
| 162.032 | 63.9 | 0.0 | 36.1 | C9H6O3 | Addition of caffeic acid Hydroxylation and addition of coumaric acid |
| 104.048 | 26.7 | 0.0 | 73.3 | C4H8O3 | NSd |
| 143.277 | 76.5 | 2.9 | 20.6 | Addition of C12H33N, and deletion of O3 | NSd |
| 162.053b | 57.9 | 5.8 | 36.3 | C6H10O5 | Addition of hexose Hydroxylation and addition of deoxyhexose |
| 86.001 | 67.9 | 10.3 | 21.8 | C3H2O3 | Addition of malonic acid |
| 162.054b | 60.0 | 9.1 | 30.9 | C6H10O5 | Addition of hexose Hydroxylation and addition of deoxyhexose |
| 456.148a | 50.0 | 9.1 | 40.9 | C17H28O14 | NSd |
| 440.153 | 47.1 | 11.8 | 41.2 | C17H28O13 | NSd |
| 17.027 | 45.2 | 14.3 | 40.5 | H3N | Addition of an amino group |
| 42.011 | 33.3 | 14.8 | 51.9 | C2H2O | NSd |
Assigned to the same elemental composition, respectively.
Assigned to the same elemental composition, respectively.
Elemental composition difference with the highest percentage in all molecular formula combinations.
Not suggested. Known chemical blocks were not suggested by putative structures or database hits.
Secondary metabolites in tomato
In addition to the frequently occurring mass differences, the tomato fruit metabolites analyzed using LC-FTICR-MS include diverse flavonoids and glycoalkaloids. Of the 869 annotated metabolites, 70 and 93 were assigned to the flavonoid and glycoalkaloid groups, respectively. The number of flavonoids increased during ripening (Table S5). In addition, peel tissues contained a larger number of flavonoids than flesh. Four chalcone and flavanone aglycones [naringenin chalcone (NGC), naringenin (NG), eriodictyol (ED) and eriodictyol chalcone (EDC)] and two flavonol aglycones [kaempferol (Kae) and quercetin (Que)] were identified by MS/MS and MS3 fragmentation patterns combined with UV/visible absorption spectra, as reported previously (Bino et al., 2005; Iijima et al., 2008). Dehydrokaempferol glycosides, previously identified in other cultivars of tomato (Le Gall et al., 2003; Moco et al., 2006), were not detected in the Micro-Tom samples.
MS/MS fragmentation patterns of the flavonoids demonstrated the occurrence of various glycosylations and acylations. Flavonoids in the chalcone/flavanone and flavonol groups showed different conjugation patterns. Conjugate moieties of NH3 (m/z 17.027) and C3H7NO2S (m/z 121.020) were associated exclusively with chalcones and flavanones. On the other hand, deoxyhexose, p- coumaroyl hexose and feruloyl hexose were associated exclusively with Kae and Que.
Possible pathway relationships for the flavonoids are illustrated based on the putative structures (Figure 5a). The modification pattern observed in the NGC pathway is quite similar to that in the EDC pathway. Likewise, the modification patterns observed in pathways starting from Kae and Que are similar to each other. The apparent similarities suggest that regulation of modification reactions may be similar between the NGC and EDC pathways and between the Kae and Que pathways. To test this, we investigated flavonoid levels in fruits of transgenic Micro-Tom lines over-expressing PAP1, an Arabidopsis transcription factor that up-regulates flavonoid pathway genes (Borevitz et al., 2000). We focused on comparison of the pairs of NGC and EDC derivatives and the pairs of Kae and Que derivatives, each of which has an identical conjugate moiety (numbered metabolites in Figure 5a). The accumulation levels of three pairs of metabolites in the NGC and EDC pathways changed in a highly correlated manner (correlation coefficient >0.6) in PAP1 over-expressing lines (Figure 5b), as did those of six pairs of metabolites in the Kae and Que pathways (Figure 5c). This suggests that pairs of genes responsible for the same modification reactions are coordinately regulated by the over-expression of PAP1. Alternatively, each pair of modifications may be catalyzed by an identical enzyme.
Figure 5.
Reaction and pathway relationships of Micro-Tom flavanoids. (a) Putative metabolic pathway for the flavonoids. Underlined letters indicate metabolites that were not detected in this study. Solid arrows indicate the occurrence of modification between the detected metabolites. Broken arrows indicate possible reactions between detected and non-detected metabolites. Hex, hexose; dHex, deoxyhexose; Glc, glucose; Rut, rutinose; Pen, pentose. (b, c) Correlations between the relative accumulation levels of (b) chalcone/flavanone metabolites and (c) flavonol metabolites in Arabidopsis PAP1- over-expressing tomato fruits (gray bars) in comparison with control fruit (black bars). Lines: C, control; 1–9, independent lines of PAP1- over-expressing Micro-Tom. Metabolites: numbers indicate the metabolites shown in (a) (highlighted by gray shading). CC, correlation coefficient. Means ± SD of three biological repeats are indicated.
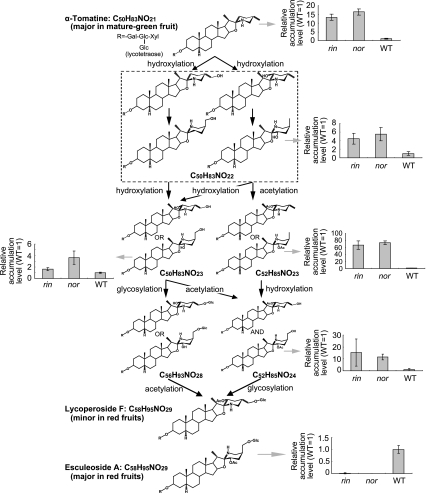
Most of the glycoalkaloids annotated in this study (Table S6) appear to be novel, as they were not found in the literature or public databases. The composition of glycoalkaloids showed tissue-dependent differences. Peel contained a larger number of glycoalkaloids than flesh. The composition of glycoalkaloids also appeared to change with ripening. The intensity of the mass peak of tomatine (m/z 1034.55303 [M+H]+) was high in fruits at the mature green and breaker stages, but very weak at the red stage, suggesting that levels of tomatine decreased during ripening. On the other hand, a number of glycoalkaloids that are larger than tomatine were detected at the red stage. According to MSn data, some of these were assigned as putative intermediate metabolites in the metabolic pathway between tomatine and esculeoside A, the major glycoalkaloid at the red stage (Fujiwara et al., 2004) (Figure 6). To test whether this pathway is regulated by ripening, we investigated the accumulation levels of the intermediates in fruit tissues (containing both peel and flesh) of non-ripening (nor) and ripening-inhibitor (rin) mutants that do not exhibit ripening-associated ethylene production. The levels of metabolites upstream of C52H85NO24 increased in nor and rin fruits in comparison with wild-type Rutgers, but the level of esculeoside A decreased remarkably (Figure 6). This indicates that the final step of esculeoside A biosynthesis is associated with developmentally regulated ripening events.
Figure 6.
Putative metabolic pathway from α-tomatine to esculeoside A. Graphs show the relative abundance of indicated metabolites (gray arrows) in nor and rin mutant fruits (containing both peel and flesh) in comparison with wild-type Rutgers (WT), the background line of the mutants. Means ± SD of three biological repeats are indicated. Esculeoside A was almost absent in the fruits of nor and rin mutants. However, other intermediate glycoalkaloids accumulated at higher levels in nor and rin than WT. The result suggests that the final step of esculeoside A biosynthesis (glycosylation of C52H85NO24) is controlled by developmentally regulated ethylene production.
Discussion
Concept of metabolite annotation
We established a metabolite annotation procedure and constructed a comprehensive metabolite annotation database to organize experimental information obtained by LC-FTICR-MS, using tomato as a model plant species. The term ‘metabolite annotation’ has been proposed previously to describe the process of labeling experiments with biological metadata (such as a description of actual experimental conditions) in order to help unravel the biological role of metabolites based on changes in their levels in response to genetic and environmental perturbation (Fiehn et al., 2005; Scholz and Fiehn, 2007). Their concept of ‘metabolite annotation’ comprises (i) mass spectral annotation and (ii) biological metadata annotation. In this study, we used the term ‘metabolite annotation’ to describe a procedure by which mass spectral information is provided to individual metabolites, thus our annotation procedure can be classified as mass spectral annotation.
The metabolite annotation procedure reported in this study is based on four novel concepts. First, we provided annotations to individual ‘metabolites’. We identified metabolite-representing peaks systematically based on the following criteria: (i) that mass signals were detected in consecutive scans to form a peak group, and (ii) that quasi-molecular ions were accompanied by isotopic ions. Second, we aimed to establish a data-driven annotation protocol for LC-MS-derived data as only a few metabolic profiling methods for LC-MS-derived data have been reported (De Vos et al., 2007; Smith et al., 2006). This is in contrast to the well-established metabolic profiling methods for GC-MS-derived data (Duran et al., 2003; Fiehn et al., 2005; Tikunov et al., 2005). Third, we provided annotations for non-volatile secondary metabolites that are difficult to detect by GC-MS, which allowed us to explore a diverse range of secondary metabolites. Fourth, we introduced a grading system to describe the experimental evidence by which the annotation was supported. It should be mentioned that the metabolite annotations provided in this study are open to future curation. For example, heuristic rules for filtering molecular formulae have been proposed recently (Kind and Fiehn, 2007). In the current study, we implemented procedures equivalent to element number filtering, LEWIS and SENIOR checks, and isotopic pattern filtering, but did not implement element ratio checks or element probability checks. Thus, curation of molecular formula annotations will be feasible by applying these rules.
Limitation in complete coverage and quantification of metabolites
In this study, tomato fruit tissues were extracted using 75% w/v methanol. This method was suitable for extracting a wide range of secondary metabolites, amino acids, sugars, nucleotides and organic acids, but did not extract non-polar metabolites such as lycopene. This demonstrates that the metabolite composition detected is inevitably biased by the choice of extraction method. Thus, an appropriate combination of multiple extraction methods is needed for complete coverage of metabolites.
For comprehensive profiling of the annotated metabolites, quantification depends on the measurement of mass signal intensity. However, differences in the mass signal intensity may be caused by a different degree of ion suppression, a phenomenon by which the intensity of a certain ion is suppressed by the presence of other ions. Even with LC separation prior to MS, several peaks co-eluted in single m/z scans. We performed semi-quantitative analyses of flavonoids and glycoalkaloids based on comparison of the relative mass signal intensities of an identical metabolite across samples (Figures 5b,c and 6). To minimize the possibility that mass signal intensity was affected by different degrees of ion suppression, we checked (i) whether the mass signal intensity is proportional to the UV/visible absorbance, (ii) whether the profile of ions co-eluted with the target ion is similar, and (iii) whether ion suppression is observed in the intensity of co-injected internal calibration standards. Further study is needed to estimate the extent to which ion suppression affects the quantification.
Novel metabolites in tomato fruit
Comparison of 869 annotated metabolites with compounds registered in public databases revealed that 494 of the annotated metabolites appear to be novel. Putative structures for the novel metabolites can be predicted from the annotations of MS/MS fragmentation data. This was particularly effective in predicting putative structures of novel flavonoids and glycoalkaloids. In the flavonoid group, an unknown moiety, C3H7NO2S (m/z 121.020), was found as conjugates with NGC, NG and ED. Its predicted molecular formula matched that of cysteine. It has been reported that cysteine forms a conjugate with epicatechin when procyanidins depolymerize in the presence of cysteine (Torres et al., 2002). However, cysteine conjugates of chalcones and flavanones have not been reported. Structural identification of the moiety will be required to understand the biosynthesis of C3H7NO2S conjugates. Modification of flavonoids has been attracting attention as the biological effects of flavonoid conjugates depend on the nature of the conjugate moieties. The tomato flavonoids found in the present study provide an experimental basis to search for novel functional flavonoids, and to elucidate unknown mechanisms of flavonoid modification. In the glycoalkaloid group, our results indicated the presence of novel glycoakaloids with m/z values larger than the maximum molecular mass (1271 Da) of tomato glycoalkaloid reported so far (Ono et al., 2006) (Table S6). Most of these novel glycoalkaloids appeared after the onset of ripening. This suggests that glycoalkaloid metabolism is active during fruit ripening, and that glycoalkaloids play unidentified physiological roles in the ripening fruit.
Carotenoids, another major secondary metabolite group in tomato, were not detected under our experimental conditions. Development of a metabolite annotation method for MS data obtained in atmospheric pressure photo-ionization mode, which efficiently ionizes non-polar metabolites including carotenoids, is currently underway.
Reaction and pathway relationships
Metabolite annotations aid our understanding of mechanisms controlling metabolism from chemical and biological points of view. From a chemical point of view, metabolite annotations provide detailed chemical information for each metabolite, which will serve as a basis for identifying unknown metabolites. From a biological point of view, metabolite annotations provide a basis for elucidating biological relationships between metabolites, such as reaction and pathway relationships.
To obtain insights into reaction relationships between metabolites, we performed mass difference analysis. Several Δ[m/z ] values occur frequently in metabolites from tomato fruit, suggesting that chemical building blocks corresponding to those Δ[m/z ] values appear frequently in tomato fruit metabolites. It should be emphasized that signal intensities were not taken into consideration in this analysis. Thus, when we state that certain Δ[m/z ] values occur frequently, this does not mean that the accumulation levels of these metabolites are high. Nevertheless, mass difference analysis combined with inspection of MS/MS spectra annotations provides an efficient way to study metabolites relating to a reaction of interest.
To understand the metabolic pathway relationships between annotated metabolites, we arranged flavonoids detected in this study into metabolic diagrams (Figure 5a). These demonstrate that the modification patterns between the NGC and EDC pathways and between the Kae and Que pathways, respectively, are similar to each other. When the flavonoid pathway was up-regulated by over-expression of PAP1, changes in the relative accumulation levels of several pairs of metabolites with identical conjugation patterns were highly correlated (Figure 5b,c). This result demonstrates that genes responsible for each pair of modification reactions are coordinately regulated by PAP1. Alternatively, identical enzymes may use both Kae and Que derivatives as substrates, as reported previously for flavonol glycosyltransferases (Jones et al., 2003; Yonekura-Sakakibara et al., 2007). For glycoalkaloids, a biosynthetic pathway from tomatine to esculeoside A (Fujiwara et al., 2004) was illustrated (Figure 6). By analyzing fruits of nor and rin mutants, we have demonstrated that the reaction step between C52H85NO24 and esculeoside A is regulated by the occurrence of ripening, which is developmentally controlled by NOR and LeMADS-RIN (Giovannoni, 2004). These results demonstrate that the metabolite annotation procedure is a powerful approach for producing hypotheses with respect to unknown metabolic pathways.
Possible link between metabolite annotations and integrated ‘omics’ study
Further insights into the regulation of metabolite biosynthesis will be obtained by the integration of metabolomics data with other ‘omics’ data. A parallel analysis of metabolites and transcripts is a promising approach to achieve this goal (Hirai et al., 2004; Nikiforova et al., 2005; Tohge et al., 2005; Urbanczyk-Wochniak et al., 2003). Another promising approach involves combination of metabolite analysis with genetic analysis such as quantitative trait loci (QTL) analysis (Keurentjes et al., 2006; Morreel et al., 2006; Schauer et al., 2006). In such approaches, the metabolite annotation plays a complementary role to the metabolic profiling in linking metabolite information to other ‘omics’ information. By contrast to quantitative metabolic profiling, annotations of mass spectral features facilitate qualitative characterization with respect to identity, structural similarity and biochemical relationships between the metabolites. This assists in inference of biological meanings from metabolic profiling combined with other ‘omics’ data. Additionally, new metabolites predicted by the metabolite annotations will be included in multi-‘omics’ pathway tools (Thimm et al., 2004; Tokimatsu et al., 2005; Zhang et al., 2005), and expand our knowledge about unknown metabolic pathways. Metabolite annotations provide firm foundations for integrating chemical information regarding metabolites into a system-level study of plant metabolism.
Experimental procedures
Plant materials
Seeds of cultivated tomato (S. lycopersicum cv. Micro-Tom) were sown in pots (500 ml) filled with a mixture of vermiculite and Powersoil (mix ratio 1:1, Kureha Chemical Industries, http://www.kureha.co.jp/ and Kanto Hiryou Industries, http://www.okumurashoji.co.jp/). Until germination, seeds were covered with plastic film and kept in the dark at 25°C. After 4 days in the dark, they were grown with a photoperiod of 16 h light (80 μmol m−2 s−1)/8 h dark at 25°C. Hyponex® (Hyponex Ltd, http://www.scotts.com/) at 1000-fold dilution was applied to plants once a week. Fruits at the mature green (G, approximately 30 days after anthesis), breaker (B, approximately 35 days after anthesis), turning (T, approximately 38–40 days after anthesis) and red (R, approximately 45–48 days after anthesis) stages were harvested. A vector construct expressing Arabidopsis PAP1 under the control of the CaMV 35S promoter (Tohge et al., 2005) was provided by K. Saito (Chiba University, Japan). Transformation of Micro-Tom was performed according to the protocol reported previously (Sun et al., 2006). Seeds of wild-type Rutgers (LA1090) and the nor (LA3013) and rin (LA3012) mutants were obtained from the C.M. Rick Tomato Genetic Resource Center (University of California, Davis, CA, USA).
Metabolite extraction
The peel and the flesh of tomato fruit were separated using a razor blade. Each sample was sliced, immediately frozen in liquid nitrogen and ground to powder using a Shake Master homogenizer (Biomedical Science, http://www.bmsci.com). Powdered samples (50–70 mg) were extracted with three volumes of methanol containing formononetin (20 μg ml−1) as an internal standard. After homogenization using a Mixer Mill MM 300 (Qiagen, http://www.qiagen.com/) at 27 Hz for 2 min twice, homogenates were centrifuged (12 000 g, 10 min, 4°C). The supernatant was filtered through 0.2 μm PVDF membrane (Whatman, http://www.whatman.com), and the filtrate was used for LC-FTICR-MS analysis.
LC-FTICR-MS analysis
An Agilent 1100 system (Agilent, http://www.agilent.com) coupled to a Finnigan LTQ-FT (Thermo Fisher Scientific; http://www.thermofisher.com) was used for LC-FTICR-MS analysis. The data were acquired and browsed using Xcalibur software version 2.0 (Thermo Fisher Scientific). Methanol extract was applied to a TSKgel column ODS-100V (4.6 × 250 mm, 5 μm; TOSOH Corporation, http://www.tosoh.com). Water (HPLC grade; solvent A) and acetonitrile (HPLC grade; solvent B) were used as the mobile phase with 0.1% v/v formic acid added to both solvents. The gradient program was as follows: 10% B to 50% B (50 min), 50% B to 90% B (20 min), 90% B (5 min) and 10% B (10 min). The flow rate was set to 0.5 ml min−1, and the column oven temperature was set at 40°C; 20 μl of each sample were injected.
To monitor HPLC elution, a photodiode array detector was used in the wavelength range 200–650 nm. The ESI setting was as follows: spray voltage 4.0 kV and capillary temperature 300°C for both positive- and negative-ionization modes. Nitrogen sheath gas and auxiliary gas were set at 40 and 15 arbitrary units, respectively. A full MS scan with internal standards was performed in the m/z range 100–1500 at a resolution of 100 000 (at m/z 400).
A mixture of internal calibration standards dissolved in 50% v/v acetonitrile was introduced by a post-column method at a flow rate of 20 μl min−1. The concentration of each standard in the mixture was as follows: for positive mode,: 10 μm lidocaine (m/z 235.18049 [M+H]+; Sigma-Aldrich, http://www.sigmaaldrich.com/), 5 μm prochloraz (m/z 376.03809 [M+H]+; AccuStandard Inc., http://www.accustandard.com), 1.2 μm reserpine (m/z 609.28066 [M+2H]2+; Sigma-Aldrich), 0.8 μm bombesin (m/z 810.41481 [M+H]+; Sigma-Aldrich), 0.4 μm aureobasidin A (m/z 1123.67778 [M+Na]+; Takara Bio Inc., http://www.takara-bio.com), 22 μm vancomycin (m/z 1448.43747 [M+H]+; MP Biomedicals Inc., http://www.mpbio.com); for negative mode: 11.2 μm 2,4-dichlorophenoxyacetic acid (m/z 218.96212 [M-H]−; Sigma-Aldrich), 3.1 μm ampicillin (m/z 348.10235 [M-H]−, Sigma-Aldrich), 0.25 μm CHAPS (m/z: 659.39468 [M+HCOO]−; Sigma-Aldrich), 1.0 μm tetra-N- acetylchitotetraose (m/z 875.32626 [M+HCOO]−; Toronto Research Chemicals, Inc., http://www.trc-canada.com), 0.6 μm aureobasidin A (m/z 1145.68676 [M+HCOO]−, Takara Bio Inc.). MS/MS and MS3 fragmentation were carried out at a normalized collision energy of 35.0% and a isolation width of 4.0 (m/z), and were obtained by ion trap mode. Relative accumulation levels of flavonoids and glycoalkaloids were estimated by dividing the peak area of the metabolite by that of internal standard (formononetin).
Chemicals
Authentic naringenin chalcone was generously provided by the Kikkoman Corporation (http://www.kikkoman.com). Esculeosides A and B were kindly provided by T. Nohara and Y. Fujiwara (Kumamoto University, Japan). Other authentic compounds were purchased from EXTRASYNTHESE (http://www.extrasynthese.com), Funakoshi Co. Ltd (http://www.funakoshi.co.jp), Sigma-Aldrich, Tokyo Chemical Industry (http://www.tci-asiapacific.com) and Wako Pure Chemical Industries Ltd (http://www.wako-chem.co.jp/).
Metabolite annotation procedure
A program written in Microsoft VC++ was used to export the raw data (XRAW) file of each single run as a text file. The output file includes retention time, scan number, m/z value and their intensities. To discriminate mass signals from baseline noise, mass signals whose intensities were more than three times the baseline level of each scan were selected. Next, m/z values of all ions in each scan were bulk-calibrated with observed m/z values of internal calibration compounds in the same scan using the computational tool DrDMASS (http://kanaya.naist.ac.jp/DrDMASS/, Oikawa et al., 2006). By using internally calibrated m/z, if them/z were obtained in more than 30% of the total mass scans, those mass signals could be regarded as artificial noise and thus excluded from further analyses. After removing noise, all data were collected as a Microsoft Excel file. The quasi-molecular ions detected with a 13C isotopic ion in the scan at an m/z value that was 1.003 greater were selected. After sorting mass signals by scan number, those detected in more than three consecutive scans were selected and grouped. If a peak group consisted of three or four mass signals, an accurate m/z value for the group was obtained as the mean m/z value for the three or four mass signals. If a peak group consisted of five or more mass signals, an accurate m/z value was obtained as the mean m/z value for the five most intense signals. For the peak group whose intensity was more than 1 000 000, m/z values for the highest intensity signals were not used for the mean value calculation. Instead, a mean value was calculated using the m/z values of mass peaks whose intensities were just below 1 000 000. Molecular formulae that matched a given accurate m/z value were determined as follows. A library of molecular formulae with all possible elemental combinations whose theoretical m/z matched the input m/z with 1 ppm tolerance was generated using elements C, H, N, O, P and S. To screen the library for chemically possible molecular formulae, all formulae were tested for whether they met following criteria (Senior, 1951): (i) the sum of valences is an even number, and (ii) the sum of valences is greater than or equal to twice the number of atoms minus 1. The accurate m/z was used for molecular formula calculation. Upper limits of 95 for C, 182 for H, 10 for N, 45 for O, 6 for P and 5 for S were used for calculation of formulae. In addition, the relative intensity of the 13C1 isotopic ion was calculated. The number of carbons in the molecular formula was estimated using the following equation:
where n represents the number of carbons. The tolerance for relative intensity was set at 5%. Chemically possible molecular formulae and the relative intensities of the isotope ions were calculated by programs written in Java. The library of molecular formulae was constructed using MySQL. A Java program was developed to search the molecular formula library for molecular formulae matching the criteria described above. Any peak group that is selected based on these criteria is defined as a metabolite. The analysis was repeated three times for each tomato fruit tissue. When a metabolite was detected in two or more repeats, it was regarded as ‘present’ in that tissue. Computational assignment of peak groups of isotopic ions to the parental metabolite was re-checked manually. Assignments of fragment ions and adduct ions to the parental metabolite were performed manually. Peak groups composed of adduct ions produced during ionization were assigned using two criteria as follows. First, it was checked whether the m/z values of ions matched theoretical m/z values of adducts ([M+Na]+, [M+K]+, [M+NH3+H]+, [M+CH3CN+H]+ (Svatos et al., 2004) and [2M+H]+). Second, retention time was checked to determine whether the adduct ions co-eluted with the proton adduct ion. In negative-ionization ESI mode, formic acid adduct ions ([M+HCOO]−) were frequently produced together with [M-H]− ions, and were assigned using the same criteria. Metabolite annotations were provided for the adduct ion species with the highest intensity, i.e. [M+H]+ and [M-H]− in positive- and negative-ionization ESI modes, respectively, for the majority of the metabolites detected in the present study (Table S2). After these manual curation processes, metabolites were designated as ‘annotated metabolites’.
Database construction
For database construction, a dataset comprised of accurate m/z values, predicted molecular formula, retention time, MS/MS data and λmax of the UV/visible absorption spectra was compiled. As MS/MS data, the m/z value, raw intensity and relative intensity of the 20 highest-intensity MS/MS fragment ions were retrieved. References for each annotated metabolite were searched for in the public databases PubChem (http://pubchem.ncbi.nlm.nih.gov/), the Dictionary of Natural Product (http://www.chemnetbase.com/scripts/dnpweb.exe?welcome-main), KNApSAcK (http://kanaya.naist.jp/KNApSAcK/), KEGG (http://www.genome.jp/kegg/kegg2.html) and MotoDB (http://appliedbioinformatics.wur.nl/moto/). To browse and search the annotation information, a web-based database (http://webs2.kazusa.or.jp/komics/) was constructed using MySQL and PHP.
Annotation grading system
To each metabolite, an annotation grade was added to describe the evidence supporting the annotations for that metabolite (Figure 2). First, annotations were classified into two grades (A/B versus C) according to whether a single molecular formula was obtained or not. Grades A and B were further classified according to whether the mass spectral attributes of the metabolites matched those of standard chemicals or not. In grade A, annotations were verified by comparison with standard chemicals. In grade B, annotations were assigned with single molecular formulae but lacked verification by standard chemicals. Annotations in grade B were classified into eight sub-grades according to the availability of MS/MS, λmax and reference information. In grade C, multiple molecular formulae were assigned to each metabolite. Annotations in grade C were classified into eight sub-grades according to the availability of MS/MS and λmax information.
Mass difference analysis
Mass difference values (Δ[m/z ]) were calculated for pairwise combinations of m/z values shown in Table S2 at the 0.001 Da interval. Δ[m/z ] values were calculated separately for m/z datasets of tomato tissue samples and for m/z datasets obtained in positive- and negative-ionization ESI modes. Δ[m/z ] values were calculated in the 500 Da range. To identify Δ[m/z ] values that occurred more frequently than others, a threshold probability was determined based on the standard deviation of the probability distribution within each sample. Probabilities of 10-, 20-, 30-, 40-, 50-, 60- and 70-fold standard deviation levels were tested, and the 40-fold standard deviation level was used as the threshold (Figure S1). MS/MS data inspection was performed manually using m/z values for the 20 fragment ions with highest intensity. To match MS/MS fragments between a pair of metabolites, the m/z tolerance was set to 0.1% as MS/MS spectra were obtained by the ion-trap mode, which is less accurate than the FTICR mode. A pairwise difference in elemental composition was calculated based on the molecular formula annotation provided in Table S2.Δ[m/z ] spikes between stages were identified using following criteria: (i) the probability was above the 40-fold standard deviation level, (ii) the frequency of the Δ[m/z ] value increased in the later stages, and (iii) the probability of the Δ[m/z ] value increased in the later stages. To obtain the chemical information of KEGG compounds, compound files were first retrieved from the KEGG ftp site (ftp://ftp.genome.jp/pub/kegg/ligand/compound/, 9 March 2007), and then compounds containing C, H and O in the molecular formula were selected. Finally, compounds with a non-redundant compound ID were chosen. The theoretical molecular weight of KEGG compounds were calculated using accurate masses of the elements C, H, N, O, P and S. Programs for calculating Δ[m/z ] and elemental composition difference were written in Perl. The program for the selection of KEGG compounds was written in Java.
Acknowledgments
We thank Toshihiro Nohara and Yukio Fujiwara (Kumamoto University) and Kikkoman Corporation for providing us with authentic compounds, Kazuki Saito (Chiba University) for providing us with the PAP1 vector construct, and the C.M. Rick Tomato Genetics Resource Center (University of California, Davis, USA) for the seeds of nor and rin. We thank Takeshi Ara (Kazusa DNA Research Institute) for critical reading of the manuscript. We also thank Miyuki Inde, Rumiko Itoh, Tsugumi Isozaki and Tsurue Aoyama (Kazusa DNA Research Institute) for technical assistance and plant care. This work was partly supported by the Research and Development Program for New Bio-Industry Initiatives, and by a grant from Kazusa DNA Research Institute.
Supplementary material
(A) The numbers of Δ[m/z] spikes detected at different threshold probability. Δ[m/z] spikes were detected in each of eight tomato fruit samples. The plotted points indicate means and SE of eight samples. (B) Decrease ratio of the number of Δ[m/z] spikes. “Decrease level” is defined as follows: [Decrease level at (n×10)-fold standard deviation threshold] = [The number of spikes detected at (n×10)-fold standard deviation threshold] / [The number of spikes detected at {(n-1)×10}-fold standard deviation threshold]. The arrow indicates an inflection point at the 40-fold standard deviation level. (C) Coefficient of variance of the numbers of Δ[m/z] spikes at different threshold probability. The arrow indicates the maximal point at the 40-fold standard deviation level. The number of spikes identified in each sample decreased with increase of threshold level (Panel A). A decrease-trend curve had an inflection point at 40-fold standard deviation level (Panel B). Additionally, coefficient of variance for the number of spikes showed maximal value at 40-fold standard deviation level, implying that considerable sample-to-sample variation of spike numbers was expected at this threshold (Panel C). Thus, we used a threshold of 40-fold standard deviation values to identify Δ[m/z] spikes.
Tissue-dependent occurrence of Δ[m/z] spikes. The numbers in boxes indicate Δ[m/z] values. (A) Δ[m/z] spikes detected in both peel and flesh samples. (B) Δ[m/z] spikes specifically detected in peel samples. (C) Δ[m/z] spikes detected preferentially in flesh samples. Abbreviations for tissues: GP, mature green peel; BP, breaker peel; TP, turning peel; RP, red peel; GF, mature green flesh; BF, breaker flesh; TF, turning flesh; RF, red flesh. P(Δ[m/z]) indicates probability of the occurrence of Δ[m/z] values.
m/z values for five major metabolites in tomato fruits before and after the internal standard calibration.
Mass spectral data and metabolite annotations of the metabolites in Micro-Tom fruit tissues.
Molecular formulae and molecular weights of KEGG CHO compounds.
Δ[m/z] spikes at the threshold probability of 40-fold standard deviation level.
Flavonoids in Micro-Tom fruit tissues.
Glycoalkaloids in Micro-Tom fruit tissues.
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aharoni A, Ric de Vos CH, Verhoeven HA, Maliepaard CA, Kruppa G, Bino R, Goodenowe DB. Nontargeted metabolome analysis by use of Fourier transform ion cyclotron mass spectrometry. OMICS. 2002;6:217–234. doi: 10.1089/15362310260256882. [DOI] [PubMed] [Google Scholar]
- Ashburner M. A biologist's view of the Drosophila genome annotation assessment project. Genome Res. 2000;10:391–393. doi: 10.1101/gr.10.4.391. [DOI] [PubMed] [Google Scholar]
- Bino RJ, Ric de Vos CH, Lieberman M, Hall RD, Bovy A, Jonker HH, Tikunov Y, Lommen A, Moco S, Levin I. The light-hyperresponsive high pigment-2dg mutation of tomato: alterations in the fruit metabolome. New Phytol. 2005;166:427–438. doi: 10.1111/j.1469-8137.2005.01362.x. [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, et al. Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol. 2006;142:1380–1396. doi: 10.1104/pp.106.088534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos RC, Moco S, Lommen A, Keurentjes JJ, Bino RJ, Hall RD. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007;2:778–791. doi: 10.1038/nprot.2007.95. [DOI] [PubMed] [Google Scholar]
- Duran A, Yang J, Wang L, Sumner LW. Metabolomics spectral formatting, alignment and conversion tools (MSFACTs) Bioinformatics. 2003;19:2283–2293. doi: 10.1093/bioinformatics/btg315. [DOI] [PubMed] [Google Scholar]
- Fiehn O, Wohlgemuth G, Scholz M. Setup and annotation of metabolomic experiments by integrating biological and mass spectrometric metadata. In: Ludäscher B, Raschid L, editors. Data Integration in Life Sciences, Lecture Notes in Computer Science, vol. 3615. Berlin/Heidelberg: Springer-Verlag; 2005. pp. 224–239. [Google Scholar]
- Fujiwara Y, Takaki A, Uehara Y, Ikeda T, Okawa M, Yamauchi K, Ono M, Yoshimitsu H, Nohara T. Tomato steroidal alkaroid glycosides, esculeosides A and B, from ripe fruits. Tetrahedron. 2004;60:4915–4920. [Google Scholar]
- Giovannoni JJ. Genetic regulation of fruit development and ripening. Plant Cell. 2004;16:S170–S180. doi: 10.1105/tpc.019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S, Okuno Y, Hattori M, Nishioka T, Kanehisa M. LIGAND: database of chemical compounds and reactions in biological pathways. Nucleic Acids Res. 2002;30:402–404. doi: 10.1093/nar/30.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. Plant metabolomics: from holistic hope, to hype, to hot topic. New Phytol. 2006;169:453–468. doi: 10.1111/j.1469-8137.2005.01632.x. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y, Suda K, Suzuki T, Aoki K, Shibata D. Metabolite profiling of chalcones and flavanones in tomato fruit. J. Japan. Soc. Hortic. Sci. 2008;77:94–102. [Google Scholar]
- Jones P, Messner B, Nakajima J, Schaffner AR, Saito K. UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 2003;278:43910–43918. doi: 10.1074/jbc.M303523200. [DOI] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LH, Jansen RC, Vreugdenhil D, Koornneef M. The genetics of plant metabolism. Nat. Genet. 2006;38:842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Fiehn O. Seven golden rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall G, DuPont MS, Mellon FA, Davis AL, Collins GJ, Verhoeyen ME, Colquhoun IJ. Characterization and content of flavonoid glycosides in genetically modified tomato (Lycopersicon esculentum) fruits. J. Agric. Food Chem. 2003;51:2438–2446. doi: 10.1021/jf025995e. [DOI] [PubMed] [Google Scholar]
- Moco S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, Vervoort J, de Vos CH. A liquid chromatography–mass spectrometry-based metabolome database for tomato. Plant Physiol. 2006;141:1205–1218. doi: 10.1104/pp.106.078428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Goeminne G, Storme V, et al. Genetical metabolomics of flavonoid biosynthesis in Populus: a case study. Plant J. 2006;47:224–237. doi: 10.1111/j.1365-313X.2006.02786.x. [DOI] [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, et al. The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiol. 2005;138:1310–1317. doi: 10.1104/pp.105.060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murch SJ, Rupasinghe HP, Goodenowe D, Saxena PK. A metabolomic analysis of medicinal diversity in Huang-qin (Scutellaria baicalensis Georgi) genotypes: discovery of novel compounds. Plant Cell Rep. 2004;23:419–425. doi: 10.1007/s00299-004-0862-3. [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Daub CO, Hesse H, Willmitzer L, Hoefgen R. Integrative gene–metabolite network with implemented causality deciphers informational fluxes of sulphur stress response. J. Exp. Bot. 2005;56:1887–1896. doi: 10.1093/jxb/eri179. [DOI] [PubMed] [Google Scholar]
- Oikawa A, Nakamura Y, Ogura T, Kimura A, Suzuki H, Sakurai N, Shinbo Y, Shibata D, Kanaya S, Ohta D. Clarification of pathway specific inhibition by FT-ICR/MS-based metabolic phenotyping studies. Plant Physiol. 2006;142:398–413. doi: 10.1104/pp.106.080317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Takara Y, Egami M, Uranaka K, Yoshimitsu H, Matsushita S, Fujiwara Y, Ikeda T, Nohara T. Steroidal alkaloid glycosides, esculeosides C and D, from the ripe fruit of cherry tomato. Chem. Pharm. Bull. (Tokyo) 2006;54:237–239. doi: 10.1248/cpb.54.237. [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie A. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell. 2001;13:11–29. doi: 10.1105/tpc.13.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Zamir D, Fernie AR. Metabolic profiling of leaves and fruit of wild species tomato: a survey of the Solanum lycopersicum complex. J. Exp. Bot. 2005;56:297–307. doi: 10.1093/jxb/eri057. [DOI] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, et al. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 2006;24:447–454. doi: 10.1038/nbt1192. [DOI] [PubMed] [Google Scholar]
- Scholz M, Fiehn O. SetupX – a public study design database for metabolomic projects. Pacific Symp. Biocomput. 2007;12:169–180. [PubMed] [Google Scholar]
- Senior JK. Partitions and their representative graphs. Am. J. Math. 1951;73:663–689. [Google Scholar]
- Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Stein L. Genome annotation: from sequence to biology. Nat. Rev. Genet. 2001;2:493–503. doi: 10.1038/35080529. [DOI] [PubMed] [Google Scholar]
- Sun HJ, Uchii S, Watanabe S, Ezura H. A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 2006;47:426–431. doi: 10.1093/pcp/pci251. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Sasaki R, Ogata Y, et al. Metabolic profiling of flavonoids in Lotus japonicus using liquid chromatography Fourier transform ion cyclotron resonance mass spectrometry. Phytochemistry. 2008;69:99–111. doi: 10.1016/j.phytochem.2007.06.017. [DOI] [PubMed] [Google Scholar]
- Svatos A, Antonchick A, Schneider B. Determination of brassinosteroids in the sub-femtomolar range using dansyl-3-aminophenylboronate derivatization and electrospray mass spectrometry. Rapid Commun. Mass Spectrom. 2004;18:816–821. doi: 10.1002/rcm.1413. [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Tikunov Y, Lommen A, de Vos CH, Verhoeven HA, Bino RJ, Hall RD, Bovy AG. A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol. 2005;139:1125–1137. doi: 10.1104/pp.105.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- Tokimatsu T, Sakurai N, Suzuki H, Ohta H, Nishitani K, Koyama T, Umezawa T, Misawa N, Saito K, Shibata D. KaPPA-view: a web-based analysis tool for integration of transcript and metabolite data on plant metabolic pathway maps. Plant Physiol. 2005;138:1289–1300. doi: 10.1104/pp.105.060525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres JL, Lozano C, Julia L, Sanchez-Baeza FJ, Anglada JM, Centelles JJ, Cascante M. Cysteinyl-flavan-3-ol conjugates from grape procyanidins. Antioxidant and antiproliferative properties. Bioorg. Med. Chem. 2002;10:2497–2509. doi: 10.1016/s0968-0896(02)00127-x. [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Luedemann A, Kopka J, Selbig J, Roessner-Tunali U, Willmitzer L, Fernie AR. Parallel analysis of transcript and metabolic profiles: a new approach in systems biology. EMBO Rep. 2003;4:989–993. doi: 10.1038/sj.embor.embor944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C, Sefkow M, Kopka J. Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry. 2003;62:887–900. doi: 10.1016/s0031-9422(02)00703-3. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Niida R, Saito K. Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 2007;282:14932–14941. doi: 10.1074/jbc.M611498200. [DOI] [PubMed] [Google Scholar]
- Zhang P, Foerster H, Tissier CP, Mueller L, Paley S, Karp PD, Rhee SY. MetaCyc and AraCyc. Metabolic pathway databases for plant research. Plant Physiol. 2005;138:27–37. doi: 10.1104/pp.105.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The numbers of Δ[m/z] spikes detected at different threshold probability. Δ[m/z] spikes were detected in each of eight tomato fruit samples. The plotted points indicate means and SE of eight samples. (B) Decrease ratio of the number of Δ[m/z] spikes. “Decrease level” is defined as follows: [Decrease level at (n×10)-fold standard deviation threshold] = [The number of spikes detected at (n×10)-fold standard deviation threshold] / [The number of spikes detected at {(n-1)×10}-fold standard deviation threshold]. The arrow indicates an inflection point at the 40-fold standard deviation level. (C) Coefficient of variance of the numbers of Δ[m/z] spikes at different threshold probability. The arrow indicates the maximal point at the 40-fold standard deviation level. The number of spikes identified in each sample decreased with increase of threshold level (Panel A). A decrease-trend curve had an inflection point at 40-fold standard deviation level (Panel B). Additionally, coefficient of variance for the number of spikes showed maximal value at 40-fold standard deviation level, implying that considerable sample-to-sample variation of spike numbers was expected at this threshold (Panel C). Thus, we used a threshold of 40-fold standard deviation values to identify Δ[m/z] spikes.
Tissue-dependent occurrence of Δ[m/z] spikes. The numbers in boxes indicate Δ[m/z] values. (A) Δ[m/z] spikes detected in both peel and flesh samples. (B) Δ[m/z] spikes specifically detected in peel samples. (C) Δ[m/z] spikes detected preferentially in flesh samples. Abbreviations for tissues: GP, mature green peel; BP, breaker peel; TP, turning peel; RP, red peel; GF, mature green flesh; BF, breaker flesh; TF, turning flesh; RF, red flesh. P(Δ[m/z]) indicates probability of the occurrence of Δ[m/z] values.
m/z values for five major metabolites in tomato fruits before and after the internal standard calibration.
Mass spectral data and metabolite annotations of the metabolites in Micro-Tom fruit tissues.
Molecular formulae and molecular weights of KEGG CHO compounds.
Δ[m/z] spikes at the threshold probability of 40-fold standard deviation level.
Flavonoids in Micro-Tom fruit tissues.
Glycoalkaloids in Micro-Tom fruit tissues.