Summary
Although the replication, expression, and maintenance of DNA are well-studied processes, the way that they are coordinated is poorly understood. Here, we report an analysis of the changing association of proteins with chromatin (the chromatin proteome) during progression through interphase of the cell cycle. Sperm nuclei were incubated in Xenopus egg extracts, and chromatin-associated proteins were analyzed by mass spectrometry at different times. Approximately 75% of the proteins varied in abundance on chromatin by more than 15%, suggesting that the chromatin proteome is highly dynamic. Proteins were then assigned to one of 12 different clusters on the basis of their pattern of chromatin association. Each cluster contained functional groups of proteins involved in different nuclear processes related to progression through interphase. We also blocked DNA replication by inhibiting either replication licensing or S phase CDK activity. This revealed an unexpectedly broad system-wide effect on the chromatin proteome, indicating that the response to replication inhibition extends to many other functional modules in addition to the replication machinery. Several proteins that respond to replication inhibition (including nuclear pore proteins) coprecipitated with the Mcm2–7 licensing complex on chromatin, suggesting that Mcm2–7 play a central role in coordinating nuclear structure with DNA replication.
Keywords: DNA, CELLCYCLE
Results and Discussion
Experimental Setup and Data Processing
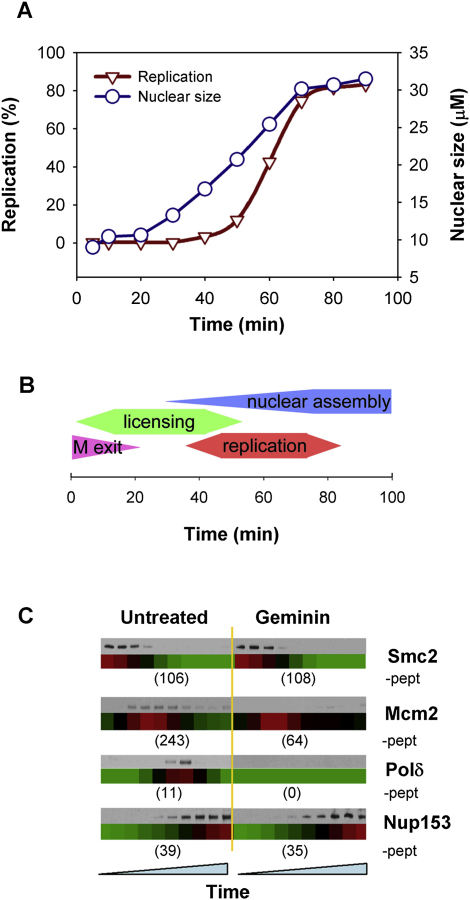
Xenopus extracts were supplemented with demembranated sperm nuclei and simultaneously released from their natural arrest in meiotic metaphase II (Figures 1A and 1B). Over the next 20–30 min, the sperm chromatin decondensed and was licensed for replication; the DNA was then assembled into interphase nuclei and the extracts entered S phase; by 80 min, most of the DNA had been replicated and the extracts entered G2 (Figure 1B) [1]. Chromatin samples, isolated by centrifugation through a sucrose cushion [2], were taken every 10 min. Associated proteins were eluted from chromatin and analyzed by mass spectrometry. At each time point, the abundance of proteins was estimated from the extracted ion chromatograms of their corresponding peptides [3, 4]. The resulting temporal profiles were subjected to smoothing and normalized so that over the time series the maximum abundance of each protein was set to a value of 1 (Figure S1A available online). We identified 606 nonredundant proteins on untreated chromatin, which were subject to further analysis.
Figure 1.
Proteomic Data Acquisition, Manipulation, and Verification
(A) Sperm chromatin was incubated in egg extract and the efficiency of DNA replication was monitored by [α32P] dATP incorporation. The mean size of approximately 20 randomly selected nuclei at each time point is also shown.
(B) Cartoon of cell-cycle events occurring during incubation of sperm nuclei in Xenopus egg extract, showing the approximate timing of mitotic exit, origin licensing (Mcm2–7 loading), nuclear assembly, and DNA replication.
(C) Chromatin was isolated in parallel to the mass-spectrometry samples and immunoblotted for the indicated proteins. Below are corresponding heat maps derived from mass spectrometry (red highest, green lowest, black intermediate). The total number of peptides identified for each protein across the entire time course is also shown.
Protein abundance is presented as a heat map, where red, black, and green indicate high, medium, and low abundance, respectively. Figure 1C shows that there is good agreement between the relative levels of proteins measured by mass spectrometry and standard immunoblotting. Our protocol cannot measure absolute amounts of proteins, or compare levels between different treatments. An approximate comparison of protein levels between experiments can be derived from the number of different peptides detected. For example, geminin reduces the amount of Mcm2 loaded onto DNA as shown by immunoblotting, and although the heat map shows a relatively unchanged pattern, the numbers of Mcm2 peptides detected is greatly reduced (Figure 1C).
Defining Temporal Groups
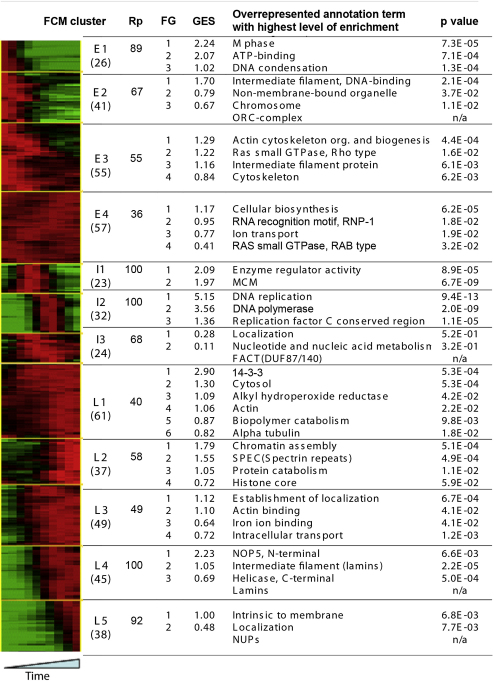
Some proteins showed only small dynamic changes on chromatin during interphase, most of which consisted of ribosomal proteins, chaperonins, and translation elongation factors, which were probably cytoplasmic contaminants. We therefore excluded from further analysis the 148 proteins with less than 15% variation on chromatin. To identify groups of proteins with similar temporal profiles of chromatin-association, we used fuzzy c mean (FCM) soft clustering [5]. Different combinations of cluster number and the noise sensitivity parameter were iteratively tested. The Mcm2–7 proteins (which peak on chromatin prior to entry into S phase) and the replication fork proteins (which peak on chromatin during S phase) could be separated when 12 clusters were used.
Figure 2 shows heat maps for all 458 chromatin proteins showing more than 15% variation in abundance sorted into the 12 FCM clusters. The 12 clusters were divided into three general types that have their peak abundance on chromatin early (E), intermediate (I), or late (L) in interphase. The early group, containing four clusters, were named E1, E2, E3, and E4 to reflect how rapidly their presence on chromatin decreased (E1 fastest, E4 slowest). The second group, where maximum abundance was at intermediate times, was represented by three clusters: I1 (containing the Mcm2–7 licensing proteins), I2 (containing replication fork proteins), and I3 (where maximal abundance was more broadly in the middle of the time course). The third group reached its maximum abundance on replicating chromatin at later times, and its five clusters were named L1–L5, reflecting the order in which they accumulated (L1 earliest, L5 latest). The composition of each FCM cluster is given in Table S3. Although the abundance data were highly reproducible between different runs (Figure S1B), there was some variability in the assignment of proteins to the different FCM groups in three independent experiments. Figure 2 and Figure S1C show that the level of reproducibility of different FCM clusters was approximately proportional to how much the protein abundance changed during the time course. Clusters E1, I1, I2, L4, and L5, where protein abundance dropped to virtually zero at certain times, were highly reproducible, whereas clusters E4 and L1, where protein abundance only ever fell to about 50% of peak values, were much less reproducible.
Figure 2.
FCM Clustering and Functional Annotation Analysis of Temporal Profiles of Polypeptides Associated with Replicating Chromatin
FCM clustering results for 458 polypeptides identified on replicating chromatin that demonstrated more than 15% variation in abundance during interphase are presented as the heat map. The color coding is as follows: green, 0.0; black, 0.5; and red, 1.0. Reproducibility level (column Rp) is defined as a percentage of cases where at least two individual profiles we present in a cluster together with corresponding averaged one. Functional annotation analysis of FCM clusters is demonstrated next to corresponding FCM cluster. The number of proteins separated into each FCM cluster is indicated in brackets. “FG” indicates functional groups identified by DAVID; “GES” indicates group enrichment score.
We next used the DAVID software [6] to collect ontological terms [7] associated with all the proteins. This revealed that all FCM clusters showed significant enrichment with particular sets of ontological terms, which in turn represented a small number of functional groups (Figure 2). The statistical significance (p value) and the group enrichment score (a measure of the significance of all annotation terms in the group) of the terms is also shown.
Functional Overview of Chromatin-Bound Polypeptides
Figure 2 suggests that each FCM cluster contains several groups of functionally related proteins that associate with and/or dissociate from chromatin with similar dynamics during cell-cycle progression. Many of the groups associate with chromatin as expected from their known function. The early E1 cluster contains “M phase” and “DNA condensation” functional groups that play roles in late mitotic events. The E2 cluster, which falls slightly slower than E1, contains a series of proteins related to chromosome structure and metabolism (E2FG1–E2FG3), as well as all essential members of the Origin Recognition Complex (ORC) that marks origins of replication [8–10]. Clusters E3 and E4 contain “Ras small GTPase, Rho type” (E3FG2) and “RAB” (E4FG4) functional groups consistent with the known role of RanGTP in nuclear pore and nuclear envelope formation [11]. The I1 cluster contains all of the Mcm2–7 proteins (functional group “MCM”), which are loaded onto chromatin after ORC and are displaced from DNA as it replicates [12–14]. The I2 cluster, which peaks during S phase, contains known replication fork proteins (“DNA replication,” “DNA polymerase,” and “RF-C”). The I3 cluster, which remained on chromatin slightly longer than the classical replication fork proteins in the I2 group, contains functional groups associated with nucleic acid metabolism that are likely to be involved in the processing of newly replicated DNA. Chromatin assembly factors and variants of histone core proteins are found in late cluster L2, as expected from the doubling of DNA content during S phase. Nuclear pore proteins, nuclear envelope proteins, and the nuclear lamins that show increasing association with chromatin throughout interphase were in the L4 and L5 clusters. Functional groups involved in intracellular transport, including nucleocytoplasmic transport, are present in late FCM cluster L3. We also identified proteins that are not classically associated with chromatin but whose temporal profiles suggested specific roles in the building of chromatin and nuclei. These include proteins associated with intermediate filaments (E2, E3, and L1), actins (E3, L1, and L3), 14-3-3 proteins (L1), and protein catabolism (L1 and L2).
Use of Geminin and Roscovitine to Inhibit Replication
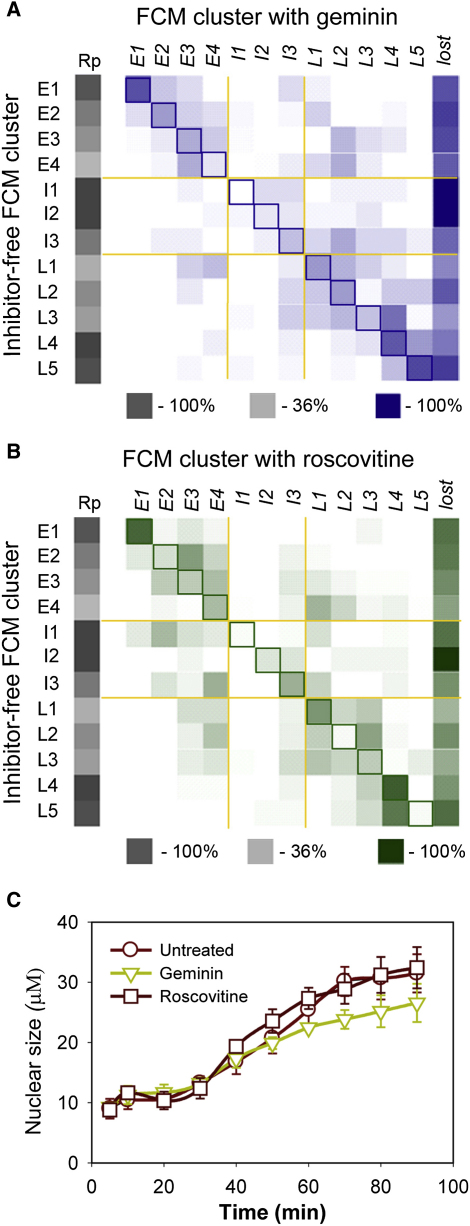
We next investigated how the inhibition of DNA replication changed the chromatin proteome. We used two different replication inhibitors. Geminin blocks the licensing of replication origins by preventing the Cdt1-mediated loading of Mcm2–7 onto chromatin [15, 16]. Roscovitine is a CDK inhibitor that blocks replication initiation at licensed origins [17, 18]. Figures 3A and 3B show how each protein identified in the replicating sample (y axis) was redistributed into temporal FCM clusters after inhibitor treatment (x axis). The highlighted diagonal represents no change to the protein behavior. Levels of cluster reproducibility on replicating chromatin are indicated in shades of gray. Inhibition of replication with either drug had a system-wide effect on the chromatin proteome, and only 15%–20% of proteins remained unaffected. Although all clusters were affected by replication inhibition, the clusters that were most affected were ones that normally showed the greatest changes in abundance during S phase, particularly the intermediate clusters (I1–I3), which are largely composed of replication proteins, but also the E4, L2, L3, and L5 clusters.
Figure 3.
System-wide Effect of Replication Inhibitors on the Chromatin Proteome
Sperm nuclei were incubated in either untreated extracts or extracts supplemented with geminin or roscovitine.
(A and B) Chromatin proteins were identified and sorted into the 12 FCM clusters. For each protein, the cluster membership was compared between the untreated sample and in the presence of either geminin (A) or roscovitine (B). Column Rp presents reproducibility level. Proteins falling along the highlighted diagonal did not change cluster membership on addition of the inhibitor. Proteins that were present in the replicating chromatin but were not detected on chromatin after addition of inhibitors are recorded in the “lost” column.
(C) Effect of replication inhibitors on nuclear size. Error bars indicate the standard deviation (SD).
Approximately 30% of the members of each FCM cluster were not detected on chromatin at all after drug treatment (“lost” column in Figures 3A and 3B). Because we have not sampled the complete set of chromatin-associated proteins, some proteins may be lost because of a random sampling problem [19]. However, in cases where a large proportion of proteins in a given cluster are lost (such as in the intermediate clusters), this is likely to be a genuine effect of the treatment.
Hierarchical Clustering in the Entire Dataset
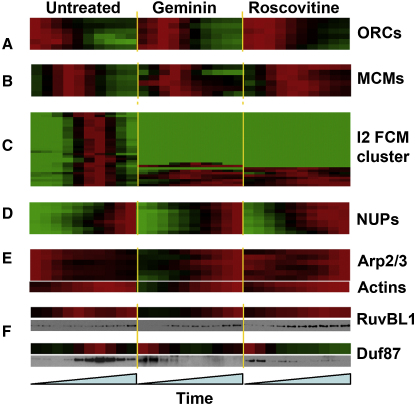
Hierarchical clustering [20] was then performed with a combined dataset from the inhibitor-free, geminin, and roscovitine experiments (Figure 4 and Figures S2 and S4). With the combined dataset, many proteins known to act together in protein complexes cluster next to each other on the same branch of the hierarchical tree (Figure 4). Examples of such clustering are the five tightly associated members of the ORC, the six members of the Mcm2–7 complex, a group of interacting nucleoporins (NUPS), and two groups of actin-related proteins. Virtually all of the replication fork proteins in the I2 cluster were lost or were greatly perturbed when replication was inhibited (Figure 4C, Figure S4F).
Figure 4.
Hierarchical Clustering of the Combined Data Set
(A–E) The combined data set was constructed by uniting data from three experimental conditions (inhibitor free, geminin, and roscovitine) for each protein. Selected groups of highly correlated proteins are shown.
(F) Chromatin was isolated in parallel to the mass-spectrometry samples and immunoblotted for DUF87 and RuvBL1. The color coding is as follows: green, 0.0; black, 0.5; and red, 1.0.
We chose two relatively uncharacterized proteins that showed interesting responses by mass spectrometry, DUF87 (part of the FACT chromatin-remodeling complex and previously implicated in Xenopus DNA replication [21]) and RuvBL1 (Pontin; part of the INO80 chromatin-remodeling complex with reported roles in DNA replication and repair [22, 23]) and raised antibodies to them. The antibodies were then used to immunoblot chromatin isolated at different times from extracts optionally treated with geminin or roscovitine. Figure 4F shows that the immunoblotting profiles resembled those obtained by mass spectroscopy, further validating our approach.
Replication Inhibition Has a System-wide Effect on the Chromatin Proteome
Figure 3 shows that inhibition of replication affects many other proteins in addition to the replication proteins of clusters I1, 2, and 3. Both geminin and roscovitine affected late cluster L5, which contains nuclear pore proteins. Closer examination suggested that a relatively uncharacterized protein, ELYS/MEL-28, was a new nuclear pore complex protein, and this was subsequently confirmed [24–26]. Inhibition of Mcm2–7 loading delays nuclear pore complex assembly (Figure 4F) and slows nuclear growth (Figure 3C). This can be explained by Mcm2–7 on chromatin stimulating the association of ELYS/MEL-28 with DNA [25]. In contrast, roscovitine accelerated the association of nucleoporins with chromatin and therefore redistributed them into the L4 cluster (Figure 4D). This may be a consequence of roscovitine preventing the release of Mcm2–7 from DNA, thereby facilitating increased loading of ELYS/MEL-28.
Nuclear actins are involved in many basic nuclear processes, such as transcription and chromatin remodeling [27]. We found the complete Arp2/3 complex (involved in actin-filament nucleation and assembly) together with actin-capping proteins grouped in the early FCM cluster E3 (Figure 2). Proteins in this group are maximally loaded on replicating chromatin at the beginning of the time course and then gradually decreased. Geminin and roscovitine changed this behavior so that no consistent decline in chromatin association occurred (Figure 4E), suggesting that the release of these proteins from chromatin is dependent on DNA replication. Conventional actins were also found as a complex in the late cluster L1 and also were delayed in their chromatin association by geminin (Figure 4E). A similar geminin-induced delay in chromatin loading of tubulins, keratins, and microfilament components (Figures S2 and S4) suggests that building of the entire nuclear structure is affected by replication licensing. Consistent with this idea, geminin (and to a lesser extent roscovitine) also affected the rate of growth of nuclei (Figure 3E).
Some proteins were only detected on chromatin in the presence of roscovitine or geminin (Figure S3). Their absence from untreated chromatin may be a consequence of the random-sampling problem [19] or may represent chromatin recruitment specific for the inhibition of replication. Of potential relevance is the appearance of a “DNA repair” functional group on chromatin treated with geminin and a “phosphorylation” group on chromatin treated with roscovitine (Table S1).
ChiP Analysis of Mcm2- and Mcm3-Interacting Proteins
The Mcm2–7 complex plays a central role in DNA replication and is differentially affected by the two inhibitors we use here. Its loading onto DNA is inhibited by geminin, whereas its activation as an essential component of the replication fork is inhibited by roscovitine. To investigate whether global effects of replication inhibition could be mediated by direct interaction with Mcm2–7, we identified proteins coprecipitating with Mcm2 or Mcm3 on chromatin (Table S2).
As expected, the complete set of Mcm2–7 proteins was the most abundant component of both the Mcm2 and Mcm3 precipitates, as well as the licensing proteins Orc1-5 and Cdc6, plus several replication fork proteins. In addition, other proteins whose abundance on chromatin was affected by replication inhibition were also coprecipitated with Mcm2 and Mcm3. These included UBF1, whose chromatin dynamics closely resemble ORC, the complete Arp2/3 complex together with capping proteins, Plk1, and all members of the chromosome passenger complex (INCENP, Dasra A, Aurora B, and Survivin [28]). A number of nucleoporins, including ELYS/MEL28, were also identified in both Mcm immunoprecipitates. The binding of these proteins to chromatin was delayed by geminin treatment (Figure 4D), suggesting a functional connection to replication licensing. These observations led us perform experiments showing that the interaction between ELYS and Mcm2–7 promotes nuclear pore complex assembly and helps coordinate DNA replication with nuclear assembly [25]. The large number of dynamic changes that occur in response to replication inhibition also support the idea that cell-cycle processes such as nuclear assembly and DNA replication do not occur independently and in isolation but are part of a highly coordinated biological system.
Acknowledgments
This work was supported by Cancer Research UK grants C303/A5434 (G.K. and P.J.G.), C303/A3106 (G.S.), C303/A3135 (J.J.B.), and C303/A7399 (J.J.B.).
Published online: May 29, 2008
Footnotes
Experimental Procedures, four figures, and three are available at http://www.current-biology.com/cgi/content/full/18/11/838/DC1/.
Contributor Information
Jason R. Swedlow, Email: jason@lifesci.dundee.ac.uk.
J. Julian Blow, Email: j.j.blow@dundee.ac.uk.
Supplemental Data
References
- 1.Blow J.J. Control of chromosomal DNA replication in the early Xenopus embryo. EMBO J. 2001;20:3293–3297. doi: 10.1093/emboj/20.13.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murnion M.E., Adams R.R., Callister D.M., Allis C.D., Earnshaw W.C., Swedlow J.R. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 2001;276:26656–26665. doi: 10.1074/jbc.M102288200. [DOI] [PubMed] [Google Scholar]
- 3.Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 4.Schulze W.X., Mann M. A novel proteomic screen for peptide-protein interactions. J. Biol. Chem. 2004;279:10756–10764. doi: 10.1074/jbc.M309909200. [DOI] [PubMed] [Google Scholar]
- 5.Futschik M.E., Carlisle B. Noise-robust soft clustering of gene expression time-course data. J. Bioinform. Comput. Biol. 2005;3:965–988. doi: 10.1142/s0219720005001375. [DOI] [PubMed] [Google Scholar]
- 6.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 7.Khatri P., Draghici S. Ontological analysis of gene expression data: Current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePamphilis M.L. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle. 2005;4:70–79. doi: 10.4161/cc.4.1.1333. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie P.J., Li A., Blow J.J. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 2001;2:15. doi: 10.1186/1471-2091-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prasanth S.G., Mendez J., Prasanth K.V., Stillman B. Dynamics of pre-replication complex proteins during the cell division cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:7–16. doi: 10.1098/rstb.2003.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walther T.C., Askjaer P., Gentzel M., Habermann A., Griffiths G., Wilm M., Mattaj I.W., Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- 12.Blow J.J., Dutta A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiorano D., Lutzmann M., Mechali M. MCM proteins and DNA replication. Curr. Opin. Cell Biol. 2006;18:130–136. doi: 10.1016/j.ceb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Moldovan G.L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Tada S., Li A., Maiorano D., Mechali M., Blow J.J. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wohlschlegel J.A., Dwyer B.T., Dhar S.K., Cvetic C., Walter J.C., Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 17.Luciani M.G., Oehlmann M., Blow J.J. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J. Cell Sci. 2004;117:6019–6030. doi: 10.1242/jcs.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meijer L., Borgne A., Mulner O., Chong J.P., Blow J.J., Inagaki N., Inagaki M., Delcros J.G., Moulinoux J.P. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 1997;243:527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu H., Sadygov R.G., Yates J.R., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 20.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okuhara K., Ohta K., Seo H., Shioda M., Yamada T., Tanaka Y., Dohmae N., Seyama Y., Shibata T., Murofushi H. A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr. Biol. 1999;9:341–350. doi: 10.1016/s0960-9822(99)80160-2. [DOI] [PubMed] [Google Scholar]
- 22.Jin J., Cai Y., Yao T., Gottschalk A.J., Florens L., Swanson S.K., Gutierrez J.L., Coleman M.K., Workman J.L., Mushegian A. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- 23.Shimada K., Oma Y., Schleker T., Kugou K., Ohta K., Harata M., Gasser S.M. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- 24.Franz C., Walczak R., Yavuz S., Santarella R., Gentzel M., Askjaer P., Galy V., Hetzer M., Mattaj I.W., Antonin W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8:165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillespie P.J., Khoudoli G.A., Stewart G., Swedlow J.R., Blow J.J. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication Licensing. Curr. Biol. 2007;17:1657–1662. doi: 10.1016/j.cub.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasala B.A., Orjalo A.V., Shen Z., Briggs S., Forbes D.J. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M., Shen X. Nuclear actin and actin-related proteins in chromatin dynamics. Curr. Opin. Cell Biol. 2007;19:326–330. doi: 10.1016/j.ceb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 28.Ruchaud S., Carmena M., Earnshaw W.C. Chromosomal passengers: Conducting cell division. Nat. Rev. Mol. Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.