Abstract
Integrins play a role in fibroblast growth factor (FGF) signaling through cross-talk with FGF receptors (FGFRs), but the mechanism underlying the cross-talk is unknown. We discovered that FGF1 directly bound to soluble and cell-surface integrin αvβ3 (KD about 1 μm). Antagonists to αvβ3 (monoclonal antibody 7E3 and cyclic RGDfV) blocked this interaction. αvβ3 was the predominant, if not the only, integrin that bound to FGF1, because FGF1 bound only weakly to several β1 integrins tested. We presented evidence that the CYDMKTTC sequence (the specificity loop) within the ligand-binding site of β3 plays a role in FGF1 binding. We found that the integrin-binding site of FGF1 overlaps with the heparin-binding site but is distinct from the FGFR-binding site using docking simulation and mutagenesis. We identified an FGF1 mutant (R50E) that was defective in integrin binding but still bound to heparin and FGFR. R50E was defective in inducing DNA synthesis, cell proliferation, cell migration, and chemotaxis, suggesting that the direct integrin binding to FGF1 is critical for FGF signaling. Nevertheless, R50E induced phosphorylation of FGFR1 and FRS2α and activation of AKT and ERK1/2. These results suggest that the defect in R50E in FGF signaling is not in the initial activation of FGF signaling pathway components, but in the later steps in FGF signaling. We propose that R50E is a useful tool to identify the role of integrins in FGF signaling.
Fibroblast growth factors (FGFs)2 constitute a family of heparin-binding polypeptides involved in the regulation of biological responses such as growth, differentiation, and angiogenesis (1–4). The FGF family currently consists of 22 members with FGF1 (acidic FGF) and FGF2 (basic FGF) the most extensively studied. The biological effects of FGFs are mediated by four structurally related receptor tyrosine kinases designated FGFR1–4. The binding of FGF to its receptor results in receptor dimerization and subsequent autophosphorylation of specific tyrosine residues within the cytoplasmic domain (1–4). Activation of the receptor allows proteins containing Src homology 2 or phosphotyrosine binding domains to bind to sequence recognition motifs in the FGFR, resulting in phosphorylation and activation of these proteins (5). This leads to the activation of intracellular signaling cascades. The main signaling cascade activated through the stimulation of FGFR is the Ras/MAPK pathway.
FGF signaling enhances multiple biological processes that promote tumor progression (6). Therapies targeting FGF receptors and/or FGF signaling not only affect the growth of the tumor cells but also modulate tumor angiogenesis (7). FGF1 and FGF2 are responsible for resistance to chemotherapeutic agents in cancer (8–11) and are also pro-inflammatory growth factors that play a role in pathological angiogenesis in chronic inflammatory diseases (12). Thus FGF signaling is a potential therapeutic target for cancer and pathological angiogenesis in chronic inflammatory diseases.
It has been proposed that cross-talk between integrins and growth factor receptors plays a critical role in growth factor signaling (13), but the specifics of the cross-talk are unclear. Integrins are a family of cell adhesion receptors that recognize extracellular (ECM) ligands and cell-surface ligands (14). Integrins are transmembrane α-β heterodimers, and at least 18 α and 8 β subunits are known (15). Integrins play an important role in anchorage-dependent cell survival and proliferation (16). Integrin-stimulated pathways are very similar to those mediated by growth factor receptors and are intimately coupled with them. Many cellular responses to soluble growth factors, such as epidermal growth factor, platelet-derived growth factor, and thrombin, are dependent upon the adherence of the cell to ECM ligands via integrins. FGF2-induced angiogenesis requires integrin signaling from the ECM (cross-talk between integrins and FGFRs). Indeed mAb against integrin αvβ3 blocks FGF2-induced angiogenesis (17, 18).
It has been reported that substrate-bound FGF2 promotes endothelial cell adhesion by directly interacting with integrin αvβ3 (19) and induces endothelial cell proliferation, motility, and the recruitment of FGFR1 in the cell substrate contact (20). However, because heat-denatured FGF2 still supports integrin binding (20), it is unclear whether this interaction is biologically relevant, or how integrins interact with FGF2. It has also been reported that FGF2 binds to fibrinogen, and this interaction enhances FGF2-mediated endothelial cell proliferation and subsequent co-localization of αvβ3 and FGFR1 (21, 22). However, because FGF1 does not bind to fibrinogen (23), the FGF-integrin cross-talk model, mediated by FGF-fibrinogen binding, cannot be applied to FGF1 and also perhaps not to other members of the FGF family. Thus we hypothesized that there may be an alternative model by which FGF1 might be able to cross-talk with the integrin signaling pathway.
In this paper, we show that integrin αvβ3 directly and specifically binds to FGF1. We found that the integrin-binding site overlaps with the heparin-binding site by using docking simulation and mutagenesis. One integrin-binding defective mutant (R50E) showed markedly reduced ability to induce cell proliferation and migration, whereas R50E induced FGFR1 phosphorylation, FRS2α phosphorylation, and AKT and ERK1/2 activation. We propose that the direct binding of integrins to FGF is a mechanism of integrin-FGFR cross-talk.
EXPERIMENTAL PROCEDURES
Materials
Recombinant soluble αvβ3 was synthesized in CHO K1 cells using the soluble αv and β3 expression constructs provided by Tim Springer (Center for Blood Research, Boston) and purified by nickel-nitrilotriacetic acid affinity chromatography as described (24). K562 erythroleukemia cells that express human integrin α1(α1-K562) were provided by Roy Lobb (Biogen, Cambridge, MA). K562 cells that express human αvβ3(αvβ3-K562) (25) were provided by Eric Brown (University of California, San Francisco). K562 cells that express human α2, α3, or α4 have been described (26). K562 cells expressing human integrin α6(α6-K562 cells) were generated as described for other K562 transfectants. Briefly, we transfected human α6 cDNA in pBJ-neo vector (provided by Vito Quaranta) and selected for G418 resistance. Stable transfectants were cloned for high expressers by cell sorting. Chinese hamster ovary (CHO) cells that express WT β1 or the β1–3-1 mutant have been described (27).
Plasmid Construction, Protein Expression, and Purification of the WT and Mutant FGF1
The human FGF1 cDNA was amplified using PCR with human placenta library as a template. A BglII restriction site was introduced at the 5′ end and an EcoRI site at the 3′ end of the cDNA fragment with following primers: 5′-GCAGATCTTTTAATCTGCCTCCAGGGAAT-3′ and 5′-GCGAATTCTTAATCAGAAGAGACTGGCAG-3′. The resulting fragments were digested with BglII and EcoRI and then subcloned into the BamHI/EcoRI sites of the pGEX-2T (Amersham Biosciences) vector. Site-directed mutagenesis was performed using the QuickChange method (28). The presence of the mutations was verified by DNA sequencing. The WT FGF1 and its mutants were expressed in Escherichia coli BL21 (DE3) and purified by glutathione affinity chromatography as described by the manufacturer's instructions (GE Healthcare). To remove endotoxin, GST-FGF1 fusion protein on glutathione-agarose was extensively washed with 1% Triton X-114 in PBS. WT, the R50E mutant, and the 3xA (E101A/Y108A/N109A) FGF1 mutant were further purified using a heparin-Sepharose column (Amersham Biosciences) after removing the GST tag by thrombin. The 4xE (K127E/K128E/K133E/R134E) mutant was purified by gel filtration. The FGF1 preparations were more than 90% homologous in SDS-PAGE.
Synthesis of the FGFR1 D2D3 Fragment
A DNA fragment encoding amino acid residues 140–365 of the immunoglobulin-like D2 and D3 domains of FGFR1 was amplified by PCR with the full-length human FGFR1 cDNA in the pcDNA3 vector (gift from Ann Hanneken, the Scripps Research Institute, La Jolla, CA) as a template. A BamHI restriction site was introduced at the 5′ end and an XhoI site at the 3′ end of the cDNA fragment with the following primers: 5′-GCGGATCCACAGATAACACCAAACCAAACC-3′, 5′-GCCTCAGTCACCTCTCTTCCAGGGCTTCC-3′. The resulting cDNA fragment was subcloned into the BamHI/XhoI sites of the vector pET21a, and transformed into BL21 (DE3). The protein was expressed as an insoluble protein and refolded as described (29). The refolded protein was purified by affinity chromatography on heparin-Sepharose to enrich the properly folded protein. Bound protein was eluted with 1 m NaCl.
Surface Plasmon Resonance (SPR) Analysis
Heparin-FGF Interaction—Biotinylated heparin (Sigma) in Biacore HBS-EP buffer (0.01 m Hepes, pH 7.4, 0.15 m NaCl, 3 mm EDTA, and 0.0005% of surfactant P20) was immobilized by a streptavidin sensor chip (Biacore SA Chip) by injecting at 2 μl/min for 15 min. One response unit difference represents about 1 pg/mm2 of the analytes on the surface matrix of the sensor chip. 5-Fold serially diluted WT, R50E, and 3xA ranging from 800 nm to 51.2 pm and 2-fold serially diluted 4xE ranging from 1.6 μm to 50 nm in HBS-EP were injected at 50 μl/min for 3 min.
Integrin-FGF Interaction—Soluble αvβ3 was immobilized on the CM5 sensor chip using a standard amine coupling procedure (30). The WT and R50E FGF1 were individually 2-fold serially diluted from 8 μm to 125 nm in HBS-P buffer (0.01 m Hepes, pH 7.4, 0.15 m NaCl, and 0.0005% of surfactant P20) with 1 mm of Mn2+, and the 3xA FGF1 was 2-fold serially diluted from 6 μm to 93.75 nm in the same buffer. Samples were injected at 50 μl/min for 1.8 min. The HBS-P buffer with 1 mm of Mn2+ was then injected at 50 μl/min for 3 min to allow the bound FGF1s to dissociate from the integrin.
FGF1-FGFR1 D2D3 Interaction—About 1500 response units of FGFR1 D2D3 was immobilized to a sensor chip CM5 by using a standard amine coupling method. WT or mutant FGF1 was 2-fold serially diluted from 800 to 50 nm in HBS-P buffer containing 1 mm of Mn2+ and injected at a flow rate 30 μl/min for 3 min. The same buffer was injected at the same flow rate for 3 min to measure the dissociation of the bound FGF1 from FGFR1 D2D3.
Docking Simulation
In the AutoDock 3.05 program, the ligand is presently compiled to a maximum size of 1024 atoms. The solvent-exposed Mg2+ octahedral vertex was left empty in the model during docking calculations. Atomic solvation parameters and fractional volumes were assigned to the protein atoms by using the AddSol utility, and grid maps were calculated by using Auto-Grid utility in AutoDock 3.05. A grid map with 127 × 127 × 127 points and a grid point spacing of 0.603 Å included the whole metal ion-dependent adhesive site-containing face of the I-like domain of β3 and the β-propeller domain containing repeats 2–4, which are large enough to accommodate the FGF1 structure. Kollman “united-atom” charges were used. AutoDock 3.05 uses a Lamarckian genetic algorithm (LGA) that couples a typical Darwinian genetic algorithm for global searching with the Solis and Wets algorithm for local searching. The LGA parameters were defined as follows: the initial population of random individuals had a size of 50 individuals; each docking was terminated with a maximum number of 1 × 106 energy evaluations or a maximum number of 27,000 generations, whichever came first; mutation and crossover rates were set at 0.02 and 0.80, respectively. An elitism value of 1 was applied, which ensured that the top ranked individual in the population always survived into the next generation. A maximum of 300 iterations per local search was used. The probability of performing a local search on an individual was 0.06, whereas the maximum number of consecutive successes or failures before doubling or halving the search step size was 4. This set of parameters was used for all dockings.
Integrin Binding Assays
Cell adhesion and soluble integrin binding assays were performed as described previously (24). Native or heat-denatured (70 °C for 10 min) FGF in 0.1 m carbonate buffer, pH 9.4, was incubated in a polystyrene 96-well non-tissue culture plate overnight at 4 °C. Unbound FGF was removed, and 200 μl of 0.1% BSA in PBS was added and incubated for 60 min at room temperature. The wells were washed with PBS, and soluble integrin αvβ3 in 50 μl in Hepes-Tyrode's buffer supplemented with 1 mm Mn2+ was added to the wells and incubated at room temperature for 60 min. Then non-bound soluble integrin was removed by rinsing the wells with the same buffer. Horseradish peroxidase (HRP)-conjugated anti-His tag mouse IgG was added to the wells and incubated for 60 min. Non-bound antibodies were removed by rinsing the wells with the same buffer, and bound integrins were quantified by measuring the absorbance at 450 nm developed from adding the substrate 3,3′,5,5′-tetramethylbenzidine of HRP.
DNA Synthesis
DNA synthesis was measured by BrdUrd incorporation. Balb 3T3 cells were plated on sterile coverslips in 6-well culture plates and serum-starved in DMEM supplemented with 0.4% FCS for 48 h. They were stimulated with 5 ng/ml WT FGF1 and its mutants for 24 h in the presence of 5 μg/ml heparin. BrdUrd (10 μg/ml) was added to the medium for the last 6 h of the incubation. Cells were then fixed with 70% ethanol and incubated with 2 n HCl. After the medium was neutralized with 0.1 m borate buffer (pH 0.5), the cells were incubated with anti-BrdUrd antibody (Pharmingen). BrdUrd-incorporated cells were stained with HRP-conjugated secondary antibody (Bio-Rad) and metal-enhanced 3,3′-diaminobenzidine substrate kit (Pierce). 3,3′-Diaminobenzidine-positive and -negative cells were counted from the digital images of three independent fields.
Proliferation Assay Using BaF3 Cells Expressing the Human FGFR1c Isoform
Mouse pro-B BaF3 cells that express human FGFR1c (BaF3-FR1c, kindly provided by David Ornitz, Washington University, St. Louis) were maintained in a medium containing 0.5 ng/ml IL-3 as described (31). For proliferation assays, cells were maintained with WT or mutant FGF1 instead of IL-3, and cell proliferation was measured by MTS assays.
Western Blot Analysis
NIH3T3 or Balb 3T3 cells were grown to confluence and starved in DMEM supplemented with 0.4% FCS for 24 h before FGF1 treatment. The starved cells were treated with WT or mutant FGF1 (5 ng/ml) in the presence of 5 μg/ml heparin for 10–15 min at 37 °C. Afterward, cells were washed twice with ice-cold PBS, and lysed with the lysis buffer (20 mm Tris-HCl, pH 8.0, 120 mm NaCl, 5 mm EDTA, 0.5% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, 10 mm NaF, 1 mm Na3VO4, 10 μg/ml aprotinin). Protein concentrations in the cell lysates were determined using the Bradford protein assay (Bio-Rad). Equal amounts of cell proteins were analyzed by SDS-PAGE in a 10% polyacrylamide gel and Western blotting. The first antibodies were anti-FGFR1, anti-phospho-FGFR1 (Tyr-653/Tyr-654, BIOSOURCE), anti-FRS2α (Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho-FRS2α (Tyr-196), anti-p44/42 ERK1/2 MAPK, anti-phospho-p44/42 ERK1/2 MAPK (Thr-202/Tyr-204), anti-AKT, or anti-phospho-AKT antibody (Cell Signaling Technology, Inc, Danvers, MA). Bound antibodies were detected with HRP-conjugated anti-mouse or anti-rabbit IgG, and ECL Western blotting detection reagents (Pierce).
In Vitro Wound Healing Assay
Balb 3T3 cells were plated into 6-well cell culture plates. Cells were allowed to grow in DMEM containing 10% FCS overnight, and then cells were washed with serum-free medium and starved for 24 h. A scratch was made across the cell layer using a pipette tip. After washing with serum-free medium twice, DMEM containing 10 ng/ml WT or mutant FGF1 together with 5 μg/ml heparin was added to the cells. The wounded areas were quantified by using ImageJ.
Chemotaxis
A polycarbonate filter of 8 μm pore size of the Transwell insert was coated with 10 μg/ml fibronectin (Sigma) overnight at 4 °C. After washing, the insert was placed into a 24-well cell culture plate, and the lower portion of the plate was filled with 600 μl of serum-free DMEM containing 5 ng/ml WT or mutant FGF1. Balb 3T3 cells (105 cells/filter) were plated on the filter and incubated at 37 °C for 24 h, and cells were visualized by crystal violet staining (0.5% crystal violet in 50 mm borate, pH 9.0, and 2% ethanol). The uncoated side of each filter was wiped with a cotton swab to remove cells that had not migrated through the filter. Chemotaxed cells were counted from the digital images of the stained cells, determining the mean number of cells counted per field. Results are expressed as means ± S.E. of the relative cell number with nonstimulated cells set as 100.
RESULTS
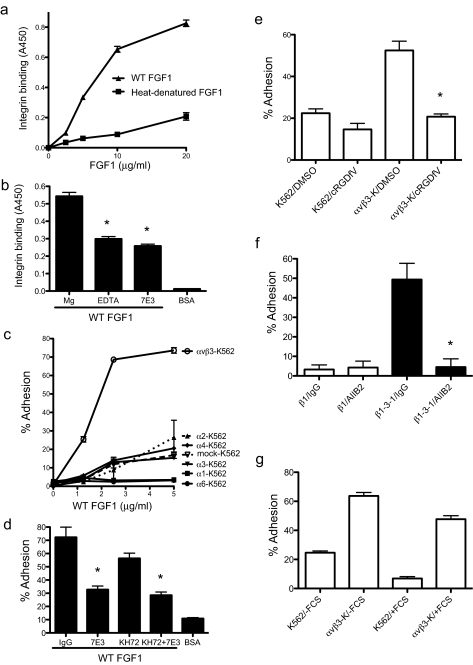
FGF1 Specifically Binds to Integrin αvβ3—We tested whether FGF1 directly interacts with integrins in vitro. We found that recombinant soluble αvβ3 bound to immobilized WT FGF1 in a dose-dependent manner but did not bind to heat-denatured FGF1 in ELISA-type integrin binding assays (Fig. 1a). These results suggest that FGF1 binds to integrin αvβ3, and this interaction requires an intact three-dimensional structure of FGF1. EDTA and mAb 7E3 (anti-β3) blocked the binding of soluble αvβ3 to FGF1, suggesting that binding to FGF1 is cation-dependent and specific to αvβ3 (Fig. 1b). An SPR analysis of the FGF1-αvβ3 interaction showed that FGF1 binds to αvβ3 at a high affinity (see below).
FIGURE 1.
Direct binding of FGF1 to integrin αvβ3. a, recombinant soluble αvβ3 bound to WT FGF1 but not to heat-denatured FGF1. Wells of 96-well microtiter plates were coated with WT or heat-denatured FGF1 at the indicated concentrations, and the remaining protein-binding sites were blocked with BSA. Recombinant soluble αvβ3 was added to the wells and incubated for 1 h at room temperature. Bound αvβ3 was determined by using HRP-conjugated anti-His6 antibody and peroxidase substrate at A450. Data are shown as means ± S.E. of triplicate experiments. b, effects of antibodies and EDTA on the binding of soluble αvβ3 to FGF1. The ELISA-type integrin-binding assay was performed as described above. Soluble αvβ3 was incubated with 10 μg/ml 7E3 or control mouse IgG for 10 min on ice prior to adding to the wells. Concentrations of MgCl2 and EDTA are 10 and 5 mm, respectively. BSA was used as a negative control. Data are shown as means ± S.E. of triplicate experiments. *, p < 0.05, n = 3 by t test. c, adhesion of K562 cells that express different integrins to FGF1. Wells of 96-well microtiter plates were coated with FGF1, and the remaining protein-binding sites were blocked with BSA. Cells were added to the wells and incubated for 1 h at 37 °C in Hepes-Tyrode's buffer containing 1 mm MnCl2, and bound cells were quantified after removing unbound cells by using phosphatase assays. Data are shown as means ± S.E. of triplicate experiments. d, inhibition of cell adhesion to FGF1 by anti-integrin antibodies. Adhesion assay was performed as described in c. Cells were incubated with 10 μg/ml control mouse IgG, mAb 7E3 (anti-β3, function blocking), or KH72 (anti-α5) for 30 min on ice prior to adding to the wells. Data are shown as means ± S.E. of triplicate experiments. *, p < 0.05 to control IgG, n = 3 by t test. e, inhibition of αvβ3-FGF1 interaction by cyclic RGDfV, a specific antagonist to αvβ3. Adhesion assay was performed as described in c. Cyclic RGDfV was used at 10 μm, and DMSO, in which stock cyclic RGDfV was solubilized, was used as a control. *, p = 0.0024 betweenαvβ3-K562 (αvβ3-K)/DMSO andαvβ3-K562/cyclic RGDfV. f, binding of theβ1–3-1 mutant to FGF1. Adhesion assay was performed as described in c. Cells were incubated with 10 μg/ml control mouse IgG or mAb AIIB2 (anti-β1, function blocking). Data are shown as means ± S.E. of triplicate experiments. *, p < 0.05 to DMSO, n = 3 by t test. g, effect of FCS to αvβ3-FGF1 interaction. Adhesion assay was performed as described in c. Assays were performed in the absence and presence of FCS (10%). Data are shown as means ± S.E. of triplicate experiments.
We next tested whether cell-surface αvβ3 binds to FGF1. We found that K562 erythroleukemic cells (α5β1+) expressing exogenous αvβ3(αvβ3-K562, α5β1+/αvβ3+) cells adhered to FGF1, but mock-transfected K562 cells showed only weak adhesion to FGF1 (Fig. 1c). This suggests that FGF1 binds to αvβ3, but not to α5β1, on the cell surface under the current assay conditions. mAb 7E3 blocked, but mAb KH72 (anti-α5) did not significantly block, the adhesion of αvβ3-K562 cells to FGF1 (Fig. 1d). Consistent with this finding, cyclic RGDfV, which is an antagonist specific to αvβ3 (32), blocked the adhesion of αvβ3-K562 cells to FGF1 (Fig. 1e). K562 cells that express several β1 integrins only weakly adhered to FGF1 (Fig. 1c). These results suggest that αvβ3 is a predominant, if not the only, integrin that binds to FGF1 in our assay conditions.
Because FGF1-αvβ3 interaction is blocked by mAb 7E3 that has been mapped in the ligand-binding site of the β3 subunit (33, 34), and blocked by cyclic RGDfV, it is suggested that FGF1 binds to the ligand-binding site of αvβ3. We previously reported that a small disulfide-linked loop in the I-like domain of β3 plays a critical role in determining ligand specificity of αvβ3 (27). To test if FGF1 binding requires the specificity loop of αvβ3, we used a β1 mutant in which a disulfide-linked loop of β1 (the specificity loop, residues 187–193) was swapped with the corresponding amino acid residues of β3 (the CYDMKTTC sequence) (the β1–3-1 mutation) (27). αvβ1–3-1 has been shown to bind to several αvβ3 ligands, including vitronectin, fibrinogen, von Willebrand factor (27, 35), and a viral surface protein (36). Nevertheless function-blocking anti-β1 mAbs such as AIIB2 blocked ligand binding to αvβ1–3-1 (27). We found that CHO cells that express β1–3-1 (β1–3-1-CHO) strongly adhered to FGF1, whereas those expressing WT β1 (β1-CHO) did not (Fig. 1f). Furthermore, anti-β1 mAb AIIB2 effectively blocked the binding of β1–3-1-CHO cells to FGF1. This suggests that the specificity loop is critical for FGF1 binding to αvβ3 and that FGF1 interacts with αvβ3 in a manner similar to those of known αvβ3 ligands (e.g. vitronectin). These results also suggest that αvβ1 and αvβ5 that are expressed in parent CHO and β1-CHO cells do not appear to be important for FGF1 binding.
It is possible that integrin αvβ3 may not interact with FGF1 in the presence of plasma adhesive proteins, because they may compete with FGF for binding to αvβ3. We found that the presence of 10% fetal bovine serum did not suppress αvβ3-specific cell adhesion to FGF1 (Fig. 1g). This consistent with the previous reports that integrin-binding sites in serum-adhesive proteins are cryptic (see “Discussion”).
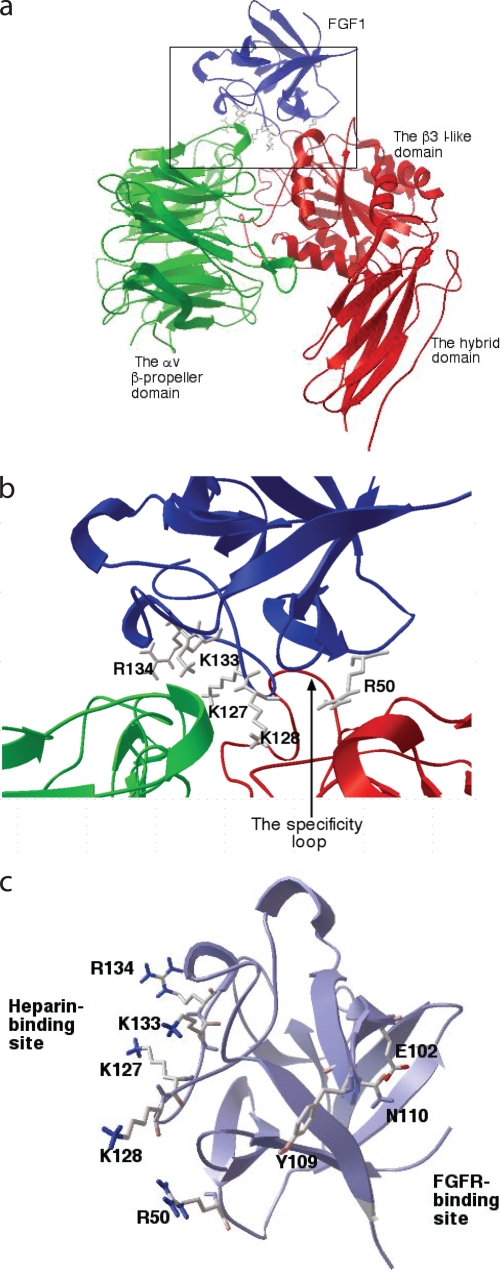
Generation of Integrin-binding Defective FGF1 Mutants—To localize the integrin-binding site in FGF1, we used a docking simulation program and site-directed mutagenesis. AutoDock is a set of docking tools widely used for predicting the pose of small ligands bound to receptors (37), and the methods are being extended to predict protein-protein complex poses (38). We performed 50 dockings of the FGF1-integrin αvβ3 interaction, each one starting with a random initial position and orientation of FGF1 (PDB code 1AXM) with respect to the integrin αvβ3 headpiece (PDB code 1L5G). The results were clustered together by positional root mean square distance into families of similar poses (data not shown). Many of the docking poses clustered well with the lowest docking energy, 26.1 kcal/mol (cluster 1). These results predict that the docking pose of cluster 1 (Fig. 2a) may represent the most probable stable FGF1 pose upon binding to αvβ3. This model predicts that the integrin-binding interface of FGF1 with integrin αvβ3 is distinct from the FGFR-binding site (39) but is close to or overlapping with the heparin-binding site (Fig. 2, b and c).
FIGURE 2.
Identification of the integrin-binding site in FGF1. a, model of FGF1-integrin interaction. Docking simulation of the interaction between FGF1 (PDB code 1AXM) and integrin αvβ3 (PDB code 1L5G) was performed as described under the “Experimental Procedures” using AutoDock3. Model a of 1AXM was used for docking. The headpiece of 1L5G was used as a receptor. The pose in the cluster 1 with the lowest docking energy -26.3 kcal/mol is shown. This pose represents the most stable pose of FGF1 when FGF1 interacts with integrin αvβ3. b, positions of amino acid residues that are selected for mutagenesis at the predicted interface between FGF1 and αvβ3. Several amino acid residues within the predicted integrin-binding site in FGF1 were selected for mutagenesis. c, positions of the amino acid residues (E102A, Y109A, and N110A) at the FGFR-binding site selected for mutagenesis. Note that the predicted integrin-binding site is distinct from the FGFR-binding site.
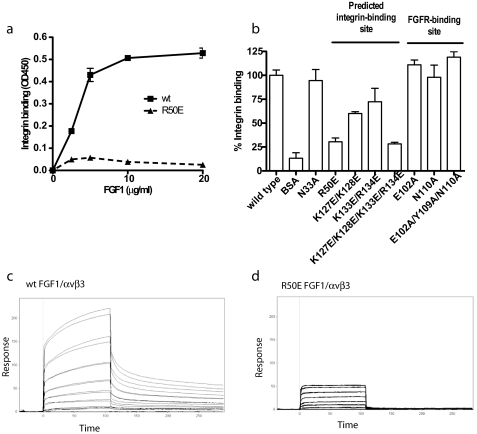
We introduced several mutations within the predicted integrin-binding site (Fig. 2b and Table 1) to localize the integrin-binding site in FGF1. We tested the ability of the mutants to bind to αvβ3 in ELISA-type binding assays. The Arg-50 to Glu (R50E) mutation effectively reduced the binding of soluble αvβ3 to FGF1 (Fig. 3a). Mutating positively charged residues (Lys-127, Lys-128, Lys-133, and Arg-134) in the predicted integrin-binding site to Glu in combination diminished the binding of soluble αvβ3 to FGF1 (Fig. 2b and Fig. 3b). Mutating all these residues simultaneously to Glu (designated the 4xE mutation) effectively diminished integrin binding. We predicted that Glu-102, Tyr-109, and Asn-110 are located in the FGFR-binding site from the FGF1-FGFR-2 complex structure (39) and mutagenesis data of FGF2 (40). Mutating these residues to Ala individually or simultaneously (designated 3xA mutant) did not affect integrin binding (Fig. 2c and Fig. 3b), whereas the 3xA mutation effectively suppressed the binding to FGFR1 (below).
TABLE 1.
Amino acid residues at the predicted interface between FGF1 and integrin αvβ3 Amino acid residues of FGF1 or αvβ3 that are within a 6-Å distance from αvβ3 or FGF1, respectively, were selected using Swiss-pdb viewer (version 3.7). Amino acid residues of FGF1 (bold) were selected for mutagenesis.
| FGF1 | αv | β3 |
|---|---|---|
| Asn-33, Gly-34, Gly-35 His-36, Arg-39, Leu-41 | Met-118 | Tyr-122, Ser-123, Met-124, Lys-125, Asp-126, Asp-127, Trp-129 |
| Asp-43, Thr-45, Val-46 | Ser-144, Gln-145, Asp-146, Ile-147, Asp-148, Ala-149, Asp-150, Gly-151 | Tyr-166 |
| Asp-47, Gly-48, Thr-49 Arg-50, Asp-51, Arg-52 | Tyr-178 | Asp-179, Met-180, Lys-181, Thr-182, Arg-214, Asn-215, Arg-216, Ala-218, Asp-251, Ala-252, Lys-253 |
| Ser-53, Asp-54 | Thr-212, Gln-214, Ala-215, Ile-216, Asp-218, Asp-219 | |
| Lys-127, Lys-128, Asn-129, Gly-130, Ser-131, Cys-132, Lys-133, Arg-134, Arg-137, Thr-138, Gly-141, Gln-142, Lys-143, Ala-144 | Arg-248 |
FIGURE 3.
Effect of FGF mutations on the binding to integrin αvβ3. a, R50E mutation of FGF1 reduced its binding to integrinαvβ3. Amino acid residues in the integrin-binding site or in the FGFR-binding site were mutated individually or in groups. WT FGF1 and the R50E mutant were coated to a plastic plate at various concentrations as indicated, and the binding of soluble αvβ3 was determined as described in Fig. 1a. The results show that the R50E mutation reduced αvβ3 binding. b, summary of the effects of FGF1 mutations on integrin binding. The results suggest that mutations in the predicted integrin-binding site of FGF1 reduced the binding of solubleαvβ3 to FGF1, but mutations in the FGFR-binding site did not. c and d, binding of WT FGF1 (c) and R50E FGF1 (d) to αvβ3 in SPR. Soluble αvβ3 was immobilized on a CM5 sensor chip. WT and R50E FGF1 were individually 2-fold serially diluted from 8 μm to 125 nm in HBS-P buffer with 1 mm of Mn2+, and the 3xA FGF1 was 2-fold serially diluted from 6 μm to 93.75 nm in the same buffer. 4xE did not show any binding to integrin (data not shown).
We assessed the ability of the FGF1 mutants to bind to αvβ3 in surface plasmon resonance (SPR) using a sensor chip immobilized with soluble αvβ3. WT FGF1 bound to αvβ3 at a high affinity (KD 1.1 μm) (Fig. 3c). The R50E mutant showed much lower Rmax value and enhanced off-rate to αvβ3, whereas it has a KD value to αvβ3 (1.31 μm) similar to WT FGF1 (Fig. 3d). The ability of the 3xA mutant to bind to αvβ3 was intact (KD 0.18 μm), but the 4xE mutant did not show binding affinity to αvβ3 (data not shown)). These results are consistent with those obtained by ELISA-type binding assays.
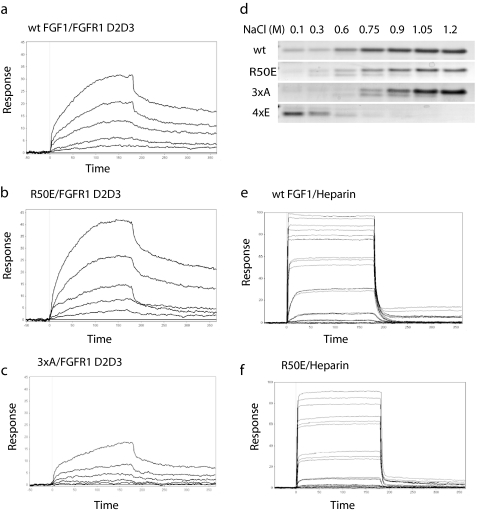
We assessed the ability of these FGF1 mutants to interact with the FGFR1 fragment (domains 2 and 3, or FGFR1 D2D3) in SPR using a sensor chip immobilized with FGFR1 D2D3. WT FGF1 (KD 30 pm) and R50E FGF1 (KD 14 pm) had similar affinity to FGFR1 (Fig. 4, a and b). The 4xE mutant showed low affinity to FGFR1 (KD 5.5 μm; data not shown). The 3xA mutant has a much lower affinity (KD 0.41 mm) (Fig. 4c).
FIGURE 4.
Effect of FGF mutations on the binding to FGFR and heparin. a–c, binding of WT (a), R50E (b), and 3xA (c) FGF1 mutant to FGFR1 D2D3 fragment in SPR. FGFR1 D2D3 was immobilized to a CM5 sensor chip. WT or mutant FGF1 was 2-fold diluted serially from 800 to 50 nm. KD is calculated as 30 pm for WT FGF1, 14 pm for R50E, 5.5 μm for 4xE, and 0.41 mm for 3xA. d, binding of R50E to heparin-Sepharose. We incubated partially purified WT and mutant FGF1 with heparin-Sepharose and eluted with increasing concentrations of NaCl. Eluted proteins were analyzed by SDS-PAGE and proteins stained with Coomassie Brilliant Blue. e and f, binding of WT FGF1 (e) and R50E FGF1 (f) to heparin in SPR. Biotinylated heparin was immobilized to a streptavidin sensor chip. 5-fold serially diluted WT, R50E, and 3xA ranging from 800 nm to 51.2 pm, and 2-fold serially diluted 4xE ranging from 1.6 μm to 50 nm in HBS-EP were injected at 50 μl/min for 3 min. KD is calculated as 16.6 nm for WT FGF1, 66 nm for R50E, and 11.5 nm for 3xA. 4xE did not show any binding to heparin (data not shown).
We assessed the ability of the mutants to bind to heparin-Sepharose. WT FGF1, and the R50E and 3xA mutants, bound strongly to heparin-Sepharose and required 1.2 m NaCl for elution, but the 4xE mutant did not show affinity to heparin (Fig. 4d). In SPR R50E (Fig. 4e) had slightly lower affinity to heparin (KD 66 nm) than WT FGF1 (KD 16.6 nm) (Fig. 4f), whereas the slightly reduced affinity of R50E to heparin did not affect its ability to induce FGFR1 activation and ERK1/2 activation (see below). The 3xA mutant (data not shown) showed affinity to heparin (KD 11.5 nm) similar to that of WT FGF1, but the 4xE did not show any affinity to heparin (data not shown).
These results are consistent with the prediction that in mutations in the predicted integrin-binding site in FGF1 compromised integrin binding and heparin binding, but those in the FGFR-binding site affected FGFR binding but did not affect integrin binding. The R50E mutation was unique in that it reduced integrin binding and induced minimal effect on heparin or FGFR binding. Thus we propose that the R50E mutant is useful to determine the potential role of integrin αvβ3 in FGF signaling. The 4xE mutation was defective in integrin, heparin, and FGFR1 binding and could be used as a control together with the 3xA mutant. Positions of these mutations in FGF1 are shown in Fig. 2, B and C.
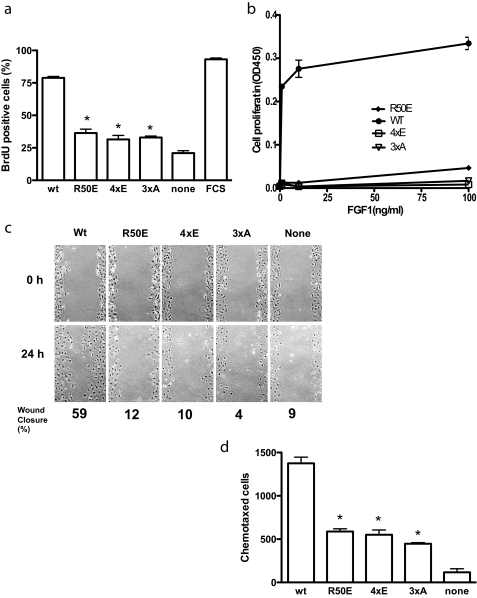
Effect of the Mutations on FGF-induced DNA Synthesis and Cell Migration—We examined the effect of the FGF1 mutations on FGF1-induced DNA synthesis in Balb 3T3 cells. The R50E, 4xE, and 3xA mutants induced much lower BrdUrd incorporation than WT FGF1 (Fig. 5a), suggesting that these mutants are defective in inducing DNA synthesis.
FIGURE 5.
Integrin-binding defective FGF1 mutants are defective in FGF signaling. a, DNA synthesis. Balb 3T3 cells were plated on coverslips in 6-well culture plates, serum-starved for 48 h, and stimulated with 5 ng/ml WT or mutant FGF1s in the presence of 5 μg/ml heparin for 24 h. BrdUrd was added to the medium for the last 6 h of the incubation. BrdUrd-positive cells were counted. Results are shown as means ± S.E. of triplicate experiments. *, the BrdUrd incorporation was lower in cells treated with R50E (p = 0.0003), 4xE (p = 0.019), and 3xA (p = 0.003) than with WT FGF1. We used 2-way analysis of variance for statistical analysis. Similar results were obtained using NIH3T3 cells. FCS, 10% FCS was added to the medium as a positive control. b, proliferation of BaF3 cells that express human FGFR1c (BaF3-FR1c). BaF3-FR1c cells were maintained for 48 h with WT or mutant FGF1 at indicated concentrations instead of IL-3, and cell proliferation was measured by MTS assays. Results are shown as means ± S.E. of triplicate experiments. c, in vitro scratch wound healing. Confluent serum-starved Balb 3T3 cells were scratched. After washing with serum-free medium, the cells were incubated in DMEM containing 5 μg/ml heparin and 5 ng/ml WT or mutant FGF1s for 24 h at 37 °C. The rate of wound healing was quantified by using ImageJ. d, chemotaxis. The bottom of the polycarbonate filter of the Transwell apparatus was coated with fibronectin. The lower chamber contained serum-free DMEM containing 5 ng/ml WT or the mutants of FGF1. Balb 3T3 cells (105 cells/filter) were plated on filter and incubated 37 °C for 24 h, and the cells were stained with crystal violet. The cells that migrated to the bottom side of the membrane were counted. Results are expressed as means of the number of chemotaxed cells in three fields. *, the number of chemotaxed cells was lower with R50E (p = 0.026), 4xE (p = 0.0004), and 3xA (p = 0.0078) than with WT FGF1. We used 2-way analysis of variance for statistical analysis.
To test the ability of the FGF1 mutants to induce cell proliferation, we used BaF3 mouse pro-B cells that express human FGFR1c (designated BaF3-FGFR1c) (41), which have been used to sensitively detect cell proliferation that is dependent on exogenous FGF1. We found that WT FGF1 robustly induced proliferation of BaF3-FGFR1c cells, whereas the R50E, 4xE, and 3xA mutants did not induce detectable cell proliferation (Fig. 5b). This is consistent with the results that these mutants were defective in inducing DNA synthesis.
Because FGF1 is a potent inducer of cell migration and chemotaxis (42, 43), we tested the ability of the mutants to induce cell migration. Using scratch wound healing assays with Balb 3T3 cells, we found that although WT FGF1 closed the wound by 59% in 24 h, the R50E, 4xE, and 3xA mutants did not close the wound in this time frame (Fig. 5c). We found that the R50E, 4xE, and 3xA mutants induced much lower chemotaxis than WT FGF1 (Fig. 5d). These results suggest that these mutants are defective in inducing cell migration.
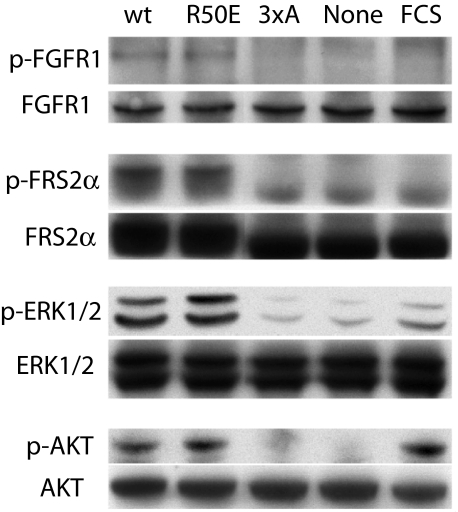
Effect of the Integrin-binding Defective Mutations on FGF1 Signaling—Next we studied whether these FGF1 mutants induce FGF signaling inside the cells. The binding of FGF to FGFR leads to tyrosine phosphorylation of the docking protein FRS2α, which functions as a major mediator of signaling via FGFR (1–4). This results in recruitment of multiple Grb2-Sos complexes leading to activation of the Ras/MAPK signaling pathway (44). The mitogenic action of the FGFs is mediated in part by the activation of ERK1/2 (45). WT FGF1 induced phosphorylation of FGFR1, FRS2α, AKT (protein kinase B), and ERK1/2. The 4xE and 3xA mutants showed little or no ability to induce FGFR1, FRS2α, or ERK1/2 activation as compared with WT FGF1 and the R50E mutant (Fig. 6), which is consistent with the finding that the 4xE and 3xA mutants are defective in binding to FGFR1. Notably, the R50E mutant induced FGFR1, FRS2α, AKT, and ERK1/2 activation to the similar extent of WT FGF1 (Fig. 6). These results suggest that the integrin-binding defective R50E FGF1 mutant can induce the initial FGFR activation and subsequent FGF signaling and that the slightly reduced heparin binding to this mutant did not affect it.
FIGURE 6.
Effect of FGF1 mutations on FGF signaling. FCS, 10% FCS. Serum-starved NIH3T3 cells were stimulated with WT or mutant FGF1s (5 ng/ml) in the presence of 5 μg/ml heparin for 10 min at 37 °C. For FGFR phosphorylation (p-FGFR1), phosphorylated FGFR1 was first immunoprecipitated from lysates (750 μl, total 2.6 mg of protein) using anti-phospho-Tyr antibodies and then blotted using anti-FGFR1 antibodies. Whole FGFR1 in cell lysates were detected with anti-FGFR1. For FRS2α, ERK1/2, and AKT, cell lysates were analyzed by Western blotting with antibodies against phospho-FRS2α, FRS2α, phospho-ERK1/2, ERK1/2, phospho-AKT, and AKT, respectively. Results with the 4xE mutant were very similar to those with the 3xA mutant (data not shown).
DISCUSSION
Although it has been reported that integrins play a critical role in FGF signaling (13), the specifics governing their interaction in this process are still unclear. In this study, we showed that integrin αvβ3 directly bound to FGF1 at an affinity comparable with those of known integrin ligands. Heat denaturation of FGF1 destroyed integrin binding, and the amino acid residues of FGF1 that are critical for integrin-binding were discontinuous, indicating that proper folding of FGF1 is required for integrin binding. Because mAb 7E3 and cyclic RGDfV blocked the FGF1-αvβ3 interaction, it is suggested that FGF1 binds to the ligand-binding site of αvβ3. We showed that the specificity loop plays a role in the binding of FGF1 to αvβ3, suggesting that FGF1 binds to αvβ3 in a manner similar to that of other αvβ3 ligands tested (27, 35, 36). We showed that 10% fetal bovine serum did not suppress αvβ3-mediated cell adhesion to FGF1. This is consistent with the reports that integrin-binding sites are cryptic in fibrinogen (46), fibronectin (47), and vitronectin (48). These findings suggest that the observed FGF1 binding to αvβ3 may happen in biological body fluid. It has been proposed that productive integrin-ligand interactions require ligand “activation” (e.g. by immobilization and proteolysis) in addition to activation of integrins (46). We recently showed that αvβ3 binds to the C-terminal fragment of the fibrinogen γ-chain but not to whole fibrinogen in solution (49).
We localized critical amino acid residues of FGF1 for binding to αvβ3 using docking simulation and mutagenesis. We found that the integrin-binding site overlaps with the heparin-binding site. These findings suggest that docking simulation and mutagenesis are useful tools for studying how integrins bind to protein ligands. The R50E mutation affected integrin binding, although its effect on heparin and FGFR binding was minimal, suggesting that this mutation primarily affected integrin binding. The R50E mutant did not induce cell proliferation and migration, whereas R50E induced FGF1 signaling (FGFR1 phosphorylation, FRS2α phosphorylation, and ERK1/2). This suggests that the integrin binding may not be required for the early activation of ERK1/2 but is required for the later steps in FGF signaling that leads to cell proliferation and migration. Indeed it has been reported that that transient ERK1/2 activation is not integrin-dependent and not related to cell proliferation, but that sustained ERK1/2 activation is integrin-dependent and directly related to cell cycle entry (50, 51). The R50E mutant of FGF1 may be a useful tool to study the role of integrins in FGF signaling. Further studies in this direction are under way.
We showed that WT FGF1 free in solution had a KD of 1.1 μm to αvβ3 and a KD of around 1 pm to FGFR1. It is thus expected that FGF1 will bind preferentially to FGFR on the cell surface. We expect that the binding of FGF1 to FGFR will facilitate the binding of FGF1 to αvβ3 because FGF1 is concentrated on the cell surface through binding to FGFR. Our results predict that the integrin-binding site of FGF1 would be still exposed upon binding to FGFR. The results in this study suggest that integrin αvβ3 is a major, if not the only, integrin that binds to FGF1. It has been reported that αvβ3 is detectable in NIH3T3 cells (52) and that αvβ3 is markedly induced when the Ras-ERK1/2 pathway is activated by Raf oncogene in NIH3T3 cells (53). The R50E mutation is close to the FGF1-FGFR1 domain D2 interface as shown in the crystal structure of the FGF1-FGFR1 complex (PDB code 1EVT). We showed that the R50E mutation did not affect the binding of FGF1 to FGFR1 D2D3. Also we showed that this mutant induced phosphorylation of FGFR1 and FRS2α and activation of ERK1/2 and AKT. It is thus unlikely that the R50E mutation suppressed FGFR binding. We propose that the primary defect in the R50E mutant is in its ability to bind to integrins.
In current models of the role of integrins in growth factor signaling, integrins interact with ECM ligands, and the resulting integrin-mediated signaling merges with growth factor-mediated signaling (54). The present results suggest that the direct binding of FGF to integrin αvβ3 is critical for proper FGF signaling and that FGF1 itself acts as an integrin ligand. If this is the case, it is likely that integrin-FGFR cross-talk may be mediated by direct binding of FGF to integrins and FGFR. We showed that FGF1 binds to integrins in the presence of 10% serum, consistent with the previous reports that integrin-binding sites in major serum adhesive proteins (e.g. fibrinogen) are cryptic in solution as discussed above. We propose that FGF-integrin interaction occurs in body fluids. Also, we propose that FGF may be concentrated on the cell surface through high affinity binding to FGFR. This would facilitate FGF-integrin interaction on the cell surface. Because FGFR and integrins bind to distinct sites in FGF1, FGFR binding to FGF would not block FGF-integrin interaction. Thus we expect that FGF may interact with integrins on the cell surface under physiological conditions even if the affinity of αvβ3 to FGF in solution in SPR is low (KD 1 μm). We showed that an integrin-binding defective FGF1 mutant (R50E) was defective in inducing cell proliferation and migration. Taken together, we propose that the direct FGF-integrin interaction on the cell surface is biologically relevant. The present results, however, do not rule out the possibility that ECM-integrin interaction is required for FGF signaling. Integrin antagonists or down-regulation of integrins block both integrin-ECM and integrin-FGF interactions, and do not distinguish the two interactions in FGF signaling. The R50E mutant of FGF1 is thus an important tool that selectively blocks FGF1-integrin interaction in FGF signaling.
Acknowledgments
We thank Anne Hanneken, David Ornitz, and Tim Springer for valuable reagents. We also thank Garrett Morris and Art Olson (Scripps Research Institute) for the help in docking simulation.
This work was supported, in whole or in part, by National Institutes of Health Grants CA113298 and CA093373 (to Y. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: FGF, fibroblast growth factor; CHO, Chinese hamster ovary; ECM, extracellular matrix; ERK, extracellular responsive kinase; FGFR, fibroblast growth factor receptor; FRS2α, FGF receptor substrate 2α; HRP, horseradish peroxidase; MIDAS, metal ion-dependent adhesive site; PTB, phosphotyrosine binding domain; SPR, surface plasmon resonance; WT, wild type; FCS, fetal calf serum; DMEM, Dulbecco's modified Eagle's medium; PDB, Protein Data Bank; PBS, phosphate-buffered saline; BSA, bovine serum albumin; MAPK, mitogen-activated protein kinase; mAb, monoclonal antibody; ELISA, enzyme-linked immunosorbent assay; IL, interleukin; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt.
References
- 1.Presta, M., Dell'Era, P., Mitola, S., Moroni, E., Ronca, R., and Rusnati, M. (2005) Cytokine Growth Factor Rev. 16 159-178 [DOI] [PubMed] [Google Scholar]
- 2.Ullrich, A., and Schlessinger, J. (1990) Cell 61 203-212 [DOI] [PubMed] [Google Scholar]
- 3.Powers, C. J., McLeskey, S. W., and Wellstein, A. (2000) Endocr.-Relat. Cancer 7 165-197 [DOI] [PubMed] [Google Scholar]
- 4.Klint, P., and Claesson-Welsh, L. (1999) Front. Biosci. 4 D165-D177 [DOI] [PubMed] [Google Scholar]
- 5.Thisse, B., and Thisse, C. (2005) Dev. Biol. 287 390-402 [DOI] [PubMed] [Google Scholar]
- 6.Grose, R., and Dickson, C. (2005) Cytokine Growth Factor Rev. 16 179-186 [DOI] [PubMed] [Google Scholar]
- 7.Eswarakumar, V. P., Lax, I., and Schlessinger, J. (2005) Cytokine Growth Factor Rev. 16 139-149 [DOI] [PubMed] [Google Scholar]
- 8.Gan, Y., Wientjes, M. G., and Au, J. L. (2006) Pharmacol. Res. 23 1324-1331 [DOI] [PubMed] [Google Scholar]
- 9.Karajannis, M. A., Vincent, L., Direnzo, R., Shmelkov, S. V., Zhang, F., Feldman, E. J., Bohlen, P., Zhu, Z., Sun, H., Kussie, P., and Rafii, S. (2006) Leukemia (Baltimore) 20 979-986 [DOI] [PubMed] [Google Scholar]
- 10.Pardo, O. E., Wellbrock, C., Khanzada, U. K., Aubert, M., Arozarena, I., Davidson, S., Bowen, F., Parker, P. J., Filonenko, V. V., Gout, I. T., Sebire, N., Marais, R., Downward, J., and Seckl, M. J. (2006) EMBO J. 25 3078-3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song, S., Wientjes, M. G., Gan, Y., and Au, J. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 8658-8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paleolog, E. M. (2002) Arthritis Res. 4 Suppl. 3, 81-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eliceiri, B. P., and Cheresh, D. A. (2001) Curr. Opin. Cell Biol. 13 563-568 [DOI] [PubMed] [Google Scholar]
- 14.Hynes, R. O. (2002) Cell 110 673-687 [DOI] [PubMed] [Google Scholar]
- 15.Shimaoka, M., and Springer, T. A. (2003) Nat. Rev. Drug Discov. 2 703-716 [DOI] [PubMed] [Google Scholar]
- 16.Schwartz, M. A., and Assoian, R. K. (2001) J. Cell Sci. 114 2553-2560 [DOI] [PubMed] [Google Scholar]
- 17.Brooks, P., Clark, R., and Cheresh, D. (1994) Science 264 569-571 [DOI] [PubMed] [Google Scholar]
- 18.Brooks, P., Montgomery, A., Rosenfeld, M., Reisfeld, R., Hu, T., Klier, G., and Cheresh, D. (1994) Cell 79 1157-1164 [DOI] [PubMed] [Google Scholar]
- 19.Rusnati, M., Tanghetti, E., Dell'Era, P., Gualandris, A., and Presta, M. (1997) Mol. Biol. Cell 8 2449-2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanghetti, E., Ria, R., Dell'Era, P., Urbinati, C., Rusnati, M., Ennas, M. G., and Presta, M. (2002) Oncogene 21 3889-3897 [DOI] [PubMed] [Google Scholar]
- 21.Sahni, A., and Francis, C. W. (2004) Blood 104 3635-3641 [DOI] [PubMed] [Google Scholar]
- 22.Sahni, A., Khorana, A. A., Baggs, R. B., Peng, H., and Francis, C. W. (2006) Blood 107 126-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahni, A., Altland, O. D., and Francis, C. W. (2003) J. Thromb. Haemost. 1 1304-1310 [DOI] [PubMed] [Google Scholar]
- 24.Takagi, J., Erickson, H. P., and Springer, T. A. (2001) Nat. Struct. Biol. 8 412-416 [DOI] [PubMed] [Google Scholar]
- 25.Blystone, S., Graham, I., Lindberg, F., and Brown, E. (1994) J. Cell Biol. 127 1129-1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleming, F. E., Graham, K. L., Taniguchi, K., Takada, Y., and Coulson, B. S. (2007) Arch. Virol. 152 1087-1101 [DOI] [PubMed] [Google Scholar]
- 27.Takagi, J., Kamata, T., Meredith, J., Puzon-McLaughlin, W., and Takada, Y. (1997) J. Biol. Chem. 272 19794-19800 [DOI] [PubMed] [Google Scholar]
- 28.Wang, W., and Malcolm, B. A. (1999) BioTechniques 26 680-682 [DOI] [PubMed] [Google Scholar]
- 29.Plotnikov, A. N., Hubbard, S. R., Schlessinger, J., and Mohammadi, M. (2000) Cell 101 413-424 [DOI] [PubMed] [Google Scholar]
- 30.Wu, P. L., Lee, S. C., Chuang, C. C., Mori, S., Akakura, N., Wu, W. G., and Takada, Y. (2006) J. Biol. Chem. 281 7937-7945 [DOI] [PubMed] [Google Scholar]
- 31.Zhang, X., Ibrahimi, O. A., Olsen, S. K., Umemori, H., Mohammadi, M., and Ornitz, D. M. (2006) J. Biol. Chem. 281 15694-15700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aumailley, M., Gurrath, M., Muller, G., Calvete, J., Timpl, R., and Kessler, H. (1991) FEBS Lett. 291 50-54 [DOI] [PubMed] [Google Scholar]
- 33.Puzon-McLaughlin, W., Kamata, T., and Takada, Y. (2000) J. Biol. Chem. 275 7795-7802 [DOI] [PubMed] [Google Scholar]
- 34.Artoni, A., Li, J., Mitchell, B., Ruan, J., Takagi, J., Springer, T. A., French, D. L., and Coller, B. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 13114-13120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokoyama, K., Zhang, X. P., Medved, L., and Takada, Y. (1999) Biochemistry 38 5872-5877 [DOI] [PubMed] [Google Scholar]
- 36.Triantafilou, K., Triantafilou, M., Takada, Y., and Fernandez, N. (2000) J. Virol. 74 5856-5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris, G. M., Goodsell, D. S., Halliday, R. S., Fig Huey, R., Hart, W. E., Belew, R. K., and Olson, A. J. (1998) J. Comp. Chem. 19 1639-1662 [Google Scholar]
- 38.Saphire, E. O., Parren, P. W., Pantophlet, R., Zwick, M. B., Morris, G. M., Rudd, P. M., Dwek, R. A., Stanfield, R. L., Burton, D. R., and Wilson, I. A. (2001) Science 293 1155-1159 [DOI] [PubMed] [Google Scholar]
- 39.Pellegrini, L., Burke, D. F., von Delft, F., Mulloy, B., and Blundell, T. L. (2000) Nature 407 1029-1034 [DOI] [PubMed] [Google Scholar]
- 40.Zhu, H., Anchin, J., Ramnarayan, K., Zheng, J., Kawai, T., Mong, S., and Wolff, M. E. (1997) Protein Eng. 10 417-421 [DOI] [PubMed] [Google Scholar]
- 41.Ornitz, D. M., Xu, J., Colvin, J. S., McEwen, D. G., MacArthur, C. A., Coulier, F., Gao, G., and Goldfarb, M. (1996) J. Biol. Chem. 271 15292-15297 [DOI] [PubMed] [Google Scholar]
- 42.Liu, J., Huang, C., and Zhan, X. (1999) Oncogene 18 6700-6706 [DOI] [PubMed] [Google Scholar]
- 43.Friesel, R. E., and Maciag, T. (1995) FASEB J. 9 919-925 [DOI] [PubMed] [Google Scholar]
- 44.Kwabi-Addo, B., Ozen, M., and Ittmann, M. (2004) Endocr.-Relat. Cancer 11 709-724 [DOI] [PubMed] [Google Scholar]
- 45.LaVallee, T. M., Prudovsky, I. A., McMahon, G. A., Hu, X., and Maciag, T. (1998) J. Cell Biol. 141 1647-1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lishko, V. K., Kudryk, B., Yakubenko, V. P., Yee, V. C., and Ugarova, T. P. (2002) Biochemistry 41 12942-12951 [DOI] [PubMed] [Google Scholar]
- 47.Ugarova, T. P., Ljubimov, A. V., Deng, L., and Plow, E. F. (1996) Biochemistry 35 10913-10921 [DOI] [PubMed] [Google Scholar]
- 48.Seiffert, D., and Smith, J. W. (1997) J. Biol. Chem. 272 13705-13710 [DOI] [PubMed] [Google Scholar]
- 49.Akakura, N., Hoogland, C., Takada, Y. K., Saegusa, J., Ye, X., Liu, F. T., Cheung, A. T., and Takada, Y. (2006) Cancer Res. 66 9691-9697 [DOI] [PubMed] [Google Scholar]
- 50.Eliceiri, B. P., Klemke, R., Stromblad, S., and Cheresh, D. A. (1998) J. Cell Biol. 140 1255-1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharrocks, A. D. (2006) Curr. Biol. 16 R540-R542 [DOI] [PubMed] [Google Scholar]
- 52.White, D. P., Caswell, P. T., and Norman, J. C. (2007) J. Cell Biol. 177 515-525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woods, D., Cherwinski, H., Venetsanakos, E., Bhat, A., Gysin, S., Humbert, M., Bray, P. F., Saylor, V. L., and McMahon, M. (2001) Mol. Cell. Biol. 21 3192-3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Juliano, R. L., Reddig, P., Alahari, S., Edin, M., Howe, A., and Aplin, A. (2004) Biochem. Soc. Trans. 32 443-446 [DOI] [PubMed] [Google Scholar]