Abstract
Collapsin response mediator protein 2 (CRMP2) binds to microtubules and regulates axon outgrowth in neurons. This action is regulated by sequential phosphorylation by the kinases cyclin-dependent kinase 5 (Cdk5) and glycogen synthase kinase 3 (GSK3) at sites that are hyperphosphorylated in Alzheimer disease. The increased phosphorylation in Alzheimer disease could be due to increases in Cdk5 and/or GSK3 activity or, alternatively, through decreased activity of a CRMP phosphatase. Here we establish that dephosphorylation of CRMP2 at the residues targeted by GSK3 (Ser-518/Thr-514/Thr-509) is carried out by a protein phosphatase 1 family member in vitro, in neuroblastoma cells, and primary cortical neurons. Inhibition of GSK3 activity using insulin-like growth factor-1 or the highly selective inhibitor CT99021 causes rapid dephosphorylation of CRMP2 at these sites. In contrast, pharmacological inhibition of Cdk5 using purvalanol results in only a gradual and incomplete dephosphorylation of CRMP2 at the site targeted by Cdk5 (Ser-522), suggesting a distinct phosphatase targets this residue. A direct comparison of dephosphorylation at the Cdk5 versus GSK3 sites in vitro shows that the Cdk5 site is comparatively resistant to phosphatase treatment. The presence of the peptidyl-prolyl isomerase enzyme, Pin1, does not affect dephosphorylation of Ser-522 in vitro, in cells, or in Pin1 transgenic mice. Instead, the relatively high resistance of this site to phosphatase treatment is at least in part due to the presence of basic residues located nearby. Similar sequences in Tau are also highly resistant to phosphatase treatment. We propose that relative resistance to phosphatases might be a common feature of Cdk5 substrates and could contribute to the hyperphosphorylation of CRMP2 and Tau observed in Alzheimer disease.
Collapsin response mediator protein 2 (CRMP2)3 was initially identified in a screen for molecules involved in collapsin-1 (also called semaphorin-3A) signaling pathways in Xenopus laevis oocytes; hence, its name (1). Since then four other CRMP family members have been identified (2, 3). All members are enriched in the central nervous system, localize to the cytoplasm and neurites of post-mitotic neurons, and exist almost exclusively as heterotetramers (4, 5). CRMP2 remains the most studied and best characterized family member, with reported cellular functions including regulation of cell surface receptor endocytosis (6), kinesin-mediated transport (7), growth cone collapse (1, 8, 9), neurite outgrowth (10-12), and microtubule dynamics (13). The latter three functions have been reported to be regulated by phosphorylation near the C terminus of CRMP2 by the brain-enriched kinases cyclin-dependent kinase 5 (Cdk5) and glycogen synthase kinase 3 (GSK3) (8, 9, 11, 12). Phosphorylation by Cdk5 at Ser-522 “primes” CRMP2 for subsequent phosphorylation by GSK3 at Ser-518/Thr-514/Thr-509. CRMP1 is also a substrate for Cdk5 and GSK3. The physiological priming kinase for subsequent GSK3-mediated phosphorylation of CRMP4 has not yet been determined, although this function can be performed by DYRK2 in vitro (14).
Interestingly, CRMP2 is hyperphosphorylated at the Cdk5 and GSK3 sites in brain tissue from human AD patients and in some mouse models of AD (15, 16). It has been shown to colocalize with neurofibrillary tangles in human AD cortex (15) and is an early event in the pathogenesis of the disease (16). Similarly, the primary constituent of neurofibrillary tangles found in the brains of AD patients, Tau, is also hyperphosphorylated in AD at sites that are targeted by Cdk5 and GSK3. These observations implicate increased activity of Cdk5 and/or GSK3 in the pathogenesis of AD. However, we and others have been unable to find evidence of altered Cdk5 or GSK3 activity in AD (14, 16-20). This may be because of post-mortem degradation issues or the sensitivity of our detection methods. Alternatively, because phosphatases are equally important for regulating phosphorylation levels, increased CRMP2 phosphorylation could occur in response to reduced phosphatase activity. Hence, we investigated the regulation of CRMP2 dephosphorylation at these important sites.
EXPERIMENTAL PROCEDURES
Materials—Production of CRMP2 cDNA constructs, GST-CRMP2 fusion proteins, and CRMP2 antibodies have been described previously (11, 14). The CRMP2(K520A) mutant was generated using the QuikChange mutagenesis kit (Stratagene), confirmed by direct sequencing, then expressed in Escherichia coli BL21 cells as a GST-tagged protein (pGEX-6 vector). Generation of Pin1 wild type and mutant constructs as well as recombinant His6-Pin1 has been described (21). The protein phosphatases PP1γ, PP2A1, and PP2C as well as His6-GSK3β were supplied by the Division of Signal Transduction Therapy, College of Life Sciences, University of Dundee. PP2A and PP2C were assayed using 32P-labeled casein. A milliunit of activity is that amount which removes 1 nmol of phosphate from phosphocasein/min (at 30 °C). Active PP2B was purchased from Promega UK Ltd as a heterocomplex of the 19-kDa calcium binding subunit and the 61-kDa catalytic subunit. One unit is that amount that removes 1 nmol of phosphate from p-nitrophenyl phosphate/min. The His6-Cdk5·p35 complex was obtained from Upstate Biotechnology. The GSK3-specific inhibitor CT99021 was prepared as described previously (14), whereas IGF1 was purchased from Invitrogen. Purvalanol, okadaic acid, calyculin A, cypermethrin, and cyclosporin A were obtained from Calbiochem. Tau-5, Ser(P)-404, and Thr(P)-231 Tau antibodies were purchased from Chemicon, whereas AT8 and Ser(P)-396 antibodies were obtained from Innogenetics and Cell Signaling, respectively.
In Vitro Phosphatase Assays—For analysis of CRMP2-Ser-522 dephosphorylation, GST-CRMP2 (2 μm) was phosphorylated using Cdk5 (2.5 milliunits/μl) in the presence of radiolabeled Mg-[γ-32P]ATP in buffer containing 50 mm Tris-HCl, pH 7.5, 0.03% (v/v) Brij-35, and 0.1% (v/v) β-mercaptoethanol (30 °C, 1 h). Cdk5 activity was inhibited by the addition of purvalanol (50 μm), then phosphatase was added at the indicated amounts to phosphorylated CRMP2 (1 μm) for up to 1 h at 30 °C. Reactions were terminated by the addition of SDS loading buffer then subjected to SDS-PAGE. Radiolabeled bands were visualized by autoradiography and excised from the gel, and the amount of 32P released from CRMP2 was determined using Cerenkov counting. For analysis of CRMP2-Ser-518/Thr-514/Thr-509 dephosphorylation, CRMP2 was phosphorylated by Cdk5 in the presence of unlabeled ATP as described above. Purvalanol was added to inhibit Cdk5 activity, then GSK3β (2.5 milliunits/μl) and radiolabeled [γ-32P]ATP was added for a further 1 h at 30 °C. GSK3β activity was inhibited by the addition of CT99021 (10 μm), then phosphatases (50 milliunits) were added for 1 h. Release of 32P from CRMP2 by phosphatases was performed as described above.
Dephosphorylation of Endogenous CRMP2 in Rat Brain Lysate—Adult rat brains were homogenized using a 10-ml glass Dounce homogenizer in lysis buffer containing 1% (v/v) Triton X-100, 50 mm Tris-HCl, pH 7.5, 0.27 m sucrose, 1 mm sodium orthovanadate, 0.1% (v/v) β-mercaptoethanol, and Complete protease inhibitor tablets (Roche Applied Science) (4 °C). After centrifugation to remove insoluble material, supernatants were collected, and protein concentrations were determined using the Bradford method (22) (Sigma-Aldrich). Lysates containing a mixture of endogenous phosphatases were diluted to 1 mg/ml and incubated on ice or at 30 °C for 1 h in the absence or presence of 1 mm MgCl2 (to re-activate PP1 (23)) and/or 100 nm calyculin A and/or 1 mm EGTA or 10 mm CaCl2. In addition, the lysate was incubated at 30 °C for 1 h as an undiluted solution (3.3 mg/ml), diluted 1/5 in lysis buffer (0.67 mg/ml), or diluted 1/25 (0.13 mg/ml). All reactions were terminated by the addition of SDS loading buffer, and proteins were visualized by Western blotting.
Cell Culture—SH-SY5Y, HEK293 cells, and primary cortical neurons isolated from P1 rat pups were maintained as previously described (11). Cells were treated with 10 μm purvalanol or 2 μm CT99021 for the times indicated or the phosphatase inhibitors okadaic acid (10 nm or 1 μm), calyculin A (10 nm), cypermethrin (10 μm), and cyclosporin A (10 μm) for up to 45 min (longer periods caused cell death). SH-SY5Y cells were incubated in serum-free Dulbecco's modified Eagle's medium overnight before the addition of 50 ng/ml IGF1. When IGF1 and inhibitors were combined, cells were pretreated with okadaic acid (10 nm), EGTA (10 mm), BAPTA-AM (10 μm), or calyculin A (10 nm) for at least 15 min, then 50 ng/ml IGF1 was added for a further 30 min. For transfection experiments, SH-SY5Y or HEK293 cells were grown in 6-cm dishes and transfected with 4 μg (total) of cDNA using Lipofectamine2000 according to the manufacturer's instructions (Invitrogen). Immunoprecipitation experiments were performed by incubating lysates with 30 μl of a 50% (v/v) slurry of anti-FLAG-agarose or microcystin-Sepharose beads (2 h, 4 °C). After extensive washing, SDS loading buffer was added to the beads, and interacting proteins or CRMP2 phosphorylation levels were detected by Western blotting.
Co-immunoprecipitation in Vitro—Equal amounts (2.5 μg) of recombinant PP1 and CRMP2 (non-phosphorylated or pre-phosphorylated by Cdk5 and GSK3β) were incubated in 150 μl of kinase buffer containing 100 nm calyculin A for 10 min at room temperature followed by 1 h at 4 °C in the presence of 25 μl of a 50% slurry of glutathione-Sepharose. The beads were washed four times, then analyzed by Western blotting.
Gel Filtration Chromatography—Rat brain lysate (0.5 ml, 6 mg/ml) was loaded onto a Superdex™ 200 HR 10/300 column equilibrated in buffer (50 mm Tris, pH 7.4, 0.03% (v/v) Brij-35, 5% (v/v) glycerol, 0.15 m NaCl, 50 mm NaF, and 5 mm Na4P2O7), and proteins were separated at 0.4 ml/min. Fractions (0.5 ml) were collected and analyzed by immunoblotting. Fractions containing CRMP2 and CRMP4 were identical (22-24). These were pooled, and protein concentrations were measured before snap-freezing in liquid nitrogen.
RESULTS
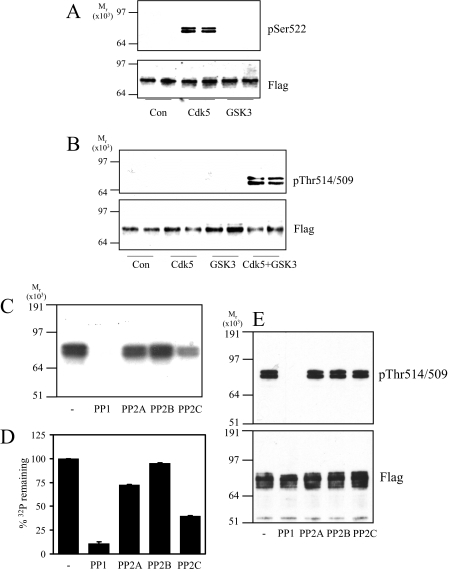
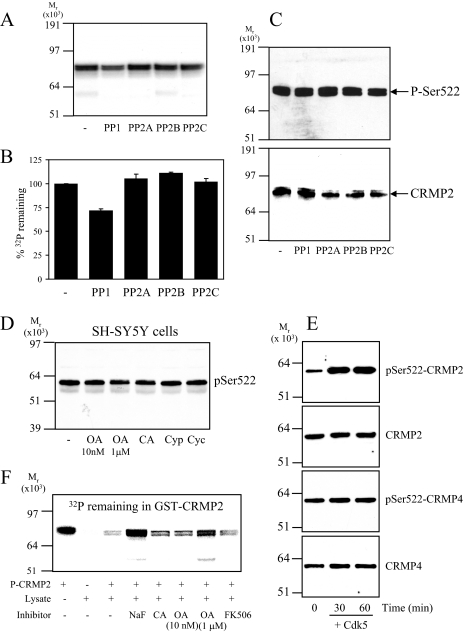
PP1 Antagonizes GSK3 Phosphorylation of CRMP2—To determine the class of phosphatase(s) that most efficiently removes phosphate from Ser-518/Thr-514/Thr-509 of CRMP2, recombinant CRMP2 was first phosphorylated at Ser-522 (primed) by Cdk5 in the presence of unlabeled ATP (Fig. 1A). GSK3 did not phosphorylate CRMP2 at Ser-522 (Fig. 1A), whereas CDK5 did not phosphorylate Thr-514/Thr-509 to any great extent (Fig. 1B). Subsequent incubation of the primed CRMP2 by GSK3β resulted in phosphorylation of Ser-518/Thr-514/Thr-509 (Fig. 1B). Previously we showed that GSK3 will not incorporate phosphate into a mutant CRMP2-S522A (11). Therefore, incubation of primed CRMP2 with radiolabeled [γ-32P]ATP, GSK3, and a Cdk5 inhibitor ensured that only Ser-518/Thr-514/Thr-509, and not the priming site (Ser-522) or any other residues on CRMP2, were radiolabeled. Equal activities (50 milliunits) of a representative of each of the four major classes of Ser/Thr protein phosphatases (PP1, PP2A, PP2B, and PP2C) were incubated with this radiolabeled CRMP2, and the amount of 32P remaining in CRMP2 was visualized by autoradiography (Fig. 1C) and measured by Cerenkov counting (Fig. 1D). In addition, release of phosphate specifically from these residues was assessed by immunoblot (Fig. 1E). CRMP2 was completely dephosphorylated by PP1, partially by PP2A and PP2C, and not at all by PP2B.
FIGURE 1.
Dephosphorylation of recombinant CRMP2 by phosphatases in vitro. A, GST-FLAG-CRMP2 was incubated alone (Con) or with Cdk5 or GSK3β in the presence of unlabeled ATP. Phosphorylation was detected using an antibody that specifically recognizes CRMP2 when phosphorylated at Ser-522 (upper panel) or an antibody that recognizes the FLAG tag (lower panel). B, GST-FLAG-CRMP2 was incubated alone (Con) or with Cdk5, GSK3β, or Cdk5 plus GSK3β in the presence of unlabeled ATP. Phosphorylation was detected using an antibody that specifically recognizes CRMP2 when phosphorylated at Thr-514/509 (upper panel) and a FLAG tag antibody (lower panel). C, GST-FLAG-CRMP2 was primed by Cdk5 in the presence of unlabeled ATP followed by incubation with GSK3β and [γ-32P]ATP. The [γ-32P]CRMP2 was then incubated with 50 milliunits of each of the phosphatases listed for 1 h. After SDS-PAGE, [γ-32P]CRMP2 was visualized by autoradiography. D, the amount of phosphate remaining in CRMP2 in C was measured by Cerenkov counting of the relevant gel pieces and is presented as a percentage of control (no phosphatase added) (n = 3). E, GST-FLAG-CRMP2 was phosphorylated by Cdk5 and GSK3β in the presence of unlabeled ATP, then incubated with 50 milliunits of the phosphatases listed for 1 h. The extent of dephosphorylation was assessed using antibodies to Thr(P)-514/509 (upper panel) and FLAG tag (lower panel) (n = 2).
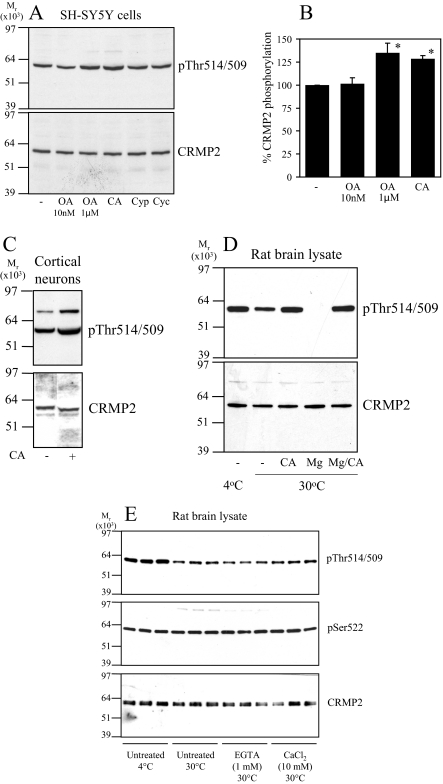
Next, SH-SY5Y neuroblastoma cells were treated with inhibitors of each class of phosphatase, and CRMP2 phosphorylation was monitored by immunoblot. A low concentration of okadaic acid (OA, 10 nm) inhibits PP2A, whereas a high concentration (1 μm) inhibits both PP2A and PP1 (dissociation constants (Ki) of 0.032 and 147 nm, respectively (24, 25)). Calyculin A (CA, 10 nm) inhibits both PP1 and PP2A, whereas cyclosporin A and cypermethrin are PP2B inhibitors. Treatment of SH-SY5Y cells with these inhibitors for 30 min (longer periods induced cell death) revealed that phosphorylation of CRMP2 at the GSK3 sites was significantly increased by treatment with CA and a high concentration of OA, but not a low concentration of OA, cyclosporin A, or cypermethrin (Fig. 2). The basal phosphorylation of CRMP2 was also increased by CA treatment of primary rat cortical neurons (Fig. 2C). Incubation of a rat brain lysate at 30 °C induced partial dephosphorylation of endogenous CRMP2 by endogenous phosphatases, which was blocked by co-incubation with CA (Fig. 2D). Dephosphorylation of CRMP2 was enhanced by addition of Mg2+ (promotes PP1 activity (23)), and this was also blocked by CA. The rate of dephosphorylation was not influenced by the presence of EGTA (a calcium chelator) or the addition of exogenous calcium (Fig. 2E). Together, these observations indicate that the major phosphatase that dephosphorylates CRMP2 at the sites targeted by GSK3 in vitro and in cells is a PP1 family member.
FIGURE 2.
Dephosphorylation of CRMP2 in cells. A, SH-SY5Y neuroblastoma cells were incubated with DMSO (control), okadaic acid (10 nm or 1 μm), calyculin A (CA, 10 nm), cypermethrin (Cyp, 10 μm), or cyclosporin A (Cyc, 10 μm) for 30 min. Cell lysates were subjected to Western blot analysis using Thr(P)-514/509 (upper panel) and total CRMP2 (lower panel) antibodies. The ratio of phospho-CRMP2/total CRMP2 in cells treated with okadaic acid or calyculin A compared with control cells is presented in B (n = 3; *, p < 0.05 relative to control (Students t test); average ± S.D.). C, primary rat cortical neurons were treated without or with calyculin A (10 nm), then cell lysates were immunoblotted as described in A. D, rat brain lysate was left on ice (lane 1) or incubated at 30 °C for 1 h (lanes 2-5) in the presence of 10 nm calyculin A (lane 3), 1 mm MgCl2 (lane 4), or 10 nm calyculin A plus 1 mm MgCl2 (lane 5). The lysates were immunoblotted as described in A. E, same as D, except lysates were incubated in the presence of 1 mm EGTA or 10 mm CaCl2 as shown.
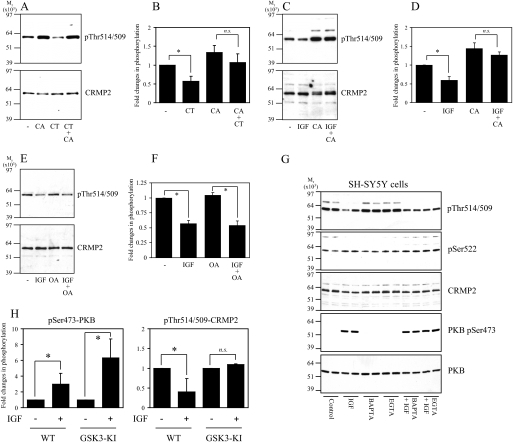
Next, SH-SY5Y cells were incubated with CT99021, a highly selective inhibitor of GSK3, or with IGF1, an established physiological inhibitor of GSK3 activity and CRMP2 phosphorylation (14), in the absence and presence of phosphatase inhibitors. Dephosphorylation of CRMP2 in response to CT99021 was blocked by co-incubating cells with CA (Fig. 3, A and B). Similarly, dephosphorylation of CRMP2 in response to IGF1 was blocked by CA (Fig. 3, C and D) but not a low concentration of OA (Fig. 3, E and F). Finally, exposing cells to extracellular (EGTA) or intracellular (BAPTA-AM) calcium chelators did not alter basal CRMP2 phosphorylation or the response of CRMP2 and protein kinase B phosphorylation to IGF1 (Fig. 3G). In theory, dephosphorylation of CRMP2 after IGF1 treatment of cells could be mediated by inactivation of GSK3 and/or activation of PP1. To investigate this further, primary cortical neurons were isolated from GSK3 “knock-in” mice, in which the regulatory phosphorylation sites on GSK3α (Ser-21) and GSK3β (Ser-9) were changed to alanine; thus, GSK3 could no longer be inhibited by IGF1 signaling (26). These were stimulated with IGF1, and the phosphorylation of CRMP2 was measured. In contrast to wild type neurons, the phosphorylation of CRMP2 was unaltered in the IGF1-treated GSK3-KI neurons despite efficient activation of the phosphatidylinositol 3-kinase-protein kinase B pathway, as shown by robust phosphorylation of protein kinase B (PKB) at Ser-473 (Fig. 3H). The same is observed after BDNF stimulation (data not shown). This demonstrates that dephosphorylation of CRMP2 (at 518/514/509) in neurons treated with growth factors results from inactivation of GSK3 and requires only basal activity of PP1.
FIGURE 3.
CT99021- and IGF1-induced dephosphorylation of CRMP2 requires PP1 activity. A, SH-SY5Y cells were incubated without (lanes 1 and 3) or with 10 nm calyculin A (lanes 2 and 4) for 15 min followed by the addition of 2 μm CT99021 (CT) for a further 30 min. Cell lysates were subjected to Western blot analysis using Thr(P)-514/509 (upper panel) and total CRMP2 (lower panel) antibodies. B, the ratio of phospho-CRMP2/total CRMP2 in cells from A (n = 3; n.s., not significant; *, p < 0.05 relative to control (Students t test); average ± S.D.). C, SH-SY5Y cells were incubated without (lanes 1 and 2) or with 10 nm calyculin A (lanes 3 and 4) for 15 min followed by the addition of 50 ng/ml IGF1 (lanes 2 and 4) for a further 30 min. Cell lysates were immunoblotted as described in A. D, the ratio of phospho-CRMP2/total CRMP2 in cells from D (n = 3). E and F, same as C and D except the calyculin A was replaced by 10 nm okadaic acid. G, same as C, except cells were treated in the absence or presence of IGF1 with 10 μm BAPTA-AM or 1 mm EGTA. PKB, protein kinase B. H, primary cortical neurons from wild type or GSK3-knock-in (KI) mice were stimulated with or without 100 ng/ml IGF1 for 1 h, then lysates were subjected to Western blot analysis as described in A. In addition, the membranes were incubated with antibodies that specifically recognize protein kinase B when phosphorylated at Ser-473 or an antibody that recognizes phosphorylated and unphosphorylated protein kinase B equally well (loading control). The phosphorylation level of protein kinase B and CRMP2 is presented relative to unstimulated controls after correction for loading (average ± S.D.; n.s., not significant; *, p < 0.002 relative to control (Student's t test); primary neuron preparations from at least seven different animals for each genotype). WT, wild type.
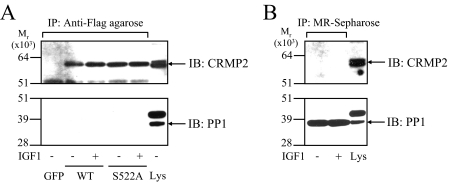
CRMP2 and PP1 Do Not Form a Stable Complex in Cells—Dephosphorylation of CRMP2 in IGF1-treated neurons was rapid (<10-15 min), so we hypothesized that PP1 might form a stable complex with CRMP2 to result in such a rapid dephosphorylation upon inactivation of GSK3. SH-SY5Y cells were transfected with FLAG-tagged CRMP2, then lysates were incubated with anti-FLAG-agarose to isolate FLAG-CRMP2 and any interacting proteins. PP1 was not detectable in the precipitates (Fig. 4A) in the presence or absence of IGF1 or upon expression of wild type or a non-phosphorylatable mutant, CRMP2-S522A (Fig. 4A). Similarly, non-transfected SH-SY5Y lysates treated with or without IGF1 were incubated with microcystin-Sepharose beads to isolate endogenous PP1 (as well as PP2A), and CRMP2 was not detectable in these precipitates (Fig. 4B). In a separate experiment, recombinant GST-CRMP2 (phosphorylated or dephosphorylated) and His6-PP1 were co-incubated in vitro then isolated using glutathione-Sepharose (CRMP2) or Ni+-Sepharose (PP1). Reciprocal pulldown analysis confirmed that there was no significant direct interaction between these two proteins irrespective of phosphorylation (supplemental Fig. 1). In addition, incubation of various dilutions of rat brain lysate at 30 °C resulted in different rates of dephosphorylation of endogenous CRMP2. If PP1 was part of a stable complex with CRMP2, the amount of CRMP2 dephosphorylation should not correlate with dilution factor. However, upon the addition of Mg2+ there was significantly greater dephosphorylation of CRMP2 at low (1/1) dilution compared with a highly diluted (1/25) lysate (supplemental Fig. 2). Together, these results show that CRMP2 and PP1 do not form a stable complex in rat brain lysate in vitro or in cells.
FIGURE 4.
PP1 does not form a stable complex with CRMP2. A, SH-SY5Y cells were transfected with equivalent amounts of cDNA encoding green fluorescent protein (GFP), FLAG-CRMP2 (WT), or FLAG-CRMP2-S522A. After 16 h the cells were serum-starved for 8 h, then treated without or with 50 ng/ml IGF1 for 15 min. The CRMP2 proteins were immunoprecipitated (IP), then subjected to Western blot (IB) analysis using total CRMP2 (upper panel) or PP1 (lower panel) antibodies. Lys denotes total SH-SY5Y transfected lysate. B, non-transfected SH-SY5Y cell lysates treated without or with IGF1 were incubated with microcystin-Sepharose to pull down endogenous PP1, then subjected to Western blot analysis as in A.
Relative Resistance of Ser-522 to Dephosphorylation—To identify the phosphatase that mediates dephosphorylation of the priming site on CRMP2 (Ser-522), dephosphorylation assays were performed in vitro as described for the GSK3 phosphorylation site analysis above. A comparatively small release of 32P from CRMP2 was achieved by PP1 (∼25%) but no release by any other phosphatase (Fig. 5, A and B). Immunoblot analysis using an antibody that specifically recognizes CRMP2 when phosphorylated at Ser-522 confirmed that only a small release of phosphate from Ser-522 could be obtained by PP1, and there was no effect of other phosphatases (Fig. 5C). Treatment of SH-SY5Y cells with phosphatase inhibitors did not significantly alter the phosphorylation level of endogenous CRMP2 at Ser-522 (Fig. 5D). A possible explanation is that Ser-522 is maximally phosphorylated in cells. To check this, CRMP was partially purified from a rat brain lysate (in the presence of phosphatase inhibitors). Fractions containing CRMP2 (as well as CRMP4) were pooled and incubated with or without Cdk5 and MgATP. Cdk5 could phosphorylate the purified CRMP2 but not CRMP4 in CRMP purified by ion exchange (Fig. 5E) or CRMP purified by size exclusion (data not shown). The increase in phosphorylation of Ser-522 upon incubation with Cdk5 rules out the possibility that CRMP2 is fully phosphorylated in vivo. However, CRMP4 phosphorylation did not increase upon incubation with Cdk5 despite being an equally good substrate for Cdk5 as CRMP2 in vitro (14). This suggests that CRMP4 is fully phosphorylated at Ser-522 in vivo.
FIGURE 5.
Dephosphorylation of Ser-522 of CRMP2. A, GST-CRMP2 was phosphorylated by Cdk5 in the presence of radiolabeled [γ-32P]ATP, then incubated for 1 h with 50 milliunits of each of the phosphatases listed. After SDS-PAGE, radiolabeled bands were visualized by autoradiography. B, the amount of phosphate remaining in GST-CRMP2 in A was detected by Cerenkov counting (n = 3). C, GST-FLAG-CRMP2 was phosphorylated by Cdk5 in the presence of unlabeled ATP, then incubated as in A. The extent of dephosphorylation was detected by Western blotting using Ser(P)-522 (upper panel) and total CRMP2 (lower panel) antibodies. D, SH-SY5Y cells were incubated with DMSO (control), okadaic acid (10 nm or 1 μm), calyculin A (10 nm), cypermethrin (10 μm), or cyclosporin (10 μm) for 30 min. Cell lysates were subjected to Western blot analysis using Ser(P)-522 antibody (for a total CRMP2 loading control, see Fig. 2A, lower panel). E, rat brain lysate (10 mg) was fractionated by anion-exchange chromatography, then CRMP2/4-containing fractions were pooled and incubated without or with Cdk5 in an in vitro kinase assay for 30 or 60 min. Reactions were terminated by the addition of SDS buffer and subjected to Western blot analysis. Membranes were probed with antibodies that recognize CRMP2 or -4 when phosphorylated at Ser-522 (Ser(P)-522) or total CRMP2 and 4 antibodies. A representative blot of two experiments is shown. F, GST-CRMP2 was phosphorylated by Cdk5 in the presence of [γ-32P]ATP, then incubated with rat brain cell lysate in the absence or presence of various phosphatase inhibitors for 3 h at 30 °C. After SDS-PAGE, the amount of radiolabeled phosphate remaining in GST-CRMP2 was visualized by phosphorimaging. A representative of two experiments is shown.
Next, a rat brain lysate prepared in the absence of phosphatase inhibitors (containing endogenous phosphatases) was incubated with GST-CRMP2 (radiolabeled at Ser-522) for 3 h. This resulted in a significant release of phosphate, which could be completely blocked by the nonspecific phosphatase inhibitor NaF, partially blocked by PP1/PP2A inhibitors OA and CA, and not blocked at all by the PP2B inhibitor FK506 (Fig. 5F). This suggests that several different types of phosphatase are able to dephosphorylate Ser-522 in a cell lysate (including PP1), albeit relatively weakly. Consistent with this hypothesis, fractionation of a rat brain lysate by gel filtration followed by incubation of the fractions with Ser(P)-522-CRMP2 revealed multiple fractions that could weakly dephosphorylate Ser-522 during prolonged incubation in vitro (data not shown).
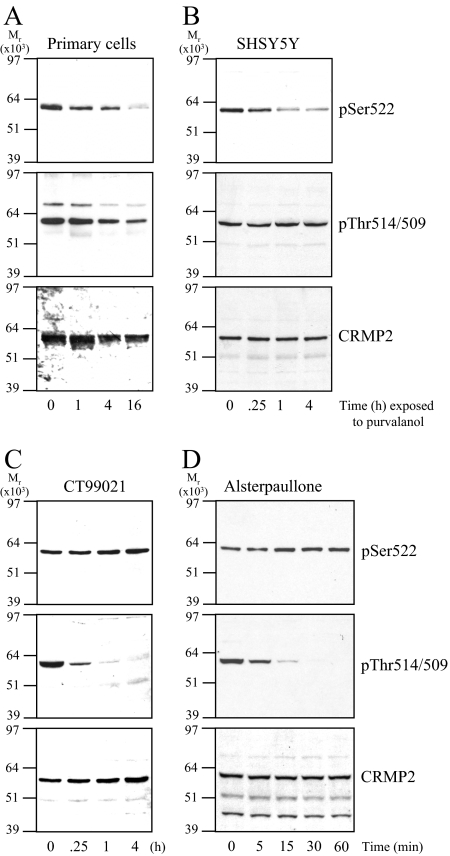
To examine whether Ser-522 could be dephosphorylated in intact rat primary cortical neurons, the cells were incubated with the Cdk5 inhibitor purvalanol, and the phosphorylation of CRMP2 was measured. Purvalanol treatment caused a gradual decrease in Ser-522 and Thr-514/509 levels without altering the CRMP2 protein levels (Fig. 6A). In SH-SY5Y cells, purvalanol treatment reduced phospho-Ser-522 levels and not CRMP2 protein levels (Fig. 6B). As expected, treatment of SH-SY5Y cells with the GSK3 inhibitor CT99021 rapidly decreased phospho-Thr-514/509 levels without affecting phospho-Ser-522 (Fig. 6C). Similarly, treatment of SH-SY5Y cells with 10 μm alsterpaullone, a near equipotent inhibitor of Cdk5 and GSK3 (27, 28), caused rapid dephosphorylation of Thr-514/509 without affecting phospho-Ser-522 (Fig. 6D). This demonstrated that although there were phospho-Ser-522 phosphatases in intact cells, this residue was relatively resistant to dephosphorylation when compared with phospho-Thr-509/514.
FIGURE 6.
Inhibition of Cdk5 reduces CRMP2 phosphorylation at Ser-522 in intact cells. A, primary rat cortical neurons were treated with 10 μm purvalanol for the times indicated. Cell lysates were subjected to Western blot analysis using Ser(P)-522 (upper panel), Thr(P)-514/509 (middle panel), and total CRMP2 (lower panel) antibodies. B, same as A, except SH-SY5Y cells were treated with purvalanol for the times indicated. C, same as B, except the SH-SY5Y cells were treated with 2 μm CT99021. D, same as B, except SH-SY5Y cells were treated with 10 μm alsterpaullone. In all cases representative blots of two experiments are shown.
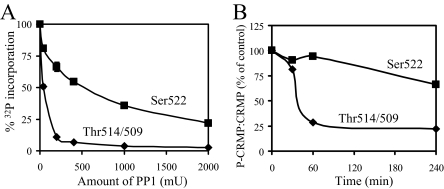
To analyze this further, we directly compared PP1-mediated dephosphorylation of CRMP2 at the Cdk5 versus GSK3 sites in vitro. Fig. 7A shows that whereas the GSK3 phosphorylation sites are almost completely dephosphorylated by 200 milliunits of PP1, Ser-522 was dephosphorylated only 35%. Even 10 times the amount of PP1 failed to completely dephosphorylate the Ser-522 site. In a separate experiment, incubation of a rat brain lysate (containing endogenous CRMP2 and phosphatases) at 30 °C caused rapid dephosphorylation of the GSK3 sites but comparatively slow dephosphorylation of the Cdk5 site (Fig. 7B). These results clearly demonstrate the relative resistance of the Ser-522 site to dephosphorylation.
FIGURE 7.
Ser(P)-522 is more resistant to dephosphorylation than Thr(P)-514/509. A, CRMP2 was phosphorylated by Cdk5 in the presence of [γ-32P]ATP (▪) or unlabeled ATP followed by GSK3β in the presence of [γ-32P]ATP (♦), then each was incubated for 1 h with PP1 at the concentrations indicated. The amount of radiolabeled phosphate remaining in CRMP2 was detected by Cerenkov counting of appropriate gel slices, and the average percentage of phosphate remaining is shown (n = 3). B, rat brain lysate was incubated at 30 °C for the times shown. Dephosphorylation of endogenous CRMP2 was determined by Western blotting using Ser(P)-522 (Cdk5 site), Thr(P)-514/509 (GSK3 sites), and total CRMP2 antibodies. After densitometric analysis, the ratio of phospho:dephospho CRMP2 was determined, and the average of two separate experiments is shown.
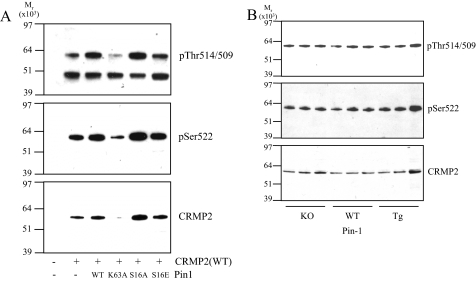
It is possible that another factor within intact cells is required for efficient dephosphorylation of the Ser-522 site. A prime candidate for such a role is Pin1. Pin1 is a peptidyl proline cis/trans isomerization enzyme that switches phosphorylated Ser/Thr-Pro motifs between cis and trans conformations (for review, see Ref. 29). This can induce subsequent dephosphorylation by phosphatases (30, 31). Reported Pin1 substrates include other AD-related proteins such as Tau (32) and β-amyloid precursor protein (33). HEK293 cells were co-transfected with CRMP2 and either wild type Pin1, an inactive mutant of Pin1 (K63A), or Pin1 mutants that display constitutive (S16A) or reduced (S16E) binding affinity for substrates (34). After 24 h, CRMP2 was immunoprecipitated using anti-FLAG-agarose and the precipitates analyzed for phosphorylation. Fig. 8A shows that co-transfection of Pin1 had variable affects on CRMP2 expression (lower panel); however, relative phosphorylation of Ser-522 (upper panel) and Thr-514/509 (middle panel) was not affected by any Pin1 construct; that is, it mirrored total CRMP2 levels (lower panel). Next, the basal level of CRMP2 phosphorylation was investigated in the cortex of mice that do not express Pin1 (knock-out (KO) (35)) or express higher than normal levels of Pin1 (transgenic over-expressor (Tg) (36)). There was no significant change in Ser-522 or Thr-514/509 phosphorylation in either Pin1-KO or Pin1-Tg mice compared with wild type littermates (Fig. 8B). Finally, in vitro phosphatase assays of Cdk5-phosphorylated CRMP2 were performed using PP1 or PP2A in the presence or absence of wild type Pin1. Pin1 was included at 2 different concentrations, either 0.4 μm (4 times less than the CRMP2 concentration, 1.5 μm) or at 6 μm (4 times more than CRMP2). We have previously found that Pin1 can associate with CRMP4 (37); therefore, these experiments were also performed using Cdk5-phosphorylated CRMP4. There was very little dephosphorylation of Ser-522 on either CRMP2 or CRMP4 using 100 milliunits of phosphatase for up to 1 h (suboptimal conditions). The addition of Pin1 did not accelerate the rate of dephosphorylation of either CRMP isoform (supplemental Fig. 3). Together, these experiments conclude that Pin1 has no affect on the rate of dephosphorylation of Ser-522.
FIGURE 8.
Pin1 does not regulate Ser-522 phosphorylation. A, HEK293 cells were co-transfected with FLAG-CRMP2 and wild type (WT) or mutant Pin1 as indicated. After 24 h, CRMP2 was isolated from lysates using anti-FLAG-agarose and subjected to Western blot analysis, and membranes were probed with Thr(P)-514/509 (upper panel), Ser(P)-522 (middle panel), or total CRMP2 (lower panel) antibodies. B, lysates from wild type, Pin1-knock-out (KO), and Pin1-OE (over-expressor) cortex were subjected to Western blot analysis, and membranes were probed as in A.
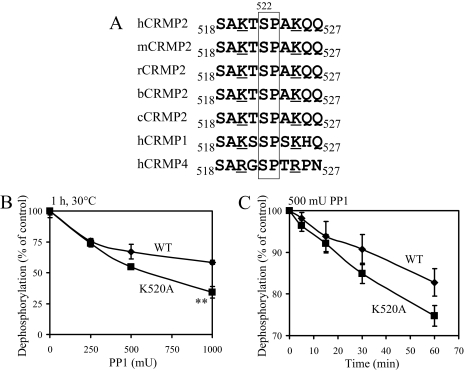
Interestingly, highly conserved and positively charged lysine residues are found both N- and C-terminal to Ser-522 (Lys-520 and Lys-525, Fig. 9A). These may help to stabilize the negatively charged phosphate at this site and protect it from dephosphorylation by phosphatases. Therefore, Lys-520, Lys-525, or both were mutated to alanine, and the rate of dephosphorylation by PP1 was compared with wild type CRMP2 in vitro. Mutation of Lys-520 had no affect on the rate of phosphorylation of Ser-522 by Cdk5 (data not shown); however, it caused an increase in the rate of dephosphorylation by PP1 in dose-response (Fig. 9B) and time-course (Fig. 9C) studies. In contrast, mutation of Lys-525 completely inhibited phosphorylation of Ser-522 by Cdk5; consequently, the phosphatase assays could not be performed. Nevertheless, results from the Lys-520 experiments suggest that removal of a basic residue close to Ser-522 renders it more susceptible to dephosphorylation in vitro.
FIGURE 9.
Lys-520 stabilizes phosphorylation at Ser-522. A, sequence alignment of residues 518-527 of CRMP2 from different species as well as CRMP1 and CRMP4 (h, human; m, mouse; r, rat; b, bovine; c, chick). Underlined residues show the highly conserved basic amino acids. Residues enclosed in the box are the Ser-522 phosphorylation site and proline 523, which are essential for phosphorylation of Ser-522. B, wild type (WT, ♦) and K520A (▪) GST-CRMP2 were phosphorylated in vitro by Cdk5 in the presence of [γ-32P]ATP, then incubated with different amounts of PP1 for 30 min or for different times with 500 milliunits PP1 (C). The amount of 32P remaining in CRMP2 was measured by Cerenkov counting, and the average percentage of phosphate remaining is shown (n = 3, **, p = 0.0013 K520 compared with wild type). mU, milliunits.
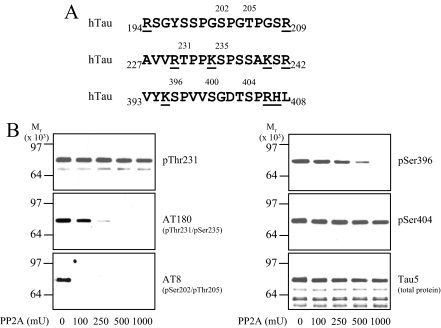
Cdk5 Phospho-sites in Tau Are Resistant to Dephosphorylation by Phosphatases—CRMP2 and Tau share a number of similarities; for example, they are both brain-enriched microtubule-binding proteins that are phosphorylated by Cdk5 and GSK3, both display abnormally high phosphorylation in AD, and both are constituents of neurofibrillary tangles (for review, see Ref. 38). We noticed that two proline-directed phospho-sites in Tau have a basic residue C-terminal that might contribute to reduce the rate of dephosphorylation; Thr(P)-231 (RTPPK) and Ser(P)-404 (DTSPRH) (Fig. 10A). Like Ser-522 in CRMP2, these are among the sites in tau reported to be hyperphosphorylated in AD (39). GST-Tau was phosphorylated by a combination of Cdk5 and GSK3, then after the addition of alsterpaullone to inhibit the kinases, different amounts of PP2A (the major Tau phosphatase (40)) were added (Fig. 10B). The reactions were then subjected to immunoblot analysis using antibodies that recognize Tau when phosphorylated at the sites mentioned above (Thr(P)-231, Ser(P)-404) as well as other proline-directed phospho-sites (Ser(P)-396, AT8 (p202/205)) and total tau (Tau5). The AT180 antibody recognizes Tau when phosphorylated at Thr-231, although this is influenced by phosphorylation at Ser-235 (41). PP2A was able to effectively remove phosphate from Ser(P)-396 and Ser(P)-202/-205. However, Ser(P)-404 was completely resistant to PP2A treatment, even up to 1000 milliunits PP2A. In addition, the sensitivity to the AT180 antibody, whose optimal epitope requires phosphorylation at both Thr-231 and Ser-235, was reduced by PP2A, whereas the Thr(P)-231 antibody showed no change in Thr-231 phosphorylation. This suggests that phosphate was removed from Ser-235 by PP2A but not Thr-231. Together, these results show that proline-directed phosphorylation sites in Tau that have a basic residue two or three amino acids C-terminal to the phosphorylation site display relatively high resistance to dephosphorylation.
FIGURE 10.
Phospho-sites on Tau preceded by basic residues display resistance to dephosphorylation by PP2A. A, amino acid sequences surrounding proline-directed phospho-sites (numbered) in human (h) Tau. Basic residues are underlined. B, wild type GST-Tau was phosphorylated in vitro by Cdk5 and GSK3β, then different amounts of PP2A were added for 1 h. Reactions were terminated by the addition of SDS loading buffer, then subjected to Western blot analysis using a total Tau antibody (Tau-5) and several phospho-specific Tau antibodies as indicated. mU, milliunits.
DISCUSSION
We have previously shown that CRMP2 is hyperphosphorylated in brain tissue from human AD patients and some mouse models of AD at residues that influence its known physiological functions (16). Although phosphorylation is increased at both the Cdk5 and GSK3 sites, we were unable to detect significant increases in Cdk5 or GSK3 activity in human AD tissue or mouse models of AD (16). This suggests that the increased phosphorylation of Ser-522 that drives hyperphosphorylation of CRMP2 in AD is not caused by dramatic increases in Cdk5 activity. Therefore, we have investigated the molecular mechanism of dephosphorylation of CRMP2. PP1-type enzymes were found to be the major phosphatase class mediating dephosphorylation of the GSK3 sites (Ser-518/Thr-514/Thr-509), whereas the specific enzyme responsible for the dephosphorylation of the Cdk5 site (Ser-522) remains elusive. Indeed it remains possible that Ser-522 may be targeted by several different phosphatases, albeit weakly, or by a rare as yet unidentified phosphatase.
Inhibition of GSK3 activity in neurons by growth factors or a specific pharmacological inhibitor resulted in rapid dephosphorylation of Thr-514/509, and this required PP1 activity. In contrast, pharmacological inhibition of Cdk5 caused a very slow rate of dephosphorylation that was incomplete, even after prolonged treatment. This highlights a major difference in the stability of the different phosphorylation sites in cells whereby GSK3 sites are dynamic with a very short half-life and are, thus, more likely to be transiently regulated. Indeed, the Cdk5 site might not be acutely regulated. Instead, it might serve to ensure reliable priming for subsequent GSK3 phosphorylation, which can be rapidly regulated by various stimuli, including growth factor signaling. Alternatively, the amount of priming by Cdk5 could serve to dictate the pool of CRMP2 available to be phosphorylated by GSK3, thus demarcating a proportion of CRMP2 that is regulated by a particular stimulus or involved in a certain cellular process (e.g. microtubule regulation).
The difference in susceptibility to phosphatases between Thr(P)-509 and Ser(P)-522 is not due to preferential binding of PP1 to CRMP. Instead, the relative resistance of Ser(P)-522 to dephosphorylation must be inherent to the sequence immediately surrounding the phosphorylation site. This is supported by our observation that a short peptide of this region that was phosphorylated by Cdk5 showed the same resistance to phosphatase treatment as the full-length protein (data not shown). Examination of the primary amino acid sequence surrounding Ser-522 showed a strict conservation of basic residues both N- and C-terminal to the phosphorylation site in CRMP1, CRMP2, and CRMP4 and in different species (Fig. 9A). We hypothesized that the positive charge on these lysine and arginine residues stabilizes the negative charge on Ser(P)-522. Indeed, mutation of Lys-520 rendered Ser(P)-522 more susceptible to dephosphorylation by PP1 (Fig. 9, B and C) without affecting the rate of Ser-522 phosphorylation by Cdk5 in vitro. In contrast, mutation of Lys-525 to Ala completely inhibited phosphorylation by Cdk5. Consequently, it was not possible to establish the relative rate of dephosphorylation of the K525A mutant. These observations are consistent with previous studies showing that a C-terminal basic residue at the +2 or +3 position is a defining feature of Cdk5 phosphorylation consensus sequences (42). Because mutation of Lys-520 to Ala increased the dephosphorylation rate only modestly, it is likely that the majority of the stabilizing effect upon Ser(P)-522 is provided by the C-terminal basic residue (Lys-525). If this is the case, then the C-terminal basic residue at the +2 or +3 position in all Cdk5 substrates is important for both substrate recognition by the kinase and resistance to the phosphatase; hence, relative resistance to phosphatases will be a common feature of many Cdk5 substrates. Supporting this, we demonstrate that proline-directed phosphorylation sites in Tau that contain a basic residue at the +2 or +3 position (Thr-231, Ser-404) have resistance to PP2A, whereas other proline-directed phospho-sites (Ser(P)202/Thr(P)-205, Ser(P)-235, and Ser(P)-396) do not (Fig. 10). In addition, treatment of cells with pharmacological inhibitors of Cdk5, such as purvalanol and roscovitine, results in either no change or weak dephosphorylation of bona fide Cdk5 substrates, almost certainly due to this relative resistance to dephosphorylation. This has major implications for programs investigating Cdk5 as a therapeutic target, as prolonged and substantial inhibition may be required for significant dephosphorylation of substrates.
Both CRMP2 and Tau are hyperphosphorylated at Cdk5 sites in AD, implicating increased Cdk5 activity as part of the disease process (see Ref. 43, although this has been disputed (17, 18)). We were unable to detect any significant changes in Cdk5 activity in tissue from human AD patients or mouse models of AD (16). However, considering that Cdk5 phosphorylation sites are relatively resistant to phosphatases, a small change in Cdk5 activity would be sufficient to cause a relatively large, gradual increase in CRMP2 and Tau phosphorylation.
Interestingly, incubation of recombinant Cdk5 and MgATP with partially purified CRMP2 and CRMP4 from rat brain caused an increase in Ser-522 phosphorylation on CRMP2 but not CRMP4. This indicates that CRMP2 is not fully phosphorylated at Ser-522 in healthy brain tissue, whereas CRMP4 is fully phosphorylated. Therefore, increased phosphorylation of CRMP2 but not CRMP4 in AD tissue (16) may be explained by the stoichiometric phosphorylation of CRMP4 at Ser-522 in healthy tissue. Considering the relative resistance of these phosphorylation sites to phosphatases, it is unclear why CRMP2-Ser-522 is not also fully phosphorylated, but in AD brain this site becomes almost stoichiometrically phosphorylated.
In summary we have shown that PP1 rapidly dephosphorylates CRMP2 at Ser-518/Thr-514/Thr-509 upon inhibition of GSK3. It remains unclear which phosphatase dephosphorylates the Cdk5 priming site (Ser-522); however, this site has a very slow turnover rate due to the relatively high resistance to classical Ser/Thr phosphatases. This resistance is at least in part due to the presence of basic residues located nearby. Therefore, resistance to dephosphorylation is likely to be common to many Cdk5 substrates and common to at least two proteins that exhibit increased phosphorylation in AD.
Supplementary Material
Acknowledgments
We thank Prof. Carol MacKintosh and Prof. Tricia Cohen (College of Life Sciences, University of Dundee) for the supply of reagents and helpful advice. We also thank Louise Ross and Adam Brown (Neurosciences Institute, University of Dundee) for help in generating cortical neuron cultures and Jenny Bain (Division of Signal Transduction Therapy, University of Dundee) for supplying pharmacological inhibitors.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1AG022082 (to K. P. L.). This work was primarily supported by The Alzheimer's Research Trust Grant ART/PG/2005/1. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1-3.
Footnotes
The abbreviations used are: CRMP, collapsin response mediator protein; AD, Alzheimer disease; Cdk5, cyclin-dependent kinase 5; GSK3, glycogen synthase kinase 3; PP1, protein phosphatase 1; PP2A, protein phosphatase 2A; CA, calyculin A; OA, okadaic acid; GST, glutathione S-transferase; IGF1, insulin-like growth factor-1.
References
- 1.Goshima, Y., Nakamura, F., Strittmatter, P., and Strittmatter, S. M. (1995) Nature 376 509-514 [DOI] [PubMed] [Google Scholar]
- 2.Wang, L. H., and Strittmatter, S. M. (1996) J. Neurosci. 16 6197-6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukada, M., Watakabe, I., Yuasa-Kawada, J., Kawachi, H., Kuroiwa, A., Matsuda, Y., and Noda, M. (2000) J. Biol. Chem. 275 37957-37965 [DOI] [PubMed] [Google Scholar]
- 4.Wang, L. H., and Strittmatter, S. M. (1997) J. Neurochem. 69 2261-2269 [DOI] [PubMed] [Google Scholar]
- 5.Fukata, Y., Kimura, T., and Kaibuchi, K. (2002) Neurosci. Res. 43 305-315 [DOI] [PubMed] [Google Scholar]
- 6.Nishimura, T., Fukata, Y., Kato, K., Yamaguchi, T., Matsuura, Y., Kamiguchi, H., and Kaibuchi, K. (2003) Nat. Cell Biol. 5 819-826 [DOI] [PubMed] [Google Scholar]
- 7.Kimura, T., Watanabe, H., Iwamatsu, A., and Kaibuchi, K. (2005) J. Neurochem. 93 1371-1382 [DOI] [PubMed] [Google Scholar]
- 8.Brown, M., Jacobs, T., Eickholt, B., Ferrari, G., Teo, M., Monfries, C., Qi, R. Z., Leung, T., Lim, L., and Hall, C. (2004) J. Neurosci. 24 8994-9004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uchida, Y., Ohshima, T., Sasaki, Y., Suzuki, H., Yanai, S., Yamashita, N., Nakamura, F., Takei, K., Ihara, Y., Mikoshiba, K., Kolattukudy, P., Honnorat, J., and Goshima, Y. (2005) Genes Cells 10 165-179 [DOI] [PubMed] [Google Scholar]
- 10.Inagaki, N., Chihara, K., Arimura, N., Menager, C., Kawano, Y., Matsuo, N., Nishimura, T., Amano, M., and Kaibuchi, K. (2001) Nat. Neurosci. 4 781-782 [DOI] [PubMed] [Google Scholar]
- 11.Cole, A. R., Knebel, A., Morrice, N. A., Robertson, L. A., Irving, A. J., Connolly, C. N., and Sutherland, C. (2004) J. Biol. Chem. 279 50176-50180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimura, T., Kawano, Y., Arimura, N., Kawabata, S., Kikuchi, A., and Kaibuchi, K. (2005) Cell 120 137-149 [DOI] [PubMed] [Google Scholar]
- 13.Fukata, Y., Itoh, T. J., Kimura, T., Menager, C., Nishimura, T., Shiromizu, T., Watanabe, H., Inagaki, N., Iwamatsu, A., Hotani, H., and Kaibuchi, K. (2002) Nat. Cell Biol. 4 583-591 [DOI] [PubMed] [Google Scholar]
- 14.Cole, A. R., Causeret, F., Yadirgi, G., Hastie, C. J., McLauchlan, H., McManus, E. J., Hernandez, F., Eickholt, B. J., Nikolic, M., and Sutherland, C. (2006) J. Biol. Chem. 281 16591-16598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida, H., Watanabe, A., and Ihara, Y. (1998) J. Biol. Chem. 273 9761-9768 [DOI] [PubMed] [Google Scholar]
- 16.Cole, A. R., Noble, W., van Aalten, L., Plattner, F., Meimaridou, R., Hogan, D., Taylor, M., LaFrancois, J., Gunn-Moore, F., Verkhratsky, A., Oddo, S., LaFerla, F., Giese, K. P., Dineley, K. T., Duff, K., Richardson, J. C., Yan, S. D., Hanger, D. P., Allan, S. M., and Sutherland, C. (2007) J. Neurochem. 103 1132-1144 [DOI] [PubMed] [Google Scholar]
- 17.Tandon, A., Yu, H., Wang, L., Rogaeva, E., Sato, C., Chishti, M. A., Kawarai, T., Hasegawa, H., Chen, F., Davies, P., Fraser, P. E., Westaway, D., and George-Hyslop, P. H. (2003) J. Neurochem. 86 572-581 [DOI] [PubMed] [Google Scholar]
- 18.Taniguchi, S., Fujita, Y., Hayashi, S., Kakita, A., Takahashi, H., Murayama, S., Saido, T. C., Hisanaga, S., Iwatsubo, T., and Hasegawa, M. (2001) FEBS Lett. 489 46-50 [DOI] [PubMed] [Google Scholar]
- 19.Swatton, J. E., Sellers, L. A., Faull, R. L., Holland, A., Iritani, S., and Bahn, S. (2004) Eur. J. Neurosci. 19 2711-2719 [DOI] [PubMed] [Google Scholar]
- 20.Pei, J. J., Tanaka, T., Tung, Y. C., Braak, E., Iqbal, K., and Grundke-Iqbal, I. (1997) J. Neuropathol. Exp. Neurol. 56 70-78 [DOI] [PubMed] [Google Scholar]
- 21.Lu, K. P., Hanes, S. D., and Hunter, T. (1996) Nature 380 544-547 [DOI] [PubMed] [Google Scholar]
- 22.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 23.Yang, S. D., Vandenheede, J. R., and Merlevede, W. (1981) FEBS Lett. 126 57-60 [DOI] [PubMed] [Google Scholar]
- 24.Takai, A., Sasaki, K., Nagai, H., Mieskes, G., Isobe, M., Isono, K., and Yasumoto, T. (1995) Biochem. J. 306 657-665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Favre, B., Turowski, P., and Hemmings, B. A. (1997) J. Biol. Chem. 272 13856-13863 [DOI] [PubMed] [Google Scholar]
- 26.McManus, E. J., Sakamoto, K., Armit, L. J., Ronaldson, L., Shpiro, N., Marquez, R., and Alessi, D. R. (2005) EMBO J. 24 1571-1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bain, J., McLauchlan, H., Elliott, M., and Cohen, P. (2003) Biochem. J. 371 199-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leost, M., Schultz, C., Link, A., Wu, Y. Z., Biernat, J., Mandelkow, E. M., Bibb, J. A., Snyder, G. L., Greengard, P., Zaharevitz, D. W., Gussio, R., Senderowicz, A. M., Sausville, E. A., Kunick, C., and Meijer, L. (2000) Eur. J. Biochem. 267 5983-5994 [DOI] [PubMed] [Google Scholar]
- 29.Lu, K. P., Finn, G., Lee, T. H., and Nicholson, L. K. (2007) Nat. Chem. Biol. 3 619-629 [DOI] [PubMed] [Google Scholar]
- 30.Zhou, X. Z., Kops, O., Werner, A., Lu, P. J., Shen, M., Stoller, G., Kullertz, G., Stark, M., Fischer, G., and Lu, K. P. (2000) Mol. Cell 6 873-883 [DOI] [PubMed] [Google Scholar]
- 31.Hamdane, M., Dourlen, P., Bretteville, A., Sambo, A. V., Ferreira, S., Ando, K., Kerdraon, O., Begard, S., Geay, L., Lippens, G., Sergeant, N., Delacourte, A., Maurage, C. A., Galas, M. C., and Buee, L. (2006) Mol. Cell. Neurosci. 32 155-160 [DOI] [PubMed] [Google Scholar]
- 32.Lu, P. J., Wulf, G., Zhou, X. Z., Davies, P., and Lu, K. P. (1999) Nature 399 784-788 [DOI] [PubMed] [Google Scholar]
- 33.Pastorino, L., Sun, A., Lu, P. J., Zhou, X. Z., Balastik, M., Finn, G., Wulf, G., Lim, J., Li, S. H., Li, X., Xia, W., Nicholson, L. K., and Lu, K. P. (2006) Nature 440 528-534 [DOI] [PubMed] [Google Scholar]
- 34.Lu, P. J., Zhou, X. Z., Liou, Y. C., Noel, J. P., and Lu, K. P. (2002) J. Biol. Chem. 277 2381-2384 [DOI] [PubMed] [Google Scholar]
- 35.Liou, Y. C., Sun, A., Ryo, A., Zhou, X. Z., Yu, Z. X., Huang, H. K., Uchida, T., Bronson, R., Bing, G., Li, X., Hunter, T., and Lu, K. P. (2003) Nature 424 556-561 [DOI] [PubMed] [Google Scholar]
- 36.Lim, J., Balastik, M., Lee, T. H., Nakamura, K., Liou, Y. C., Sun, A., Finn, G., Pastorino, L., Lee, V. M. Y., and Lu, K. P. (2008) J. Clin. Investig. 118 1877-1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rembutsu, M., Soutar, M. P., Aalten, L. V., Gourlay, R., Hastie, C. J., McLauchlan, H., Morrice, N. A., Cole, A. R., and Sutherland, C. (2008) Biochemistry 47 2153-2161 [DOI] [PubMed] [Google Scholar]
- 38.Ballatore, C., Lee, V. M., and Trojanowski, J. Q. (2007) Nat. Rev. Neurosci. 8 663-672 [DOI] [PubMed] [Google Scholar]
- 39.Hasegawa, M., Morishima-Kawashima, M., Takio, K., Suzuki, M., Titani, K., and Ihara, Y. (1992) J. Biol. Chem. 267 17047-17054 [PubMed] [Google Scholar]
- 40.Goedert, M., Jakes, R., Qi, Z., Wang, J. H., and Cohen, P. (1995) J. Neurochem. 65 2804-2807 [DOI] [PubMed] [Google Scholar]
- 41.Goedert, M., Jakes, R., Crowther, R. A., Cohen, P., Vanmechelen, E., Vandermeeren, M., and Cras, P. (1994) Biochem. J. 301 871-877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Songyang, Z., Lu, K. P., Kwon, Y. T., Tsai, L. H., Filhol, O., Cochet, C., Brickey, D. A., Soderling, T. R., Bartleson, C., Graves, D. J., DeMaggio, A. J., Hoekstra, M. F., Blenis, J., Hunter, T., and Cantley, L. C. (1996) Mol. Cell. Biol. 16 6486-6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patrick, G. N., Zukerberg, L., Nikolic, M., de la Monte, S., Dikkes, P., and Tsai, L. H. (1999) Nature 402 615-622 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.