Abstract
Metabolism of nitroglycerin (GTN) to 1,2-glycerol dinitrate (GDN) and nitrite by mitochondrial aldehyde dehydrogenase (ALDH2) is essentially involved in GTN bioactivation resulting in cyclic GMP-mediated vascular relaxation. The link between nitrite formation and activation of soluble guanylate cyclase (sGC) is still unclear. To test the hypothesis that the ALDH2 reaction is sufficient for GTN bioactivation, we measured GTN-induced formation of cGMP by purified sGC in the presence of purified ALDH2 and used a Clark-type electrode to probe for nitric oxide (NO) formation. In addition, we studied whether GTN bioactivation is a specific feature of ALDH2 or is also catalyzed by the cytosolic isoform (ALDH1). Purified ALDH1 and ALDH2 metabolized GTN to 1,2- and 1,3-GDN with predominant formation of the 1,2-isomer that was inhibited by chloral hydrate (ALDH1 and ALDH2) and daidzin (ALDH2). GTN had no effect on sGC activity in the presence of bovine serum albumin but caused pronounced cGMP accumulation in the presence of ALDH1 or ALDH2. The effects of the ALDH isoforms were dependent on the amount of added protein and, like 1,2-GDN formation, were sensitive to ALDH inhibitors. GTN caused biphasic sGC activation with apparent EC50 values of 42 ± 2.9 and 3.1 ± 0.4 μm in the presence of ALDH1 and ALDH2, respectively. Incubation of ALDH1 or ALDH2 with GTN resulted in sustained, chloral hydrate-sensitive formation of NO. These data may explain the coupling of ALDH2-catalyzed GTN metabolism to sGC activation in vascular smooth muscle.
The antianginal drug nitroglycerin (GTN)2 causes vasodilation through NO-mediated activation of sGC and cGMP accumulation in vascular smooth muscle (1). In virtually all cells and tissues, the slow reaction of GTN with thiols, in particular GSH, yields 1,2- and 1,3-GDN together with stoichiometric amounts of inorganic nitrite (2), but thiol-triggered GTN metabolism is not associated with GTN bioactivation except the still poorly understood reaction of GTN with l-cysteine that leads to formation of a bioactive species with NO-like properties (3). Recently we have identified ascorbate as another endogenous reductant that causes non-enzymatic GTN bioactivation, but the reaction is slow and may not contribute significantly to the hemodynamic effects of GTN under normal physiological conditions (4).
Besides these non-enzymatic reactions, a number of enzymatic pathways catalyzing GTN bioactivation have been described, but it has remained unclear whether there is a distinct enzyme that mediates the vascular effects of GTN (1). Several years ago, ALDH2 (mitochondrial aldehyde dehydrogenase, EC 1.2.1.3), the main mitochondrial isoform of the ALDH superfamily, was proposed as a new candidate to fulfill this function (5). The characteristics of ALDH2-catalyzed GTN metabolism, including relatively high GTN affinity (low micromolar) and selective formation of 1,2-GDN, rendered this pathway very promising, and several laboratories reported that pharmacological inhibition or gene deletion of ALDH2 led to significantly impaired GTN-induced vasorelaxation and vascular cGMP accumulation (6–10). In addition, treatment of volunteers with an ALDH inhibitor attenuated the increase in forearm blood flow caused by GTN infusion to a similar degree as observed in East Asian subjects expressing a low activity mutant of ALDH2, indicating that this pathway contributes to the hemodynamic effects of GTN in humans (11).
There are three early reports on inhibition of ALDH by organic nitrates (12–14), but the pharmacological implication of these observations had not been considered before the discovery of ALDH2-catalyzed GTN bioactivation by the Stamler laboratory in 2002 (5). Mukerjee and Pietruszko (14) showed that the cytosolic and mitochondrial ALDH isoforms are inactivated by isosorbide dinitrate in a mechanism-based manner, whereby the reversibility of enzyme inactivation by 2-mercaptoethanol suggested the involvement of sulfhydryl oxidation. Later studies confirmed and extended these observations, showing that the presence of a reductant is essential for sustained ALDH2-catalyzed GTN metabolism and bioactivation (5, 9). Intriguingly, the most abundant cellular reductant, GSH, is ineffective, raising the question for the identity of the physiological reductant. A promising candidate is dihydrolipoic acid, which was shown to restore ALDH2 activity in GTN-exposed blood vessels (15). However, enzyme reactivation required relatively high concentrations of the reductant, and evidence for a role of endogenous dihydrolipoic acid in GTN bioactivation is still missing. This issue is of considerable pharmacological interest because vascular depletion of the endogenous ALDH2 reductant may at least partially explain the phenomenon of nitrate tolerance, i.e. the loss of vascular sensitivity to GTN upon continuous application because of impaired GTN bioactivation (16).
Notwithstanding the large body of evidence for a significant contribution of ALDH2 to vascular GTN bioactivation, it is still unclear how the ALDH2 reaction, yielding inorganic nitrite as final product, is linked to sGC activation. One possibility would be that nitrite formation is coupled to a nitrite reductase pathway associated with the mitochondrial respiratory chain (17). In support of this hypothesis, isolated mitochondria were found to reduce nitrite to NO in the presence of respiratory substrates at low oxygen tension (18), presumably via activation of cytochrome c oxidase (19). In a recent study we confirmed these observations but found no correlation between mitochondrial respiratory rate and GTN-triggered sGC activation,3 indicating that mitochondrial nitrite reduction is not essential for GTN bioactivation to occur. In search of alternative pathways, we have suggested that ALDH2 itself may catalyze formation of NO from GTN (9). However, this proposal was based on preliminary data and has not been seriously considered by others so far. Therefore, we carried out a detailed study to see whether GTN metabolism catalyzed by purified ALDH2 does indeed result in NO formation and activation of sGC. In addition, we wished to clarify whether the ability to catalyze GTN bioactivation is a specific feature of ALDH2. Interestingly, we found that both ALDH1 (cytosolic aldehyde dehydrogenase, ALDH1A1, EC 1.2.1.36) and ALDH2 are able to convert GTN to NO, although low GTN affinity of ALDH1 may limit the contribution of this isoform to GTN bioactivation in vivo.
EXPERIMENTAL PROCEDURES
Materials—Bovine lung sGC was purified as described previously (20). Sephacryl S-300 HR and [α-32P]GTP (400 Ci/mmol) were obtained from GE Healthcare Europe GmbH (Vienna, Austria). Nitropohl® ampoules (G. Pohl-Boskamp GmbH. & Co., Hohenlockstedt, Germany) containing 4.4 mm GTN in 250 mm glucose were obtained from a local pharmacy; dilutions were made in 50 mm triethanolamine HCl buffer. DEA/NO and ODQ (Alexis Corp., Lausen, Switzerland) were purchased via Eubio (Vienna). DEA/NO was dissolved and diluted in 10 mm NaOH. Stock solutions of ODQ (10 mm) were prepared in dimethyl sulfoxide and diluted with 25% dimethyl sulfoxide in H2O (v/v). Oxyhemoglobin was prepared by reduction of bovine hemoglobin (Sigma) with sodium dithionite as described previously (21). Daidzin was synthesized as described (22) and dissolved in 50 mm triethanolamine HCl at maximally 160 μm (because of limited solubility). All other chemicals were from Sigma-Aldrich GmbH (Vienna).
ALDH Expression and Purification—Human liver ALDH1 and ALDH2 were expressed in Escherichia coli BL21(DE3) as described previously (23), using pT7.7 as the host vector for both cDNAs. ALDH2 was purified by p-hydroxyacetophenone affinity chromatography as described (24). For final purification, the protein was loaded on a size-exclusion chromatography column (Sephacryl S-300; 100 × 2.5 cm), which had been equilibrated in 100 mm MES buffer (pH 6.5) containing 0.2 mm MgCl2, and eluted with the same buffer at 1 ml/min.
ALDH1 was purified according to the same protocol but without the final size-exclusion chromatography step. Active fractions eluted from the p-hydroxyacetophenone column were pooled and dialyzed against 100 mm MES buffer (pH 6.5) containing 0.2 mm MgCl2 and 1 mm DTT.
Determination of ALDH Activity and Enzyme Kinetics—ALDH activity was measured as formation of NADH from NAD+ in the presence of varying concentrations of acetaldehyde by monitoring the increase in absorbance at 340 nm (ε340 = 6.22 mm–1 cm–1) at 25 °C (25). The incubation buffer (pH 9.0) was either 100 mm sodium pyrophosphate (ALDH1) or 50 mm sodium pyrophosphate containing 10 mm MgCl2 (ALDH2). As reported previously (26), we observed that Mg2+ ions slightly reduced the dehydrogenase activity of ALDH1 (by ∼20%) but increased the respective activity of ALDH2 (by ∼20%). The esterase activities of both isoforms were increased by 20–30% in the presence of Mg2+ ions (data not shown). Esterase activity was measured by monitoring formation of p-nitrophenolate from varying concentrations of p-nitrophenyl acetate (ε400 = 16 mm–1 cm–1) in the presence of 100 mm sodium pyrophosphate buffer (pH 7.5). The other ingredients were the same as described for determination of dehydrogenase activity. Enzyme kinetic parameters were calculated by fitting the data according to saturation kinetics using the Kaleidagraph® software.
Determination of 1,2- and 1,3-GDN Formation by Radio Thin Layer Chromatography—The rate of GTN metabolism yielding 1,2- and 1,3-GDN was determined according to a described protocol (9). Briefly, purified ALDH1 or ALDH2 (4 μg of each) was incubated with 14C-labeled GTN (2 μm; ∼50,000 dpm) at 37 °C for 10 min in a final volume of 200 μl of 50 mm phosphate buffer (pH 7.4) containing 3 mm MgCl2, 2 mm GSH, 2 mm DTT, 1 mm NAD+, 1 mm EDTA, 1 mm EGTA, and unlabeled GTN as required to obtain the indicated final concentrations. The amount of added radioactivity was increased 3-fold for determination of reaction rates at GTN concentrations ≥0.3 mm. Enzyme inhibitors (chloral hydrate and the ALDH2-selective compound daidzin) were present as indicated in the text and figure legends. Reaction products were extracted twice with 1 ml of diethyl ether, separated by thin layer chromatography, and quantified by liquid scintillation counting. Blank values were determined in the absence of protein under identical conditions and subtracted. Results are expressed as nmol of product per min and mg of protein. Mean values ± S.E. were calculated from reaction rates measured as duplicates in three independent experiments.
Determination of sGC Activity—Purified bovine lung sGC (50 ng) was incubated at 37 °C for 10 min in a final volume of 100 μl with the indicated concentrations of donor compounds (GTN, NaNO2, and DEA/NO) in the absence and presence of purified ALDH1 or ALDH2 or BSA as indicated in the text and figure legends. Assay mixtures contained 50 mm triethanolamine HCl (pH 7.4), 5 mm MgCl2, 0.5 mm [α-32P]GTP (∼250,000 cpm), 1 mm cGMP, and 2 mm DTT. Where indicated, chloral hydrate (1 or 10 mm), daidzin (0.1 mm), oxyhemoglobin (0.1 mm), flavin adenine dinucleotide (0.1 mm), or ODQ (0.1 mm) was additionally present. Reactions were terminated by the addition of 0.45 ml of zinc acetate (120 mm) and 0.45 ml of sodium bicarbonate (120 mm), followed by isolation and quantification of [32P]cGMP as described previously (27). Blank values were determined in the absence of sGC.
Determination of GTN-derived NO in the Presence of Purified ALDH Isoforms—NO formation was measured with a Clark-type electrode (World Precision Instruments, Berlin, Germany), calibrated daily with acidified nitrite as described previously (28), in 50 mm triethanolamine HCl (pH 7.4) containing 1,000 units of superoxide dismutase, 2 mm DTT, and GTN (1 mm and 0.1 mm with ALDH1 and ALDH2, respectively). The incubation volume was 1 ml. Where indicated, measurements were performed in the presence of the nucleotide cofactors NAD+ or NADH (2 mm each) or the ALDH inhibitor chloral hydrate (10 mm). After equilibration at 37 °C, purified ALDH1 (0.5 mg) or ALDH2 (0.25 mg), which had been adjusted to 37 °C for 10 min, was added. The NO scavenger oxyhemoglobin (10 μm final concentration) was added in some experiments to confirm that the observed signals did indeed reflect formation of NO radical.
Analysis of ALDH-bound Metals—For the determination of metals in ALDH1 and ALDH2, protein solutions (100 μl, ∼80 μm) were mixed with nitric acid (1% (v/v) final concentration) and vigorously shaken for 10 min. The samples were screened for metals with an inductively coupled plasma mass spectrometer (Agilent 7500ce, Waldbronn, Germany) in the semiquantitative mode. Metal content of buffer was subtracted as blank. The concentrations of Fe, Cu, Mn, Mo, and Zn were <1 μm. Other metals were not detectable.
RESULTS
Basic Characterization of Purified ALDH Isoforms—The proteins obtained from the E. coli expression system were >95% pure as judged by polyacrylamide gel electrophoresis (data not shown). The enzyme kinetic parameters determined for the dehydrogenase and esterase activities of ALDH1 and ALDH2 (Table 1), including the significantly lower Km values for aldehyde and ester substrates and higher specific activity of ALDH2, agree well with values reported previously (25).
TABLE 1.
Enzyme kinetic parameters of purified recombinant human ALDH1 and ALDH2
Dehydrogenase and esterase activities were determined photometrically in the presence of increasing concentrations of acetaldehyde and p-nitrophenyl acetate, respectively, as described under “Experimental Procedures.” Data are mean values ± S.E. of the parameters determined for three different enzyme preparations.
|
Dehydrogenase activity
|
Esterase activity
|
|||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| μm | nmol × min-1 × mg-1 | μm | nmol × min-1 × mg-1 | |
| ALDH1 | 200 ± 14 | 1,060 ± 20 | 0.69 ± 0.03 | 155 ± 4 |
| ALDH2 | 0.33 ± 0.005 | 4,000 ± 30 | 0.18 ± 0.01 | 210 ± 9 |
ALDH-catalyzed GTN Metabolism—Determination of 1,2- and 1,3-GDN formation by radio thin layer chromatography revealed that both ALDH isoforms catalyze GTN metabolism (Fig. 1). ALDH1 (Fig. 1A) converted 2 μm GTN into 1,2- and 1,3-GDN with rates of 3.0 ± 0.16 and 0.66 ± 0.03 nmol × min–1 × mg–1, respectively. In the presence of this low GTN concentration, ALDH2 (Fig. 1B) exhibited ∼3-fold higher activity with respect to 1,2-GDN formation (9.00 ± 0.28 nmol × min–1 × mg–1). Formation of 1,3-GDN was very low (0.18 ± 0.01 nmol × min–1 × mg–1), confirming the selective conversion of GTN into the 1,2-isomer under these conditions (5, 9). As expected, chloral hydrate inhibited GTN metabolism by both isoforms, whereas daidzin showed marked selectivity for ALDH2. As revealed by the GTN concentration dependence of the reactions shown in Fig. 2, the two isoforms exhibited similar maximal 1,2-GDN-forming activities of 30–40 nmol × min–1 × mg–1 but differed significantly in GTN affinity and product distribution. Formation of 1,2-GDN by ALDH1 (Fig. 2A) and ALDH2 (B) was half-maximal at ∼200 and 20 μm GTN, respectively, apparently reflecting the lower Km values of ALDH2 measured with dehydrogenase and esterase substrates (see Table 1). As reported previously (5, 9), specific 1,2-GDN formation by ALDH2 was lost at increasing GTN concentrations, although formation of the 1,3-isomer did not exceed 1,2-GDN formation even at 1 mm GTN. In contrast, the ALDH1-catalyzed reaction resulted in predominant formation of the 1,3-isomer at ≥1 mm GTN. In fact, the rate of 1,2-GDN formation was significantly decreased at the highest GTN concentration tested (2.8 mm). Formation of 1,3-GDN was not affected by substrate-competitive inhibitors, indicating that it does not involve binding of GTN to the catalytic sites of the enzymes.
FIGURE 1.
GTN metabolism by ALDH1 (A) and ALDH2 (B). Purified ALDH1 and ALDH2 (4 μg of each) were incubated at 37 °C for 10 min in the presence of 2 μm [14C]GTN, 3 mm MgCl2, 2 mm GSH, 2 mm DTT, 1 mm NAD+, 1 mm EDTA, and 1 mm EGTA. The enzyme inhibitors chloral hydrate (CH) and daidzin were present as indicated. 1,2- and 1,3-GDN were extracted and quantified by radio thin layer chromatography as described under “Experimental Procedures.” Data represent mean values ± S.E. of three independent experiments.
FIGURE 2.
Dependence on GTN concentration of 1,2- and 1,3-GDN formation by ALDH1 (A) and ALDH2 (B). Purified ALDH1 and ALDH2 (4 μg of each) were incubated at 37 °C for 10 min in the presence of 3 mm MgCl2, 2 mm GSH, 2 mm DTT, 1 mm NAD+, 1 mm EDTA, 1 mm EGTA, and the indicated concentrations of [14C]GTN. 1,2- and 1,3-GDN were extracted and quantified by radio thin layer chromatography as described under “Experimental Procedures.” Data represent mean values ± S.E. of three independent experiments.
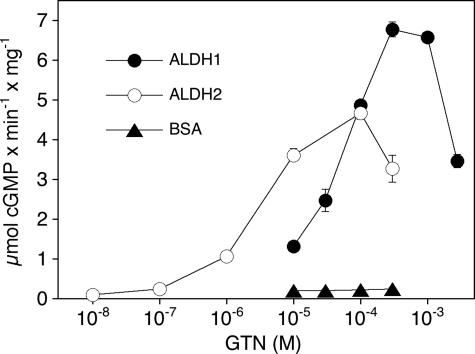
ALDH-catalyzed GTN Bioactivation Assayed as Activation of sGC—Purified sGC was used for highly sensitive detection of NO-related bioactivity associated with ALDH-catalyzed GTN metabolism. Taking into account the different GTN affinities of the two ALDH isoforms, GTN was present at final concentrations of 100 and 10 μm in the assays with ALDH1 and ALDH2, respectively. As shown in Fig. 3, cGMP synthesis was not considerably increased over basal in the presence of increasing amounts of BSA that was used as a protein control (specific activity varied between 0.11 ± 0.02 and 0.36 ± 0.05 μmol of cGMP × min–1 × mg–1). In the absence of GTN, neither ALDH1 nor ALDH2 affected cGMP formation (see Fig. 5 below for GTN dependence). However, the combined presence of GTN and ALDH1 or ALDH2 led to significant sGC activation. The effects of the ALDH isoforms were dependent on the amount of added protein. Maximal rates of cGMP formation were observed with 50–100 μg of ALDH1 or 25 μg of ALDH2; sGC activity reached a plateau or even decreased at higher ALDH concentrations and never exceeded 30% of the rates measured with maximally NO-stimulated sGC.
FIGURE 3.
Effects of ALDH1 and ALDH2 on activation of sGC by GTN. Purified sGC (50 ng) was incubated at 37 °C for 10 min in the presence of [α-32P]GTP (0.5 mm, ∼250,000 cpm), 5 mm MgCl2, 1 mm cGMP, and 2 mm DTT with the indicated amounts of purified ALDH1, ALDH2, or BSA. GTN concentration was 10 μm (ALDH2 and BSA) or 100 μm (ALDH1). [32P]cGMP was isolated and quantified as described under “Experimental Procedures.” Data are mean values ± S.E. of three independent experiments.
FIGURE 5.
Effect of GTN concentration on ALDH-mediated sGC activation. Purified sGC was incubated with ALDH1 (50 μg) or ALDH2 (25 μg) or BSA (50 μg) as described in the legend to Fig. 3 in the presence of the indicated concentrations of GTN. [32P]cGMP was isolated and quantified as described under “Experimental Procedures.” Data are mean values ± S.E. of three independent experiments.
To see whether ALDH-catalyzed GTN bioactivation involves the catalytic site of the proteins, we tested the effects of the established ALDH inhibitors chloral hydrate and daidzin (Fig. 4A). To account for slight inhibition of NO stimulation of sGC (∼10%), the effects of the added compounds are shown as the percent of maximal sGC activity determined under identical conditions in the presence of 10 μm DEA/NO. Although the non-selective compound chloral hydrate (1 mm) inhibited GTN-induced sGC activation mediated by both ALDH1 and ALDH2, daidzin selectively blocked the effect of ALDH2, strongly suggesting that binding of GTN to the substrate sites of the ALDH isoforms is essential for bioactivation. The potential involvement of NO binding to the regulatory heme group of sGC was tested with the heme site sGC inhibitor ODQ (29, 30), the superoxide-generating compound flavin adenine dinucleotide (31, 32), and the established NO scavenger oxyhemoglobin. As shown in Fig. 4B, all three agents completely inhibited ALDH-mediated sGC activation by GTN.
FIGURE 4.
Effects of enzyme inhibitors and NO scavengers on ALDH-mediated sGC activation by GTN. Purified sGC was incubated with purified ALDH1 (50 μg) or ALDH2 (25 μg) in the presence of 100 μm (ALDH1) or 10 μm (ALDH2) GTN as described in the legend to Fig. 3. The ALDH inhibitors chloral hydrate (CH) or daidzin (A), the sGC inhibitor ODQ, the superoxide-generating compound flavin adenine dinucleotide (FAD), and the NO scavenger oxyhemoglobin (OxyHb)(B) were present as indicated. [32P]cGMP was isolated and quantified as described under “Experimental Procedures.” Data are expressed as percent of maximal sGC activity obtained with 10μm DEA/NO (A) or specific sGC activity (B) and represent mean values ± S.E. of three independent experiments.
Fig. 5 shows the GTN concentration dependence of cGMP formation in the presence of ALDH1 and ALDH2. The effects of GTN were biphasic with apparent EC50 values of 42 ± 2.9 μm (ALDH1) and 3.1 ± 0.4 μm (ALDH2). The ∼10-fold higher potency of GTN to stimulate sGC in the presence of ALDH2 agrees well with the metabolism experiments shown in Fig. 2, suggesting a higher affinity of the nitrate for the mitochondrial isoform. GTN (10–300 μm) had no effect in the presence of BSA, which served as a protein control. Activation of sGC by 0.1 mm GTN was not affected by the trace metal chelator diethylenetriaminepentaacetic acid (data not shown).
ALDH-catalyzed GTN Bioactivation Resulting in Formation of NO—Inhibition of ALDH-mediated sGC activation by oxyhemoglobin and superoxide (generated via flavin adenine dinucleotide-triggered O2 reduction) suggested the involvement of free NO radical in GTN bioactivity. Therefore, we attempted to measure GTN-derived NO with a Clark-type electrode. In preliminary experiments, we observed relatively small, transient NO signals that were markedly enhanced and prolonged in the presence of superoxide dismutase (data not shown). To effectively prevent this apparent superoxide-mediated NO decay, all NO assays were performed in the presence of 1,000 units/ml of superoxide dismutase. Other experimental conditions were chosen to maximize GTN bioactivation according to the sGC data. As shown in Fig. 6, addition of ALDH1 (A) or ALDH2 (B) to GTN-containing assay mixtures resulted in the appearance of pronounced NO signals with initial rates of 1.68 ± 0.06 and 1.76 ± 0.012 μm × min–1 × mg–1, respectively. The maximal NO concentrations measured with ALDH1 and ALDH2 were 0.47 ± 0.01 and 0.33 ± 0.001 μm, respectively. No detectable signals were observed in the presence of chloral hydrate (10 mm). Chloral hydrate did not affect NO signals derived from DEA/NO decomposition (data not shown). The apparent NO formation mediated by both isoforms was enhanced by NAD+, which stimulates GTN metabolism. The nucleotide increased both initial rates of NO formation (to 8.40 ± 0.08 and 4.08 ± 0.08 μm × min–1 × mg–1 ALDH1 and ALDH2, respectively) and NO peak concentrations (0.90 ± 0.007 μm ALDH1 and 0.57 ± 0.003 μm ALDH2). We reasoned that NO formation may involve nitrite reduction (see below) and therefore tested NADH as a potential reducing cofactor. NADH enhanced NO formation in terms of initial rates and peak concentrations, but the effects were less pronounced than those observed with NAD+. Signals elicited by ALDH1 or ALDH2 decayed rapidly upon addition of oxyhemoglobin (Fig. 6C), indicating that the signals reflected formation of free NO radical.
FIGURE 6.
Formation of GTN-derived NO in the presence of ALDH isoforms. Purified ALDH1 (0.5 mg; A) or ALDH2 (0.25 mg; B) was incubated at 37 °C with GTN (1 mm and 0.1 mm, respectively) in the absence and presence of NAD+ and NADH (2 mm each) or chloral hydrate (CH; 10 mm). NO formation was monitored with a specific electrode as described under “Experimental Procedures.” C shows the effect of oxyhemoglobin, added at the time points indicated by arrows to give a final concentration of 10 μm, on the NO signals obtained from ALDH1 and ALDH2 incubated with GTN as described for A and B in the absence of nucleotides. The effect on NO derived from DEA/NO is shown as the control. The traces shown are representative for three independent experiments.
ALDH-mediated Bioactivation of Inorganic Nitrite—Because ALDH2 reduces GTN to inorganic nitrite (5), we speculated that the observed NO formation could be due to intrinsic nitrite reductase activity of the ALDH isoforms. To test this hypothesis, we used nitrite instead of GTN in sGC assays as potential ALDH substrate. To minimize thiol-mediated non-enzymatic bioactivation of nitrite, the assays were performed in the absence of DTT. As shown in Fig. 7, sGC exhibited a specific activity of 0.87 ± 0.014 μmol × min–1 × mg–1 in the presence of 10 mm nitrite. This activity was increased ∼2- or 4-fold in the additional presence of ALDH1 or ALDH2, respectively. Chloral hydrate (10 mm) had no effect on sGC activity in the presence of nitrite alone but completely inhibited the effects of ALDH1 and ALDH2, excluding nonspecific protein effects.
FIGURE 7.
Activation of sGC by inorganic nitrite in the presence of purified ALDH1 and ALDH2. Purified sGC (50 ng) was incubated at 37 °C for 10 min with [α-32P]GTP (0.5 mm, ∼250,000 cpm), 5 mm MgCl2, 1 mm cGMP, and 10 mm NaNO2 in the absence (control) or presence of purified ALDH1 and ALDH2 (50 μg each). Chloral hydrate (CH) was present at a final concentration of 10 mm. Data are mean values ± S.E. of three independent experiments.
DISCUSSION
The present study demonstrates that GTN metabolism catalyzed by purified ALDH isoforms is associated with formation of free NO radical, resulting in pronounced sGC activation. Considering that enzymatic nitrite reduction by the respiratory chain does not appear to contribute significantly to mitochondrial GTN bioactivation,3 these results may provide a simple and conclusive explanation for previous reports showing that ALDH2-catalyzed GTN metabolism triggers accumulation of cGMP in blood vessels (5, 9). The initial rates of NO formation measured in the presence of NAD+ (8.4 and 4.0 nmol × min–1 × mg–1) suggest that NO formation accounts for ∼10% of total GTN turnover under our experimental conditions (cf. Fig. 2). However, the transient time course of the NO signals that we observed even in the presence of 1,000 units/ml of superoxide dismutase indicates that the true rates might have been underestimated because of additional, unknown NO consumption pathways. This is even more relevant for the degree of sGC activation, which had to be measured in the absence of superoxide dismutase and NAD+ to avoid interference of these agents with the enzyme assay. Thus, specific NO-forming activity and, in particular, sGC activation measured under our experimental conditions most likely reflect the lower limit of the true efficiency of ALDH-catalyzed GTN bioactivation.
The pronounced effect of superoxide dismutase on detectable GTN-derived free NO (data not shown) suggests significant generation of superoxide in our assays. An unavoidable superoxide source may be GTN itself, which was shown to trigger superoxide formation in blood vessels, thereby limiting vascular NO bioavailability (33–35). In addition, we found that DTT, which was present to minimize mechanism-based oxidative ALDH inactivation, caused rapid consumption of authentic (DEA/NO-derived) NO that was prevented by superoxide dismutase.4 This apparent superoxide generation may be due to DTT autoxidation described in detail recently (36). Generation of superoxide is certainly one important factor limiting the degree of sGC activation by GTN in our experiments and may partly explain the sharply biphasic GTN concentration-response curves shown in Fig. 5. In addition, as studied in detail previously (3), oxidation of sGC-bound heme by high concentrations of GTN may counteract NO-mediated sGC activation. However, neither of these mechanisms appears to sufficiently explain limited sGC activation by a fixed concentration of GTN in the presence of increasing amounts of ALDH (Fig. 3). Because the time course of NO release from DEA/NO was not affected by addition of the proteins (data not shown), ALDH-catalyzed GTN bioactivation may have been compromised under our experimental conditions by a mechanism-based pathway that remains to be identified.
Two lines of evidence suggest that GTN bioactivation involves the catalytic site of ALDH. First, GTN-triggered sGC activation and NO formation were blocked by established substrate-competitive ALDH inhibitors. In line with the known selectivity profile of the compounds (22, 25), chloral hydrate attenuated GTN bioactivation by both ALDH1 and ALDH2, whereas daidzin selectively interfered with the ALDH2-catalyzed reaction. Second, GTN concentration-response curves revealed an ∼10-fold higher potency of the nitrate to activate sGC in the presence of ALDH2 than with ALDH1. This difference in the apparent GTN binding affinities is in line with the differences in the Km values of the two isoforms for aldehyde and ester substrates (see Table 1).
Our data with ALDH1 unequivocally demonstrate that the catalytic conversion of GTN into 1,2- and 1,3-GDN is not a specific feature of ALDH2, suggesting a basic reaction mechanism common to both ALDH isoforms. Despite similar characteristics of ALDH1- and ALDH2-catalyzed GTN metabolism with respect to stimulation by NAD+ and preferential formation of 1,2-GDN at low GTN concentrations, there were also clear differences apparent. Besides the ∼10-fold higher GTN concentrations required for half-maximal activity of ALDH1, product distribution differed significantly between the two isoforms. In contrast to ALDH2, ALDH1 preferentially produced 1,3-GDN at saturating GTN concentrations, and the rate of 1,2-GDN formation was reduced by ∼50% at the highest GTN concentration tested. Together with preliminary findings indicating that ALDH1-catalyzed GTN metabolism is less dependent on the presence of thiols than the ALDH2-catalyzed reaction,5 these results point to subtle differences in the catalytic properties of the two isoforms. Future studies on the structure and function of wild-type and mutated ALDH isoforms should clarify the molecular basis of these differences.
The observation that catalysis of GTN bioactivation is not a specific feature of ALDH2 is certainly interesting with respect to ALDH enzymology, but the pharmacological relevance of this finding is unclear. The low apparent GTN affinity of ALDH1 is expected to limit the contribution of this isoform to GTN bioactivation in patients treated with therapeutically relevant low doses of the nitrate. Nevertheless, ALDH1 or other isoforms capable of catalyzing this reaction could contribute to the so-called “high Km” pathway of GTN action (17) that persists upon ALDH2 inhibition (6, 9) or gene deletion (10). Interestingly, we found that GTN bioactivation catalyzed by rat liver microsomes was sensitive to chloral hydrate but not to daidzin (9). Collectively, the currently available data suggest that ALDH1 and possibly other isoforms of the enzyme could contribute to the low affinity component of GTN action in blood vessels.
At present we can only speculate about the mechanism underlying the observed formation of free NO radical in the course of ALDH-catalyzed GTN metabolism. The significantly enhanced activation of sGC by inorganic nitrite in the presence of the purified ALDH isoforms shows that the enzymes are capable of converting nitrite to NO that activates sGC. Inhibition of nitrite bioactivation by chloral hydrate excludes nonspecific effects of the proteins and suggests that NO formation takes place within the catalytic sites of the enzymes. However, ALDH-mediated activation of sGC by nitrite was only partial and required extremely high nitrite concentrations, seriously questioning the relevance of this reaction for formation of GTN-derived NO. Nevertheless, because the sGC assays were performed under suboptimal conditions, i.e. in the absence of NAD+ and superoxide dismutase because of interference of these agents with the assay, the true efficiency of the reaction may be considerably higher. We are currently establishing a cGMP reporter assay with rat lung fibroblasts (37) to circumvent those limitations of the sGC assay. The high nitrite concentration that was required for ALDH-mediated sGC activation may reflect low affinity of exogenous nitrite to the catalytic site but does not exclude that GTN-derived nitrite becomes efficiently converted to NO in situ in the course of the enzymatic reaction. Thus, the available data are not sufficient to definitively rule out nitrite as an intermediate. If NO were indeed formed from GTN-derived nitrite, this could occur either via intrinsic nitrite reductase activity of ALDH or disproportionation of nitrite after protonation in the catalytic site.
Preliminary data obtained with mutated ALDH2 indicate that the reactive cysteine residue in the active site (Cys302) is essential for NO formation and sGC activation.5 Therefore, the initial step of NO formation may be a nucleophilic attack of GTN at Cys302, yielding a thionitrate intermediate. As shown in Scheme 1, this intermediate could be converted to NO through two hypothetical pathways. (i) Rearrangement to a sulfenyl nitrite intermediate leads to homolysis to yield NO and sulfenyl radical. However, this pathway would result in irreversible inactivation of the enzyme after one turnover because of expected dimerization of sulfenyl radicals, and it does not explain why GTN bioactivation is triggered by free cysteine but not by cysteine-containing peptides (e.g. GSH) or proteins (e.g. BSA). (ii) Metal-catalyzed reaction of the thionitrate intermediate (38) with an adjacent cysteine residue would result in formation of NO and a disulfide. This pathway accommodates reversible inactivation of ALDH2 by GTN as well as the inactivity of proteins not containing vicinal cysteine residues in appropriate conformation, but the lack of effect of diethylenetriaminepentaacetic acid argues against the involvement of a redox-active trace metal. However, further work with various metal chelators is required to definitively settle this issue.
SCHEME 1.
Hypothetical pathways of ALDH-catalyzed NO formation from GTN.
It has been shown that vascular GTN metabolism results in production of hydroxylamine, suggesting that HNO may be formed as an intermediate of GTN bioactivation (39). The NO-like properties of HNO donors such as Angeli's salt may be explained by 1-electron oxidation of nitroxyl to NO by superoxide dismutase or transition metal ions (40). Although the trace metal chelator diethylenetriaminepentaacetic acid had no significant effect on GTN-triggered sGC activation, and the purified ALDH proteins did not contain significant amounts of bound trace metals, we cannot rule out HNO as an intermediate of GTN bioactivation.
Taken together, the present results demonstrate that ALDH1 and ALDH2 convert GTN into significant amounts of NO. It is hard to judge whether this direct formation of NO explains GTN-induced cGMP formation in vivo. Of note, we found that <5% of maximal (NO-stimulated) cGMP accumulation in blood vessels is sufficient for maximal vascular relaxation (9). Therefore, suboptimal activation of sGC by GTN as observed here in the presence of purified ALDH isoforms may fully account for the ALDH-dependent vascular effects of the nitrate in vivo. Nevertheless, we cannot exclude the existence of additional pathways linking ALDH-catalyzed GTN metabolism to sGC activation in vascular smooth muscle cells. Further studies are warranted to clarify the biological relevance of our findings and the molecular mechanism underlying ALDH-catalyzed NO formation.
Author's Choice—Final version full access.
This work was supported by Grants W901 DK Molecular Enzymology, P16690, and P20669 from the Fonds zur Föderung der Wissenschaftlichen Forschung in Austria (to B. M.), Deutsche Forschungsgemeinschaft Grant KO1157/4-1 (to D. K. and M. R.), and the Endowment for Research in Human Biology, Boston, Massachusetts (to W. M. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GTN, nitroglycerin; GDN, glycerol dinitrate; BSA, bovine serum albumin; DEA/NO, 2,2-diethyl-1-nitroso-oxyhydrazine; DTT, dithiothreitol; GSH, glutathione; MES, 2-(N-morpholino)ethanesulfonic acid; ODQ, 1H-[1,2,4]oxadiazolo[4,3-α]quinoxalin-1-one; sGC, soluble guanylate cyclase.
A. Kollau, M. Beretta, M. Russwurm, D. Koesling, W. M. Keung, K. Schmidt, and B. Mayer, submitted for publication.
K. Schmidt and B. Mayer, unpublished observations.
M. Beretta, W. M. Keung, and B. Mayer, unpublished observations.
References
- 1.Fung, H. L. (2004) Annu. Rev. Pharmacol. Toxicol. 44 67–85 [DOI] [PubMed] [Google Scholar]
- 2.Bennett, B. M., McDonald, B. J., Nigam, R., and Simon, W. C. (1994) Trends Pharmacol. Sci. 15 245–249 [DOI] [PubMed] [Google Scholar]
- 3.Gorren, A. C. F., Russwurm, M., Kollau, A., Koesling, D., Schmidt, K., and Mayer, B. (2005) Biochem. J. 390 625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kollau, A., Beretta, M., Gorren, A. C. F., Russwurm, R., Koesling, D., Schmidt, K., and Mayer, B. (2007) Mol. Pharmacol. 72 191–196 [DOI] [PubMed] [Google Scholar]
- 5.Chen, Z. Q., Zhang, J., and Stamler, J. S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 8306–8311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiFabio, J., Yanbin, J., Vasiliou, V., Thatcher, R. J., and Bennett, B. M. (2003) Mol. Pharmacol. 64 1109–1116 [DOI] [PubMed] [Google Scholar]
- 7.Sydow, K., Daiber, A., Oelze, M., Chen, Z. P., August, M., Wendt, M., Ullrich, V., Mülsch, A., Schulz, E., Keaney, J. F., Stamler, J. S., and Münzel, T. (2004) J. Clin. Investig. 113 482–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, J., Chen, Z. P., Cobb, F. R., and Stamler, J. S. (2004) Circulation 110 750–755 [DOI] [PubMed] [Google Scholar]
- 9.Kollau, A., Hofer, A., Russwurm, M., Koesling, D., Keung, W. M., Schmidt, K., Brunner, F., and Mayer, B. (2005) Biochem. J. 385 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Z., Foster, M. W., Zhang, J., Mao, L., Rockman, H. A., Kawamoto, T., Kitagawa, K., Nakayama, K. I., Hess, D. T., and Stamler, J. S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 12159–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackenzie, I. S., Maki-Petaja, K. M., McEniery, C. M., Bao, Y. P., Wallace, S. M., Cheriyan, J., Monteith, S., Brown, M. J., and Wilkinson, I. B. (2005) Arterioscler. Thromb. Vasc. Biol. 25 1891–1895 [DOI] [PubMed] [Google Scholar]
- 12.Komura, S. (1974) Acta Pharmacol. Toxicol. 35 145–154 [DOI] [PubMed] [Google Scholar]
- 13.Towell, J., Garthwaite, T., and Wang, R. (1985) Alcohol Clin. Exp. Res. 9 438–442 [DOI] [PubMed] [Google Scholar]
- 14.Mukerjee, N., and Pietruszko, R. (1994) J. Biol. Chem. 269 21664–21669 [PubMed] [Google Scholar]
- 15.Wenzel, P., Hink, U., Oelze, M., Schuppan, S., Schaeuble, K., Schildknecht, S., Ho, K. K., Weiner, H., Bachschmid, M., Münzel, T., and Daiber, A. (2007) J. Biol. Chem. 282 792–799 [DOI] [PubMed] [Google Scholar]
- 16.Münzel, T., Daiber, A., and Mülsch, A. (2005) Circ. Res. 97 618–628 [DOI] [PubMed] [Google Scholar]
- 17.Chen, Z., and Stamler, J. S. (2006) Trends Cardiovasc. Med. 16 259–265 [DOI] [PubMed] [Google Scholar]
- 18.Kozlov, A. V., Staniek, K., and Nohl, H. (1999) FEBS Lett. 454 127–130 [DOI] [PubMed] [Google Scholar]
- 19.Castello, P. R., David, P. S., McClure, T., Crook, Z., and Poyton, R. O. (2006) Cell Metab. 3 277–287 [DOI] [PubMed] [Google Scholar]
- 20.Russwurm, M., and Koesling, D. (2005) Methods Enzymol. 396 492–501 [DOI] [PubMed] [Google Scholar]
- 21.Schmidt, K., Klatt, P., and Mayer, B. (1994) Biochem. J. 301 645–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keung, W. M., and Vallee, B. L. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 1247–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng, C. F., Wang, T. T., and Weiner, H. (1993) Alcohol Clin. Exp. Res. 17 828–831 [DOI] [PubMed] [Google Scholar]
- 24.Ghenbot, G., and Weiner, H. (1992) Protein Expr. Purif. 3 470–478 [DOI] [PubMed] [Google Scholar]
- 25.Klyosov, A. A., Rashkovetsky, L. G., Tahir, M. K., and Keung, W. M. (1996) Biochemistry 35 4445–4456 [DOI] [PubMed] [Google Scholar]
- 26.Ho, K. K., Allali-Hassani, A., Hurley, T. D., and Weiner, H. (2005) Biochemistry 44 8022–8029 [DOI] [PubMed] [Google Scholar]
- 27.Schultz, G., and Böhme, E. (1984) in Methods of Enzymatic Analysis (Bergmeyer, H. U., Bergmeyer, J., and Graβl, M., eds) pp. 379–389, Verlag Chemie, Weinheim, Germany
- 28.Mayer, B., Klatt, P., Werner, E. R., and Schmidt, K. (1995) J. Biol. Chem. 270 655–659 [DOI] [PubMed] [Google Scholar]
- 29.Garthwaite, J., Southam, E., Boulton, C. L., Nielsen, E. B., Schmidt, K., and Mayer, B. (1995) Mol. Pharmacol. 48 184–188 [PubMed] [Google Scholar]
- 30.Schrammel, A., Behrends, S., Schmidt, K., Koesling, D., and Mayer, B. (1996) Mol. Pharmacol. 50 1–5 [PubMed] [Google Scholar]
- 31.Pfeiffer, S., Lass, A., Schmidt, K., and Mayer, B. (2001) J. Biol. Chem. 276 34051–34058 [DOI] [PubMed] [Google Scholar]
- 32.Schrammel, A., Gorren, A. C. F., Schmidt, K., Pfeiffer, S., and Mayer, B. (2003) Free Rad. Biol. Med. 34 1078–1088 [DOI] [PubMed] [Google Scholar]
- 33.Dikalov, S., Fink, B., Skatchkov, M., Stalleicken, D., and Bassenge, E. (1998) J. Pharmacol. Exp. Ther. 286 938–944 [PubMed] [Google Scholar]
- 34.Fink, B., Dikalov, S., and Bassenge, E. (2000) Free Rad. Biol. Med. 28 121–128 [DOI] [PubMed] [Google Scholar]
- 35.Münzel, T., Li, H., Mollnau, H., Hink, U., Matheis, E., Hartmann, M., Oelze, M., Skatchkov, M., Warnholtz, A., Duncker, L., Meinertz, T., and Förstermann, U. (2000) Circ. Res. 86 E7–E12 [DOI] [PubMed] [Google Scholar]
- 36.Rocha, J. B. T., Lissner, L. A., Puntel, R. L., Fachinetto, R., Emanuelli, T., Nogueira, C. W., and Soares, F. A. A. (2005) Biol. Pharm. Bull. 28 1485–1489 [DOI] [PubMed] [Google Scholar]
- 37.Ishii, K., Sheng, H., Warner, T. D., Förstermann, U., and Murad, F. (1991) Am. J. Physiol. 261 H598–H603 [DOI] [PubMed] [Google Scholar]
- 38.Thatcher, G. R., Nicolescu, A. C., Bennett, B. M., and Toader, V. (2004) Free Rad. Biol. Med. 37 1122–1143 [DOI] [PubMed] [Google Scholar]
- 39.Booth, B. P., Tabrizi-Fard, M. A., and Fung, H. L. (2000) Biochem. Pharmacol. 59 1603–1609 [DOI] [PubMed] [Google Scholar]
- 40.Miranda, K. M., Paolocci, N., Katori, T., Thomas, D. D., Ford, E., Bartberger, M. D., Espey, M. G., Kass, D. A., Feelisch, M., Fukuto, J. M., and Wink, D. A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 9196–9201 [DOI] [PMC free article] [PubMed] [Google Scholar]