Abstract
Platyhelminth parasites are a major health problem in developing countries. In contrast to their mammalian hosts, platyhelminth thiol-disulfide redox homeostasis relies on linked thioredoxin-glutathione systems, which are fully dependent on thioredoxin-glutathione reductase (TGR), a promising drug target. TGR is a homodimeric enzyme comprising a glutaredoxin domain and thioredoxin reductase (TR) domains with a C-terminal redox center containing selenocysteine (Sec). In this study, we demonstrate the existence of functional linked thioredoxin-glutathione systems in the cytosolic and mitochondrial compartments of Echinococcus granulosus, the platyhelminth responsible for hydatid disease. The glutathione reductase (GR) activity of TGR exhibited hysteretic behavior regulated by the [GSSG]/[GSH] ratio. This behavior was associated with glutathionylation by GSSG and abolished by deglutathionylation. The Km and kcat values for mitochondrial and cytosolic thioredoxins (9.5 μm and 131 s–1, 34 μm and 197 s–1, respectively) were higher than those reported for mammalian TRs. Analysis of TGR mutants revealed that the glutaredoxin domain is required for the GR activity but did not affect the TR activity. In contrast, both GR and TR activities were dependent on the Sec-containing redox center. The activity loss caused by the Sec-to-Cys mutation could be partially compensated by a Cys-to-Sec mutation of the neighboring residue, indicating that Sec can support catalysis at this alternative position. Consistent with the essential role of TGR in redox control, 2.5 μm auranofin, a known TGR inhibitor, killed larval worms in vitro. These studies establish the selenium- and glutathione-dependent regulation of cytosolic and mitochondrial redox homeostasis through a single TGR enzyme in platyhelminths.
The control of parasitic infections, which are a major cause of disability, mortality, and economic losses in many developing countries, remains as one of the most important challenges for medicine in the 21st century (1). In the case of the phylum platyhelminthes (flatworms), which include the causative agents of schistosomiasis (bilharzia) and hydatid disease, pharmacotherapy with praziquantel has met great success in the treatment of infection. However, drug resistance is a serious issue as it has been the case for other antiparasitic drugs (2). In the case of platyhelminths, this may have severe consequences, because praziquantel is the only drug that is readily available for large scale treatment of these infections (3). Thus, the need for new drugs and/or vaccines is of great importance. In recent years, evidence has accrued that the selenocysteine (Sec)2-containing enzyme thioredoxin glutathione reductase (TGR) is essential for platyhelminth parasites and has emerged as a rational target for chemotherapy and/or immunotherapy (4–8). In most organisms, including the mammalian hosts of platyhelminths, cellular redox homeostasis, antioxidant defenses, and supply of reducing equivalents to several targets and essential enzymes rely on two major pathways: the glutathione (GSH) and the thioredoxin (Trx) systems, which have overlapping and differential targets and functions (9, 10). In contrast, platyhelminth parasites lack conventional thioredoxin reductase (TR) and glutathione reductase (GR), and hence conventional Trx and GSH systems (4, 6, 7). Instead, they rely exclusively on linked thioredoxin-glutathione systems, with TGR being the key enzyme that provides reducing equivalents to both pathways. Another feature of the linked systems in platyhelminths is that cytosolic and mitochondrial TGR derive from a single gene and have identical sequence, once the leader peptide of the mitochondrial variant is removed (5). In the mammalian hosts, different thioredoxin reductase isozymes function in the cytosol and the mitochondria (11, 12), TGR expression is largely restricted to testis (13), and GR exists as a distinct gene (14). In sum, the dissimilar arrangements of redox pathways as compared with their hosts, the lack of back-up systems, and the fact that parasitic organisms are subjected not only to the endogenous oxidative stress, but also to the oxidative challenge imposed by the host's immune system, provide a strong rationale to target platyhelminth TGRs. Recent studies support this idea: inhibition of TGR expression by RNA interference caused death of the platyhelminth parasite Schistosoma mansoni, and auranofin, a potent inhibitor of TGR and Sec-containing TRs (15), caused a partial cure in experimental Schistosoma infection (8).
TGR possesses a fusion of conventional TR domains with a glutaredoxin (Grx) domain (13, 16). TGR, like GR and TR, is a homodimer, with monomers oriented in a head-to-tail manner. Based on biochemical data, the current model of the mechanism of reaction for TGR proposes that electrons flow from NADPH to FAD, to the C156XXXXC redox center (numeration according to Echinococcus granulosus TGR), to the C-terminal GC595UG (U is Sec) redox center of the second subunit, and finally to the C31XXC redox center of the Grx domain of the first subunit. The fully reduced enzyme can reduce either oxidized Trx using the C-terminal active site GCUG, or GSSG through the CXXC redox center of the Grx domain (13, 17). Recently, a crystallographic structure of an S. mansoni C-terminally truncated TGR (GCstop) has been solved. Based on the residual GR activity of the mutant, the authors proposed an alternative view in which GSSG could be reduced directly by the CXXXXC redox center of TR domains (18).
In the current study, we have characterized the linked thioredoxin-glutathione system of the platyhelminth E. granulosus, the causative agent of hydatid disease. We demonstrated the occurrence of functional linked systems in both cytosol and mitochondria. The analysis of activities of TGR mutants revealed that the Grx domain is required for the GR activity, but does not affect the TR activity; in contrast, both Trx- and glutathione-dependent activities require selenocysteine (Sec) residue. Our results also indicate that [GSSG]/[GSH] ratio regulates TGR activities and strongly suggest that glutathionylation/deglutathionylation is involved in this regulation. In addition, we show that larval worms are killed by very low concentrations of auranofin, a TGR inhibitor, and discuss our results in light of the current models that have been put forward to explain the GR activity of TGR.
EXPERIMENTAL PROCEDURES
Cloning of Mitochondrial Isoforms of Trx and TGR as N-terminal Fusions to EGFP
To analyze the subcellular localization of the putative mitochondrial variants of Trx and TGR, constructs were generated using pEGFP-N2 (Clontech). In the case of Trx, the sequence was retrieved from Partigen (cluster EGC03292), and the entire coding region, including the leader peptide, was cloned as an in-frame fusion to EGFP. In the case of TGR the N-terminal fragment of mitochondrial TGR, containing the leader peptide followed by the Grx domain of TGR, was cloned as an EGFP fusion. In both cases, a Kozak consensus sequence was included in the forward primer for initiation of translation at the first AUG codon. For transient expression, mouse NIH 3T3 cells (ATCC) were cultured in DMEM supplemented with 10% fetal bovine serum in the presence of 100 units/ml penicillin and 50 units/ml nystatin. Transfections were carried out in 35-mm glass bottom culture dishes using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Transfection mix was prepared using 3 μg of plasmid DNA (mitochondrial Trx construct, mitochondrial TGR construct, or pEGFP-N2, used as a control) and 6 μl of Lipofectamine per dish. Transfections were carried out in Opti-MEM (Invitrogen) for 8 h. The transfection medium was replaced with a DMEM culture medium containing MitoTracker Red CM-H2XRos (Molecular Probes), a marker of the mitochondrial compartment; cells were incubated for 30 min and then washed twice with DMEM. Transiently transfected cells were detected by confocal microscopy (Bio-Rad, MRC1024ES laser scanning microscope).
Cloning, Expression, and Purification of Recombinant TGR and Its Mutants
Different constructs were made for expression of wild-type TGR (TGRGCUG) (where U is Sec, and GCUG is the C-terminal tetrapeptide) and the following mutants: Sec596 to Cys (TGRGCCG), Sec596 to stop (TGRGC*), Sec596 to Cys, and Cys595 to Sec (TGRGUCG), as well as a mutant lacking the entire Grx domain of TGR (TRGCUG). In all cases mRNA from trizoled E. granulosus protoscoleces (larval worms) was used as a template for reverse transcription and PCR, using ThermoScript reverse transcriptase (Invitrogen) and Pfu (Fermentas), respectively. Forward and reverse gene-specific primers were derived from the previously published TGR sequence (5). In the case of mutant TGRs, the reverse primers were modified appropriately. For Sec-encoding constructs (TGRGCUG, TGRGUCG, and TRGCUG), further engineering of the reverse primers was needed to specify Sec, because the UGASec codon requires recoding by an Sec insertion sequence (SECIS) element present in the selenoprotein mRNAs. Thus, for these constructs, the reverse gene-specific primer contained, at the 5′-end, the SECIS element of Escherichia coli formate dehydrogenase H at a 10-nucleotide distance from the UGASec codon (sequences of primers for every construct are detailed in supplemental Table S1). This strategy with a bacterial-type SECIS has been previously used for C-terminal Sec incorporation in E. coli (19). The amplified products were first cloned into pGEM-T-easy (Promega), and the construct sequences were verified prior to subsequent subcloning into pET28a (Novagen). Constructs were used to transform E. coli BL21(DE3) cells, or, in the case of selenoprotein constructs, BL21(DE3) cells previously transformed with pSUABC, a plasmid that supports high level expression of genes involved in Sec synthesis and decoding (selA, selB, and selC) (19). Expression of recombinant proteins was carried out following the protocol described in a previous study (20), which has been optimized for expression of selenoproteins. Essentially, induction of recombinant proteins was carried out with 100 μm isopropyl 1-thio-β-d-galactopyranoside at late exponential phase (A600 = 2.4), during 24 h at 24 °C. Recombinant clones were grown in modified LB media according to a previous study (21), supplemented with 0.1 g/liter cysteine and 0.37 g/liter methionine (22), in the presence of kanamycin (50 μg/ml), and chloramphenicol (33 μg/ml); the latter was used only in the case of bacterial cultures harboring the TGRGCUG, TGRGUCG, and TRGCUG constructs. At the time of induction the culture was supplemented with 5 μm sodium selenite, 20 μg/ml riboflavin, 20 μg/ml pyridoxine, and 20 μg/ml niacin according to a previous study (21). For recombinant TGRs that did not contain Sec (TGRGCCG and TGRGC*) the same protocol was followed, except that the plasmid pSUABC was not used. The bacterial cultures were centrifuged, and the pellets were resuspended in modified nickel-nitrilotriacetic acid lysis buffer (300 mm NaCl, 50 mm sodium phosphate, 20 mm imidazole, pH 7.2) containing 1 mm phenylmethylsulfonyl fluoride and 1 mg/ml lysozyme, and sonicated (10 pulses of 1 min with 1-min pauses). The lysates were centrifuged for 1 h at 30,000 × g, and supernatants were applied to a nickel-nitrilotriacetic acid column (Qiagen), washed with 300 mm NaCl, 50 mm sodium phosphate, 30 mm imidazole, pH 7.2, and eluted with 250 mm imidazole. The protein-containing fractions were applied to PD10 desalting columns (GE Healthcare) using phosphate-buffered saline, 150 mm potassium chloride, 50 mm sodium phosphate, pH 7.2. Fractions containing the recombinant proteins were stored at –70 °C before use. Total protein concentration and FAD content were determined spectrophotometrically at 280 (ε = 54.24 mm–1 cm–1) and 460 nm (ε = 11.3 mm–1 cm–1), respectively. The selenium content of selenoproteins was determined by atomic absorption using a Plasma Emission Spectrometer (Jarrell-Ash 965 ICP) in Chemical Analysis Laboratory, University of Georgia. The purity of the recombinant proteins was analyzed by running 10% SDS-PAGE gels, under reducing conditions, and by size-exclusion chromatography on a Superose 12 column (GE Healthcare).
Cloning, Expression, and Purification of E. granulosus Recombinant Mitochondrial and Cytosolic Trx Forms
mRNAs encoding cytosolic and mitochondrial E. granulosus Trxs were amplified by reverse transcription-PCR from total larval worm mRNA as described above. Specific forward and reverse primers for cytosolic and the predicted mature mitochondrial Trx were derived from previously published sequences (23) and from Partigene (cluster EGC03292), respectively. The amplified products were first cloned into pGEM-T-easy (Promega), sequenced and subsequently subcloned into pET28a (Novagen) using appropriate restriction enzymes. Constructs were used to transform E. coli BL21(DE3) host cells. Expression of recombinant proteins was carried out following the standard protocol for expression of recombinant proteins. Essentially, recombinant clones were grown on LB in the presence of kanamycin, and induction of recombinant proteins was carried out with 100 μm isopropyl 1-thio-β-d-galactopyranoside at early exponential phase (A600 = 0.5), for 3 h at 37 °C. The bacterial cultures were centrifuged, and the recombinant proteins were purified and desalted as described above for TGR, except that all buffers had pH 7.8. Fractions containing the recombinant proteins were stored at –70 °C prior to use. Protein concentration was determined spectrophotometrically at 280 nm (ε = 7.6 and 6.1 mm–1 cm–1 for cytosolic and mitochondrial Trx, respectively). The purity of the recombinant proteins was analyzed by running 15% SDS-PAGE gels under reducing conditions, and by size exclusion chromatography on a Superdex 75 column (GE Healthcare).
Metabolic Labeling of Selenoproteins
To label cells with 75Se, E. coli cells carrying the different constructs were grown at 37 °C until A600 reached 0.4, and the culture was supplemented with ∼50 μCi 75Se (as freshly neutralized sodium selenite, specific activity of 1000 Ci/mmol, Research Reactor Facility, University of Missouri, Columbia, MO). After an additional 30 min, isopropyl 1-thio-β-d-galactopyranoside was added to each cell culture at a final concentration of 100 μm. After 3 h of induction at 37 °C, cells were collected, washed, and lysed by boiling in SDS-PAGE sample buffer containing 50 mm 1,4-dithiothreitol (DTT). Cell lysates were then subjected to SDS-PAGE followed by transfer of proteins onto a polyvinylidene difluoride membrane. 75Se signal was visualized with a phosphorimaging device (Fuji).
Enzymatic Assays
Insulin Reduction Assay for Trx Activity—The efficient reduction of two interchain disulfides of insulin catalyzed by Trx in the presence of DTT was used as a measure of Trx activity, according to a previous study (24). The reaction was followed by the increase in absorbance at 650 nm due to the precipitation of free insulin B-chain. The 0.8-ml reaction mixtures contained 0.33 mm DTT, 130 μm insulin, and 2 mm EDTA in 100 mm potassium phosphate buffer, pH 7.0. Runs with DTT alone were performed as controls.
DTNB Reduction Assay for TR Activity—The reduction of 5,5′-dithiobis (2-dinitrobenzoic acid) (DTNB) with concomitant NADPH oxidation was determined by the increase in absorbance at 412 nm due to formation of 5′-thionitrobenzoic acid at 25 °C (25). The 0.8-ml reaction mixtures contained 0.2 mm NADPH, 5 mm DTNB, and 10 mm EDTA in 100 mm potassium phosphate buffer, pH 7.0.
Insulin Reduction Assay for TR Activity—The Trx-coupled assay of TR activity takes advantage of the NADPH-dependent reduction of Trx by TR, which is followed by the decrease in absorbance at 340 nm; in this assay, excess of insulin is used as an electron sink to maintain a constant concentration of oxidized Trx (25). The 0.8-ml reaction mixtures contained 0.2 mm NADPH, 1 mm EDTA, 0.5 mg/ml insulin, and E. granulosus cytosolic or mitochondrial Trx (concentrations ranged from 0 to 80 μm and from 0 to 140μm, respectively), in 50 mm potassium phosphate buffer, pH 7.0. The kinetic parameters of TGR with its physiological substrates, cytosolic and mitochondrial Trx, were determined from Michaelis-Menten plots of vo (derived from time-course experiments) against substrate concentration.
GR Assay—The GR activity was assayed as NADPH-dependent reduction of oxidized glutathione (GSSG), which is followed as the decrease in absorbance at 340 nm (26). The 0.8-ml reaction mixture contained 0.125 mm NADPH, 1 mm GSSG, 1 mm EDTA in 100 mm potassium phosphate buffer, pH 7.0.
All enzymatic assays were carried out in a Cary 50 (Varian) spectrophotometer at 25 °C. Analyses of the kinetic data were performed using ORIGIN software (OriginLab).
Mass Spectrometry Analysis—Wild-type TGR samples (10 nm concentration) were incubated with GSSG in the presence or absence of 0.125 mm NADPH at molar ratios under which hysteresis was or was not observed (1 mm or 30 μm GSSG, respectively) in 100 mm potassium phosphate buffer, pH 7.0, containing 1 mm EDTA, and immediately passed through a PD10 desalting column (GE Healthcare). Protein-containing fractions were digested with trypsin and subjected to analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (4800 Analyzer, Applied Biosystems). The mass spectrometry analysis was carried out at the Institut Pasteur, Montevideo.
In Vitro Culture of Larval Worms—50,000 protoscoleces, obtained from asceptical punction of a single hydatid cyst from bovine lung, were washed several times with phosphate-buffered saline and then incubated at 37 °C, 5% CO2, in DMEM supplemented with antibiotics and 20 mm HEPES, pH 7.4. Cultured protoscoleces were treated with 1, 2.5, 5, and 10 μm auranofin or with the vehicle (DMSO), in the presence or absence of 100 μm hydrogen peroxide. Protoscoleces were observed under the microscope, and viability was assessed by exclusion of the vital dye eosin.
RESULTS
Mitochondrial Localization of Trx and TGR Forms—Sequence analyses suggested the occurrence of both cytosolic and mitochondrial forms of TGR and Trx, with TGR forms generated from a single gene, and two genes for Trx forms. The results of transient expression in mammalian NIH 3T3 cells of the predicted mitochondrial forms of Trx and TGR are shown in Fig. 1. Both EGFP fusion proteins co-localized with MitoTracker, indicating that the signal peptides of these proteins direct the fusions to the mitochondrial compartment. No obvious staining of the cytosol or other subcellular compartments was observed. TGR has been previously shown to be present in the mitochondrial subcellular fraction of a larval worm aqueous extract (5); however, the mitochondrial location of Trx was previously limited to in silico predictions in platyhelminths (7).
FIGURE 1.
Subcellular localization of GFP-fused mitochondrial TGR and mtTrx. NIH3T3 cells were transiently transfected either with the non-recombinant pEGFP vector (A) or with the pEGFP-derived constructs carrying mitochondrial TGR (B) and mtTrx (C) N-terminally fused to GFP. Images were obtained at 8 h post-transfection using an I-confocal microscope. A set of three panels is shown for each construct. Left panels show green fluorescence corresponding to transiently expressed GFP fusion proteins. Center panels show the red fluorescence of the mitochondrial dye (MitoTracker). The right panels show merged images from left and center panels.
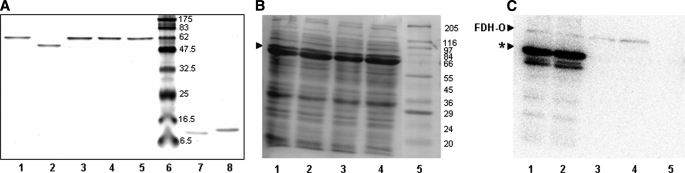
TGR Can Provide Electrons to Both Cytosolic and Mitochondrial Trx Forms—Prior to determining the enzymatic parameters of TGR with its physiological substrates, the quality of every recombinant protein was assessed in several ways. First, the purity of TGR and its mutants, and of cytosolic and mitochondrial Trx forms, was determined by SDS-PAGE under reducing conditions (Fig. 2A) and by size exclusion chromatography (data not shown). In the case of selenoproteins, Sec incorporation was evaluated by metabolic labeling of the bacterial cultures with 75Se. The results are shown in Fig. 2 (B and C); specific labeling at the expected molecular weight was observed exclusively in the bacterial lysates expressing selenoproteins, indicating that full-length selenoproteins were synthesized. In addition, the selenium content of recombinant selenoproteins was determined. Sec incorporation was close to 10% in all recombinant selenoproteins (9.2% for wild-type TGRGCUG, 7.4% for inverted TGRGUCG, and 8.7% for TRGCUG). Taken together, all these data indicated that the strategy was successful to produce full-length TGR in a bacterial host, although higher percentages of Sec incorporation (up to 50%) have been previously reported using this methodology for other selenoproteins (19). Because only a fraction of the TGR molecules incorporate Sec (due to prevalent termination of translation at Sec UGA codons), active protein concentrations of selenoproteins were corrected according to their selenium content.
FIGURE 2.
Analysis of recombinant proteins. A, SDS-PAGE analysis of purified recombinant proteins. After purification on a nickel-nitrilotriacetic acid column and desalting, recombinant proteins were run on a 12.5% polyacrylamide gel. 1 μg of each recombinant protein was loaded on the gel. Lanes 1–5, TGRGCUG, TRGCUG, TGRGUCG, TGRGCCG, TGRGC*, respectively; lane 6, molecular weight markers; lane 7, mtTrx; and lane 8, cTrx. The positions of molecular weight marker are indicated on the right. B and C. 75Se incorporation into recombinant TGRs. BL21(DE3) cells expressing TGRGCUG, TGRGUCG, TGRGCCG, and TGRGC* were induced with 100 μm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C. 50 μCi of 75Se were added to 10-ml cultures 30 min before induction. Total cell protein samples were resolved by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. B, Coomassie Blue staining of the polyvinylidene difluoride membrane. C, 75Se detection by phosphorimaging device analysis. Lanes 1–4, 10 μl of total cell protein samples from cells expressing recombinant TGRGCUG, TGRGUCG, TGRGCCG, and TGRGC*, respectively; lane 5, molecular weight marker. FDH-O, formate dehydrogenase O (110 kDa), the single selenoprotein expressed by E. coli under aerobic conditions. The bands around 66 kDa are indicated by an asterisk on lanes 1 and 2 on the right panel and correspond to 75Se-labeled Sec-containing recombinant TGRs. Lower molecular mass bands on these lanes probably correspond to secondary initiation or degradation products of these Sec-containing recombinants. The absence of bands on lanes 3 and 4 indicates no unspecific selenium incorporation was detected. The positions of molecular weight markers are indicated on the right.
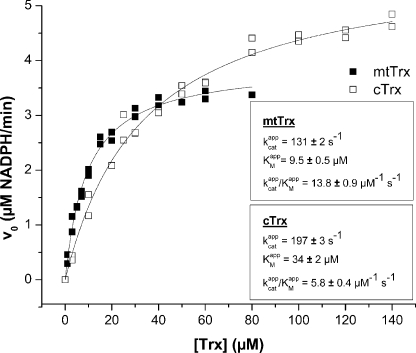
The activities of recombinant TGR and Trx were initially assessed independently of each other, using the DTNB and insulin reduction assays (see “Experimental Procedures”), respectively. Both recombinant enzymes displayed activity in these independent assays (data not shown). Then, using the insulin coupled assay, the kinetic parameters of TGR with its physiological substrates, cytosolic and mitochondrial Trx, were determined from Michaelis-Menten plots of vo against substrate concentration (Fig. 3). Km and kcat values were of the same order for both Trxs (Fig. 3, Table 1). The catalytic efficiency of TGR was (13.8 ± 0.9) × 106, and (5.8 ± 0.4) × 106 m–1 s–1 for mitochondrial and cytosolic Trx, respectively.
FIGURE 3.
Kinetic parameters of TGRGCUG with mtTrx and cTrx. Apparent Km and kcat of TGRGCUG with mtTrx and cTrx were obtained using the Trx-coupled assay. The initial reaction velocities (v0) at different Trx concentrations were measured at a constant and saturating NADPH concentration (200 μm) and a constant enzyme concentration (0.5 nm). Plots of v0 versus substrate concentration for mtTrx and cTrx are shown. The data were fitted to the Michaelis-Menten equation using Origin 7.5 software. Apparent Km and vmax were obtained from these fittings and apparent kcat was calculated from apparent vmax. Apparent Km, kcat, and kcat/Km values are indicated. The enzyme (TGRGCUG) concentration used for kcat calculation was corrected according to its selenium content.
TABLE 1.
Kinetic parameters of wild-type TGR and its mutants
Apparent kinetic parameters for mtTrx and cTrx (physiological TGR substrates) were obtained by varying the mtTrx and cTrx concentrations at a constant and saturating concentration of NADPH and constant enzyme concentrations. Apparent kcat values for DTNB were determined from the slope of the initial velocities (v0) versus enzyme concentration plots (Fig. 4). The enzyme concentrations used for kcat calculations for selenoproteins (TGRGCUG, TRGCUG, and TGRGUCG) considered the actual concentrations of active enzymes (i.e. their values were corrected according to their selenium content).
| Parameter | Substrate | TGRGCUG | TRGCUG | TGRGUCG | TGRGCCG |
|---|---|---|---|---|---|
| Apparent Km (μm) | mtTrx | 9.5 ± 0.5 | 13.0 ± 0.6 | 12.0 ± 0.6 | 14.3 ± 0.6 |
| cTrx | 34 ± 2 | ||||
| Apparent kcat (s-1) | mtTrx | 131 ± 2 | 80 ± 2 | 6.4 ± 0.2 | 0.530 ± 0.007 |
| cTrx | 197 ± 3 | ||||
| DTNB | 118 ± 3 | 60 ± 3 | 6.0 ± 0.5 | 0.63 ± 0.03 | |
| Apparent kcat/Km (μm-1s-1) | mtTrx | 13.8 ± 0.9 | 6.1 ± 0.4 | 0.27 ± 0.03 | 0.037 ± 0.002 |
| cTrx | 5.8 ± 0.4 |
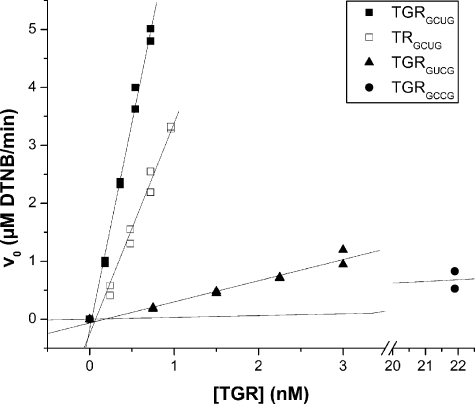
Sec but Not the Grx Domain Is Essential for TR Activity—To assess the role of the Grx domain and of Sec at the GCUG C-terminal redox center of TGR in the catalysis, we generated a set of TGR forms: wild-type TGR (TGRGCUG), TRGCUG (without the Grx domain), Sec596 to stop mutant (TGRGC*), Sec596 to Cys mutant (TGRGCCG), and Cys595 to Sec and Sec596 to Cys double mutant (TGRGUCG). Analysis of TR activity with the DTNB assay (shown in Fig. 4 and summarized in Table 1) revealed that TRGCUG and wild-type TGR have similar kcat. TGRGC* had negligible activity even at 500 nm enzyme concentration (data not shown). TGRGCCG had a kcat more than two orders of magnitude lower than that of wild-type TGR. The double mutant with Sec and Cys at inverted positions, TGRGUCG, had a kcat one order of magnitude higher than that of the Sec596 to Cys mutant. We next evaluated the kinetic parameters of the mutants with mitochondrial Trx. The results are summarized in Table 1. The catalytic efficiency (kcat/Km) of the Sec to Cys mutant was 2.5 orders of magnitude lower than that of the wild-type TGR, whereas the double mutant was approximately one order of magnitude higher than the Cys mutant (Table 1). Both TR assays indicated that, although the Grx domain did not affect the TR activity, the C-terminal Sec residue was essential for this activity. Interestingly, a Sec residue at the resolving position of the C-terminal redox center could partially compensate for the loss of activity due to Sec to Cys mutation.
FIGURE 4.
TR activity of TGR mutants. The TR activities of wild-type and mutant TGRs were compared using the DTNB assay. The assay was carried out at constant and saturating concentrations of DTNB and NADPH (5 mm and 200 μm, respectively) and different concentrations of each enzyme. The plots of initial velocities (v0) versus enzyme concentration are shown. The selenoenzyme (TGRGCUG, TRGCUG, and TGRGUCG) concentrations used for kcat calculations were corrected according to their selenium contents.
GR Activity Exhibits Hysteretic Behavior That Is Dependent on the Ratio of Oxidized and Reduced Glutathione—A hysteretic behavior (i.e. the existence of a lag time before catalysis takes place) of the GR activity of TGR was previously reported for the TGR of Taenia crassiceps (another platyhelminth parasite) (6). This behavior has not been reported for E. granulosus, S. mansoni, or mammalian TGR (5, 13, 27). We observed that E. granulosus TGR exhibited hysteretic behavior for its GR activity, which became evident at TGR concentrations below 15 nm (Fig. 5A) or high GSSG concentrations (Fig. 5B). Although the results were analogous to those described for T. crassiceps TGR, the E. granulosus TGR exhibited less marked hysteresis (i.e. lower lag times for similar experimental conditions). Rendón et al. (6) described that preincubation of TGR with GSH abolishes the hysteretic behavior of the enzyme. We found that preincubation was not needed to relieve GR hysteresis of E. granulosus TGR (Fig. 5C). Although the lag time correlated directly with the concentration of GSSG, and inversely with the concentration of GSH, the maximal slope of the curves did not significantly change once the enzyme was fully active (see Fig. 5, B and C). We next assayed whether Trx can relieve GR hysteresis and found that 20 μm Trx, without preincubation, abolished the hysteretic behavior (Fig. 5D).
FIGURE 5.
Hysteretic behavior of GR activity of TGR. Full-time courses obtained using different assay conditions are shown. In all cases the reactions were started by the addition of TGRGCUG at the indicated final concentrations. A, effect of enzyme concentration. Assays were performed at varying TGRGCUG concentrations and constant NADPH and GSSG concentrations (100 μm and 1 mm, respectively). B, effect of GSSG concentration. GSSG concentration was varied at constant NADPH and enzyme concentrations (100 μm and 10 nm, respectively). Note that at 31 and 62 μm GSSG reactions come to their end at higher A340, i.e. before depleting NADPH, because GSSG becomes the limiting reagent. C, effect of GSH concentration. GSH was included at various concentrations while maintaining constant GSSG, NADPH, and enzyme concentrations (1 mm, 125 μm, and 10 nm, respectively). D, effect of Trx concentration. cTrx was added at different concentrations to reaction mixtures containing 1 mm GSSG, 125 μm NADPH, and 10 nm TGRGCUG. The TGRGCUG concentration considered was corrected according to its selenium content.
GR Activity Is Reversibly Regulated by Glutathionylation/Deglutathionylation—Rendón et al. (6) have postulated that TGR must possess two GSH-binding sites, one of them regulatory, with high affinity for reduced GSH. We examined binding of E. granulosus TGR to a glutathione matrix and found that neither oxidized nor NADPH-reduced TGR associated with GSH-agarose. Because the [GSSG]/[GSH] ratio controls the activity of the enzyme, we reasoned that TGR may be glutathionylated by GSSG at high concentrations and deglutathionylated by GSH. Thus, we incubated 10 nm TGR with NADPH and 1 mm GSSG for 1 min (conditions at which the enzyme is still under hysteresis, see Fig. 5B) and for 10 min (the enzyme is no longer under hysteresis, see Fig. 5B). We then subjected the enzymes to tryptic digest and mass spectrometry analysis. The results indicated that TGR is glutathionylated at two Cys residues: Cys88 and Cys354 after 1-min incubation with 1 mm GSSG and NADPH (supplemental Fig. S1). Absence of NADPH did not prevent glutathionylation. In contrast, after 10-min incubation with 1 mm GSSG and NADPH, TGR was found to be deglutathionylated (supplemental Fig. S1). In similar experiments, no glutathionylation was found when 10 nm TGR was incubated for 1 min with 30 μm GSSG (a concentration under which there is no hysteresis, see Fig. 5B) with or without NADPH. In all cases, neither the Cys residues belonging to the Grx active site nor those of the CXXXXC catalytic redox center of TGR were detected as glutathionylated peptides; instead, they were detected to be forming disulfides. Some Cys-containing peptides, including the Sec-containing peptide could not be detected. Altogether the results indicated that the enzyme is glutathionylated at high GSSG concentrations and suggest that it becomes deglutathionylated once the enzyme is active.
We then tested whether the TR activity was preserved in TGR at conditions at which the GR activity was under hysteresis. Because it is not possible to measure the TR activity of TGR in the presence of GSSG, we incubated 10 nm TGR with 1 mm GSSG for 1 min (conditions under which there is GR hysteresis and glutathionylation) and applied the glutathionylated TGR to a PD10 desalting column to remove GSSG. We measured the TR activity of this treated TGR by both the insulin-coupled (Fig. 6A) and DTNB assays (data not shown) finding no hysteretic behavior. In contrast, the GR activity of the treated TGR was under hysteresis at low GSSG concentrations (condition under which there is no hysteresis if the enzyme was not pretreated) (Fig. 6B). In addition, we examined whether Grx domain contributed to glutathionylation. For this purpose, we evaluated glutathionylation and TR activity of TRGCUG at high [GSSG]. We found that the TR activity was unaffected at high [GSSG] using the Trx-coupled assay (Fig. 6C); however, the peptide containing Cys354 was not detected in TRGCUG (either native or glutathionylated).
FIGURE 6.
TR activity of TGRGCUG and TRGCUG analyzed at the hysteresis conditions for GR activity. A, the TR activities of untreated and glutathionylated TGRGCUG were compared using the Trx-coupled assay. B, the GR activities of untreated and glutathionylated TGRGCUG were compared at 100 μm GSSG. In both A and B the enzyme preparations were assayed at 1 nm TGR concentration and 150 μm NADPH. It should be noted that, to calculate the volume of enzyme preparation that ought to be used in the assay, glutathionylated TGR was assumed to be 2-fold diluted following desalting. This approximation could explain the slightly smaller slopes observed for this enzyme in both assays, as compared with the untreated one. C, the TR activity of TRGCUG was evaluated using the Trx-coupled assay both in the absence and presence of high concentration (1 mm) GSSG. The enzyme was assayed at 1 nm final concentration and 150 μm NADPH. The selenoenzyme (TGRGCUG and TRGCUG) concentrations considered were corrected according to their selenium contents.
Both the Grx Domain and the Sec Residue Are Essential for GR Activity—We further investigated whether the GR activity was affected by the Grx domain and the Sec residue at the C-terminal redox center. TRGCUG, TGRGCCG, and TGRGC* did not display significant activity at the standard conditions of the assay (i.e. 1 mm GSSG), even at high concentrations of enzymes (200 nm). Because the phenomenon of hysteresis observed for the GR activity is dependent not only on the enzyme concentration, but also on GSSG and GSH concentrations, we evaluated the activity of these mutants at high enzyme concentrations (200 nm) and different concentrations of GSSG and in the presence of GSH. None of these enzymes exhibited significant activity at low GSSG concentrations (30 μm) (Fig. 7A), and addition of 1 mm GSH did not affect the enzymatic activity (data not shown). Altogether, these results indicated that the C-terminal and Grx redox centers are essential for GR activity. Interestingly, and similar to what was observed for the TR activity, the double mutant TGRGUCG exhibited significant GR activity (Fig. 7B). We then studied the effect of GSH addition on the GR activity of the double mutant. At high concentration of GSSG (1 mm), the addition of 1 mm GSH abolished hysteresis (Fig. 7C). Finally, we examined whether a combination of functional TR domains (TRGCUG) with functional Grx domain (TGRGC*) displays GR activity. No activity was observed at high concentrations of both proteins, indicating that electron transfer between these two proteins is not efficient. This also rules out the possibility that the ∼10-fold Grx excess with respect to functional TR domains present in TGRGCUG and TGRGUCG (due to the prevalent truncated forms present in these preparations) can affect the GR activity of these selenoproteins.
FIGURE 7.
GR activity of TGR mutants. Time courses obtained for the GR activity of wild-type TGR and its mutants are shown. In all cases the reaction was started by the addition of the enzymes at the indicated final concentrations. A, comparison of the GR activity of TGRGCUG, TGRGCCG, TGRGC*, and TRGCUG at 31 μm GSSG and 125 μm NADPH. B, the GR activity of TGRGUCG at different enzyme concentrations was evaluated at 31 μm GSSG and 125 μm NADPH. C, the effect of GSH addition on the hysteretic behavior of TGRGUCG was studied by including GSH at 1 mm in a reaction mixture containing 1 mm GSSG, 125 μm NADPH, and 25 nm TGRGUCG. The enzyme concentrations for selenoproteins (TGRGCUG, TRGCUG, and TGRGUCG) were corrected according to their selenium contents.
Auranofin, a TGR Inhibitor, Kills Larval Worms at Low Concentrations—Because glutathione and thioredoxin activities depend on TGR in both compartments, mitochondria and cytosol, we evaluated, in animal culture, the effects of auranofin on protoscoleces (larval worms). All protoscoleces were dead 12 h after addition of 10 μm auranofin. The disruption of protoscoleces tegument integrity was the first microscopic sign of the drug effect; latter changes included disorganization of the protoscolex parenchyma (Fig. 8). At 5 μm auranofin, 20% of protoscoleces were dead at 12 h, and 100% at 30 h. At 2.5 μm auranofin, all protoscoleces were dead after 48 h, whereas at 1 μm auranofin, 90% of protoscoleces survived, but development toward cyst was severely compromised. We then evaluated the effect of auranofin on protoscoleces subjected to oxidative stress (100 μm hydrogen peroxide). A lower percentage of protoscoleces, only 60%, survived at 1 μm auranofin.
FIGURE 8.
Auranofin effect on E. granulosus larval worms. Protoscoleces were incubated in vitro at 37 °C, 5% CO2 in DMEM with 10 μm auranofin, a TGR inhibitor, or its vehicle (DMSO) as a control. A, control protoscoleces after 30 h of culture. B, treated protoscoleces after 12 h of culture (all protoscoleces were dead, note the disorganization of the parenchyma and the loss of the hooks or the entire crown of hooks). C, treated protoscoleces after 30 h of culture. The scale bar on C corresponds to 100 μm.
DISCUSSION
Previous studies have shown that TGR is an essential enzyme in platyhelminth parasites and an attractive target for drug and/or vaccine development (8). In this study, we provide further evidence that validates TGR as a key platyhelminth molecule to target. We have previously shown that cytosolic and mitochondrial variants of TGR from E. granulosus derive from a single gene and have the same amino acid sequence, once the leader peptide of the mitochondrial isoform is removed. We now show, by transient expression in eukaryotic cells that the mitochondrial variant of TGR and a putative mitochondrial Trx from E. granulosus localize to the mitochondrial compartment when assessed by confocal microscopy. No experimental evidence for mitochondrial co-localization of platyhelminth TGR and Trx had been previously reported. We thus also proved that TGR provides electrons to E. granulosus cytosolic and mitochondrial Trxs as well as to GSSG, indicating the existence of TGR-dependent functional thiol systems in cytosol and mitochondria. Furthermore, TGR functions equally well with both Trx isozymes, with a catalytic efficiency of 107 m–1 s–1, similar to the values reported for other TGRs and TRs. The Km and kcat values for Trxs are higher than those reported for mammalian TRs (∼3 μm and 40 to 60 s–1, respectively) (15, 28, 29). However, Km values of the TGRs of the phylogenetically closest organisms to E. granulosus (T. crassiceps and S. mansoni) are also higher than those for mammalian TRs. The reported kcat values for these TGRs are lower than the reported herein for E. granulosus TGR, but it should be noted that no corrections by protein selenium content have been done in those cases (6, 8).
During characterization of the GR activity of E. granulosus TGR we observed that at low concentrations the enzyme exhibited hysteretic behavior. Furthermore, we showed that the [GSSG]/[GSH] ratio controlled the activity of the enzyme: the lag time correlated directly with GSSG concentration and inversely with GSH concentration. Hysteretic behavior has been associated with changes in conformation and/or oligomerization in response to substrates, products, or modifiers (30). We did not detect changes in the oligomerization state of TGR upon incubation of the enzyme with high GSSG concentrations by size exclusion chromatography (data not shown). Our results strongly indicated that the observed hysteretic phenomenon correlated to the glutathionylation state of TGR as summarized in Fig. 9. Further evidence that the hysteretic behavior of TGR was due to glutathionylation derived from the fact that, once GSSG was removed using a desalting column, the glutathionylated enzyme was hysteretic even at low GSSG concentrations. Of the two Cys residues found to be glutathionylated, Cys88 is present in vertebrate TGRs and in Grx, whereas Cys354 is present only in E. granulosus TGR (supplemental Fig. S2). Interestingly, Cys88 is close to the mobile linker of Grx-TR domains (100–105), and Cys354 is located close to a solvent accessible mobile loop (359–363) (18). Glutaredoxins have been shown to catalyze deglutathionylation of target proteins (31–33). Thus, an additional issue relates to whether deglutathionylation is autocatalyzed by TGR. We observed deglutathionylation of TGR by GSH in the absence of NADPH (data not shown). This suggests that deglutathionylation under these conditions can occur, but catalysis by the Grx domain cannot be ruled out. Because TGR catalyzes numerous thiol-(selenol)-based reactions, and at the same time its activity is controlled by the [GSSG]/[GSH] ratio, it constitutes an interesting model to study enzymatic regulation by glutathionylation/deglutathionylation, as well as substrate inhibition and product activation phenomena.
FIGURE 9.
Hysteretic behavior and glutathionylation of TGR. At high GSSG concentrations, the GR activity of TGR exhibited hysteretic behavior (filled squares); TGR was found to be glutathionylated under hysteresis and deglutathionylated once hysteresis was abolished. At low GSSG concentrations, the GR activity did not exhibit hysteretic behavior (open squares), and the enzyme was not glutathionylated. The figure also shows that hysteretic behavior is favored by high [GSSG]/[GSH] ratios or low enzyme concentrations and is relieved at low [GSSG]/[GSH] ratios or high enzyme concentrations.
The fact that hysteresis is found at high GSSG concentrations raises the question of whether this phenomenon takes place in vivo. It should be noted that parasites are under oxidative stress mainly due to the host's immune response. This generates not only GSSG, but also thiyl radicals, sulfenic acids, and S-nitrosylated glutathione and protein intermediates, which have been proposed as glutathione donors and acceptors for protein-SSG formation, respectively (34). Glutathionylation of proteins has been proposed to regulate diverse intracellular signaling pathways, particularly in mammalian cells under oxidative stress (9, 31, 34) and to provide a protective mechanism for the damaging effects of oxidative agents (34, 35). Our results indicated that TGR activity is preserved for reduction of Trx during oxidative stress; under these conditions TGR would be inactivated for the GR function, whereas the TR function would remain unaffected.
To gain further insights into the catalytic mechanism of TR and GR activities of TGR, we generated a set of constructs designed to study the roles of its C-terminal redox center and the N-terminal Grx domain. Consistent with the results obtained for mammalian TGR (17), we found that the Sec residue of TGR is essential for both activities. The Sec to stop mutant (a truncated mutant at the penultimate amino acid) has negligible GR and TR activities. Furthermore, a Sec to Cys mutation also results in a remarkable loss of TR activity and almost complete loss of GR activity. These results indicated that in E. granulosus the overall redox homeostasis is controlled by TGR and is dependent on selenium. A striking observation was that the additional mutation at the C-terminal redox center (TGRGUCG) partially compensated for the loss of activity caused by the single Sec to Cys mutation (TGRGCCG). This recovery of activity was observed with all substrates studied: DTNB, thioredoxin, and GSSG. Selenoprotein redox active sites are characterized by the presence of Sec at the nucleophilic (attacking) position, and a Cys residue is usually observed at the resolving position of the catalytic mechanism (17, 36–38). The higher nucleophilicity and low pKa of the selenol group of Sec (39) are thought to confer Sec a catalytic advantage over Cys at the attacking position. In addition, recent evidence supports the model that Sec is the leaving group during reduction of the C terminus during the catalytic cycle (38). No natural selenoprotein with Sec at the resolving position has been described (40), and it is assumed that Sec would not confer a significant advantage to the catalytic efficiency when present at this position. Furthermore, a semisynthetic mammalian TR with an inverted C-terminal active site resulted in 100-fold decrease in catalytic activity (comparable to the Sec to Cys mutant) (38). Our results suggest that Sec not only provides a catalytic advantage over Cys at the nucleophilic position, but a Cys to Sec mutation at the resolving position can also enhance enzymatic activity compared with the corresponding Cys form. The fact that Sec is superior to Cys at the resolving position of the redox active center may also apply to other selenoproteins and the corresponding thiol oxidoreductases. It would be interesting to explore this possibility in the context of engineering Cys-containing enzymes for enhanced catalysis.
Regarding the role of the Grx domain in TGR functions, the comparison of wild-type TGR and TR revealed that the Grx domain does not positively or negatively affect the TR activity of the enzyme, indicating that the Grx domain neither assists nor hinders the TR activity of TGR. In contrast, the TR module of TGR has no GR activity, and thus both the Grx and TR domains are needed for this activity.
Our data clearly indicate that efficient GSSG reduction requires both the C-terminal Sec-containing redox center and the N-terminal Grx domain, in agreement with the model originally proposed for mammalian TGR (13), and later supported by characterization of the enzyme (17). This model put forward that electrons can flow from the C-terminal GCUG redox center directly to Trx, or to the N-terminal CXXC redox center of the Grx domain and finally to GSSG. This implies that a conformational switch must exist to allow either Trx or the “in built” Grx domain to receive electrons from the C-terminal redox center. An alternative electron pathway for GSSG reduction has recently been suggested based on the crystallographic structure of a C-terminally truncated S. mansoni TGR. To account for the residual GR activity of the truncated enzyme, the authors proposed that GSSG could be reduced directly by the CXXXXC redox center of TR domains (18). The fact, that auranofin, an inhibitor of Sec-containing TRs and TGRs, inhibits not only TR but also GR activities of S. mansoni TGR, suggests that the proposed alternative electron pathway is marginal and is unlikely to be physiologically relevant (8, 27).
Finally, we performed in vitro studies on E. granulosus larval worms to evaluate the effects of auranofin. Concentrations as low as 2.5 μm of auranofin kill all larval worms within 48 h, and lower concentrations severely hampered in vitro development of larval worms toward cyst. The effect of the drug on larval worms challenged with hydrogen peroxide was even more marked. Low concentrations of auranofin have also been reported to rapidly kill juvenile and adult S. mansoni in culture. Mammalian cells, however, are able to survive higher concentrations of auranofin (8).
Data available indicate that in platyhelminth parasites the biochemical scenario of thiol-disulfide redox homeostasis differs greatly from that of their mammalian hosts. In platyhelminths, the overall redox homeostasis is controlled by TGR and is dependent on selenium. The substitution of conventional Trx and GSH systems by linked systems makes TGR (the molecular link for Trx and glutathione-dependent functions) a target molecule for drug or vaccine development. In this study, we demonstrated the existence of functional linked systems in both mitochondria and the cytosol that depends on a single TGR. Thus, targeting this enzyme can be safely expected to compromise the overall cellular redox homeostasis in these organisms.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of pSUABC from Dr. Elias Arnér (Karolinska Institutet). We also thank Lic. Bruno Manta for technical assistance and Dr. Beatriz Alvarez, Dr. Alvaro Díaz, and Dr. Cecilia Fernández for helpful discussions. We also thank the Institut Pasteur, Montevideo (Unidad Tecnológica de Bioquímica y Proteómica Analíticas), for technical assistance with mass spectrometry.
This work was supported, in whole or in part, by National Institutes of Health Grant GM065204 (to V. N. G.). This work was also supported by Fogarty International Research Collaboration Award-NIH Grant TW006959 (to V. N. G.) and by the Uruguayan Research Council (Grants PDT 29-171 and PDT 63-105 to G. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
Footnotes
The abbreviations used are: Sec, selenocysteine; DMEM, Dulbecco's modified Eagle's medium; DTNB, 5,5′-dithiobis(2-dinitrobenzoic acid); DTT, dithiothreitol; EGFP, enhanced green fluorescent protein; GR, glutathione reductase; Grx, glutaredoxin; GSSG, glutathione(oxidizedform); TGR, thioredoxinglutathione reductase; TR, thioredoxin reductase; Trx, thioredoxin; SECIS, selenocysteine insertion sequence; mtTrx, mitochondrial Trx; cTrx, cytosolic Trx.
References
- 1.WHO (2006) Preventive Chemotherapy in Human Helminthiasis: Coordinated Use of Antihelminthic Drugs in Control Interventions: a Manual for Health Professionals and Programme Managers, WHO Press, World Health Organization, Geneva, Switzerland
- 2.Mansour, T. (2002) in Chemotherapeutic Targets in Parasites: Contemporary Strategies (Mansour, T., ed) pp. 58–89, Cambridge University Press, Cambridge
- 3.Doenhoff, M. J., and Pica-Mattoccia, L. (2006) Expert Rev. Anti Infect. Ther. 4 199–210 [DOI] [PubMed] [Google Scholar]
- 4.Alger, H. M., and Williams, D. L. (2002) Mol. Biochem. Parasitol. 121 129–139 [DOI] [PubMed] [Google Scholar]
- 5.Agorio, A., Chalar, C., Cardozo, S., and Salinas, G. (2003) J. Biol. Chem. 278 12920–12928 [DOI] [PubMed] [Google Scholar]
- 6.Rendón, J. L., del Arenal, I. P., Guevara-Flores, A., Uribe, A., Plancarte, A., and Mendoza-Hernández, G. (2004) Mol. Biochem. Parasitol. 133 61–69 [DOI] [PubMed] [Google Scholar]
- 7.Salinas, G., Selkirk, M. E., Chalar, C., Maizels, R. M., and Fernández, C. (2004) Trends Parasitol. 20 340–346 [DOI] [PubMed] [Google Scholar]
- 8.Kuntz, A. N., Davioud-Charvet, E., Sayed, A. A., Califf, L. L., Dessolin, J., Arnér, E. S., and Williams, D. L. (2007) PLoS Med. 4 e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandes, A. P., and Holmgren, A. (2004) Antioxid. Redox Signal. 6 63–74 [DOI] [PubMed] [Google Scholar]
- 10.Winyard, P. G., Moody, C. J., and Jacob, C. (2005) Trends Biochem. Sci. 30 453–461 [DOI] [PubMed] [Google Scholar]
- 11.Turanov, A. A., Su, D., and Gladyshev, V. N. (2006) J. Biol. Chem. 281 22953–22963 [DOI] [PubMed] [Google Scholar]
- 12.Miranda-Vizuete, A., Damdimopoulos, A. E., Pedrajas, J. R., Gustafsson, J.-A., and Spyrou, G. (1999) Eur. J. Biochem. 261 405–412 [DOI] [PubMed] [Google Scholar]
- 13.Sun, Q.-A., Kirnarsky, L., Sherman, S., and Gladyshev, V. N. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 3673–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelner, M. J., and Montoya, M. A. (2000) Biochem. Biophys. Res. Commun. 269 366–368 [DOI] [PubMed] [Google Scholar]
- 15.Gromer, S., Arscott, L. D., Williams, C. H., Jr., Schirmer, R. H., and Becker, K. (1998) J. Biol. Chem. 273 20096–20101 [DOI] [PubMed] [Google Scholar]
- 16.Sun, Q.-A., Yalin, W., Zappacosta, F., Jeang, K.-T., Lee, B. J., Hatfield, D. L., and Gladyshev, V. N. (1999) J. Biol. Chem. 274 24522–24530 [DOI] [PubMed] [Google Scholar]
- 17.Sun, Q. A., Su, D., Novoselov, S. V., Carlson, B. A., Hatfield, D. L., and Gladyshev, V. N. (2005) Biochemistry 44 14528–14537 [DOI] [PubMed] [Google Scholar]
- 18.Angelucci, F., Miele, A. E., Boumis, G., Dimastrogiovanni, D., Brunori, M., and Bellelli, A. (2008) Proteins, in press [DOI] [PubMed]
- 19.Arnér, E. S., Sarioglu, H., Lottspeich, F., Holmgren, A., and Bock, A. (1999) J. Mol. Biol. 292 1003–1016 [DOI] [PubMed] [Google Scholar]
- 20.Rengby, O., Johansson, L., Carlson, L. A., Serini, E., Vlamis-Gardikas, A., Karsnas, P., and Arnér, E. S. (2004) Appl. Environ. Microbiol. 70 5159–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bar-Noy, S., Gorlatov, S. N., and Stadtman, T. C. (2001) Free Radic. Biol. Med. 30 51–61 [DOI] [PubMed] [Google Scholar]
- 22.Müller, S., Heider, J., and Böck, A. (1997) Arch. Microbiol. 168 421–427 [DOI] [PubMed] [Google Scholar]
- 23.Chalar, C., Martinez, C., Agorio, A., Salinas, G., Soto, J., and Ehrlich, R. (1999) Biochem. Biophys. Res. Commun. 262 302–307 [DOI] [PubMed] [Google Scholar]
- 24.Holmgren, A. (1979) J. Biol. Chem. 254 9627–9632 [PubMed] [Google Scholar]
- 25.Arnér, E. S., Zhong, L., and Holmgren, A. (1999) Methods Enzymol. 300 226–239 [DOI] [PubMed] [Google Scholar]
- 26.Carlberg, I., and Mannervik, B. (1985) Methods Enzymol. 113 484–490 [DOI] [PubMed] [Google Scholar]
- 27.Alger, H. M., Sayed, A. A., Stadecker, M. J., and Williams, D. L. (2002) Int. J. Parasitol. 32 1285–1292 [DOI] [PubMed] [Google Scholar]
- 28.Holmgren, A. (2006) in Selenium: Its Molecular Biology and Role in Human Health, 2nd Ed. (Hatfield, D. L., Berry, M. J., and Gladyshev, V. N., eds) pp. 183–194, Springer, New York
- 29.Zhong, L., and Holmgren, A. (2000) J. Biol. Chem. 275 18121–18128 [DOI] [PubMed] [Google Scholar]
- 30.Frieden, C. (1970) J. Biol. Chem. 245 5788–5799 [PubMed] [Google Scholar]
- 31.Beer, S. M., Taylor, E. R., Brown, S. E., Dahm, C. C., Costa, N. J., Runswick, M. J., and Murphy, M. P. (2004) J. Biol. Chem. 279 47939–47951 [DOI] [PubMed] [Google Scholar]
- 32.Johansson, C., Lillig, C. H., and Holmgren, A. (2004) J. Biol. Chem. 279 7537–7543 [DOI] [PubMed] [Google Scholar]
- 33.Lillig, C. H., Potamitou, A., Schwenn, J. D., Vlamis-Gardikas, A., and Holmgren, A. (2003) J. Biol. Chem. 278 22325–22330 [DOI] [PubMed] [Google Scholar]
- 34.Gallogly, M. M., and Mieyal, J. J. (2007) Curr. Opin. Pharmacol. 7 381–391 [DOI] [PubMed] [Google Scholar]
- 35.Niwa, T. (2007) J. Chromatogr. B 855 59–65 [DOI] [PubMed] [Google Scholar]
- 36.Biterova, E. I., Turanov, A. A., Gladyshev, V. N., and Barycki, J. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15018–15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckenroth, B., Harris, K., Turanov, A. A., Gladyshev, V. N., Raines, R. T., and Hondal, R. J. (2006) Biochemistry 45 5158–5170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eckenroth, B. E., Lacey, B. M., Lothrop, A. P., Harris, K. M., and Hondal, R. J. (2007) Biochemistry 46 9472–9483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gromer, S., Johansson, L., Bauer, H., Arscott, L. D., Rauch, S., Ballou, D. P., Williams, C. H., Jr., Schirmer, R. H., and Arnér, E. S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 12618–12623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gladyshev, V. N. (2006) in Selenium: Its Molecular Biology and Role in Human Health, 2nd Ed. (Hatfield, D. L., Berry, M. J., and Gladyshev, V. N., eds) pp. 90–110, Springer, New York
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.