Abstract
Syndecan 1 is the major proteoglycan produced by epithelial cells. It is strategically localized at the plasma membrane to participate in growth factor signaling and cell-cell and cell-matrix interactions. Its expression may modulate the properties of epithelial lineage tumor cells in which it is generally down-regulated compared with nontumor progenitors. The present study examined the regulation of syndecan 1 in prostate epithelial cells by n-3 polyunsaturated fatty acids. In prostate tissue of mice, syndecan 1 immunostaining was demonstrated in epithelial cells throughout each gland. In animals fed an n-3 polyunsaturated fatty acid-enriched diet, syndecan 1 mRNA was increased in all prostate glands. In the human prostate cancer cell line, PC-3, delivery of exogenous n-3 (but not n-6) fatty acids resulted in up-regulation of syndecan 1 expression. This effect was mimicked by a peroxisome proliferator-activated receptor (PPAR) γ agonist, troglitazone, and inhibited in the presence of a PPARγ antagonist and in cells transfected with dominant negative PPARγ cDNA. Using a luciferase gene driven either by a PPAR response element or by a DR-1 site present in the syndecan 1 promoter, reporter activation was increased by n-3 low density lipoprotein, docosahexaenoic acid, and troglitazone, whereas activity of a luciferase gene placed downstream of a mutant DR-1 site was unresponsive. These findings indicate that syndecan 1 is up-regulated by n-3 fatty acids by a transcriptional pathway involving PPARγ. This mechanism may contribute to the chemopreventive properties of n-3 fatty acids in prostate cancer.
The syndecan family of transmembrane heparan sulfate proteoglycans has roles in cell-cell and cell-matrix interactions, growth factor signaling, and lipoprotein catabolism (1). Syndecan 1 (SDC-1)2 is known to play a role in cell adhesion (2, 3), inhibit matrix metalloproteinases (4), and decrease invasion of tumor cells (5, 6). SDC-1 ectodomain was shown to induce apoptosis in myeloma cells (6), and our studies suggest a similar property in breast cancer cells.3 Therefore, changes in the expression of SDC-1 may have profound effects on cell behavior. In studies of various cancers, loss of SDC-1 expression has been associated with tumor progression and aggressiveness (7-13). In contrast, an elevation in SDC-1 expression in stromal cells was associated with tumor progression (14) and poor clinical outcome (15-17). In human prostate cancer, there are conflicting data. High expression of SDC-1, observed in normal prostate epithelial cells, was lost in localized and locally invasive prostate cancer (18). Similarly, compared with benign prostate hyperplasia (BPH) and low grade premalignant intraepithelial neoplasia, SDC-1 expression was reduced in prostate carcinoma and lost entirely in poorly differentiated cases (19). However, a tissue microarray analysis demonstrated SDC-1 expression associated with high Gleason grade and early recurrence (20). Therefore, a clear picture of changes in SDC-1 expression, regulation, and role in tumorigenesis has yet to emerge.
Although still controversial, evidence suggests that dietary fat intake may play a role in the development of prostate cancer with diets enriched in fish oil or its n-3 polyunsaturated fatty acids (PUFA) being chemoprotective. A recent review of epidemiological evidence indicated an inverse association between fish oil consumption and advanced/metastatic prostate cancer or prostate cancer mortality (21). In addition, the Health Professionals' Follow-up study, a large prospective U.S. cohort study, identified significant inverse relationships between intakes of marine fatty acid that were strongest for metastatic cancers (22, 23). Fish oil contains the long-chain PUFA, eicosapentaenoic acid (EPA (20:5, n-3)), and docosahexaenoic acid (DHA (22:6, n-3)). Serum PUFA in patients with prostate cancer and BPH versus cancer-free individuals demonstrated EPA and DHA levels in the order of cancer-free > BPH > prostate cancer (24). Reduced prostate cancer risk was associated with high erythrocyte EPA and DHA (25), and in prostate tissue reduced EPA and DHA levels were associated with cancer compared with BPH (26) and advanced stage disease (27). These data together suggest that not only may n-3 PUFA protect against primary tumor development but that dietary supplementation with n-3 PUFA may prevent or delay metastases in prostate cancer patients.
Few studies have addressed cellular mechanisms that link dietary PUFA and prostate cancer. Androgen-mediated growth of LNCaP cells (28) and androgen-independent growth of PC-3 and DU-145 prostate cancer cells was inhibited by n-3 PUFA and stimulated by n-6 PUFA (29). Recently, DHA was shown to induce apoptosis in PC-3 cells (30). The opposing effects of n-3 and n-6 PUFA are thought to reflect differences in eicosanoid metabolism (29, 31), which may influence transcription factor activation, gene expression, and signal transduction (32). Our previous studies have shown that, in human breast cancer cells, n-3 PUFA inhibit growth, induce apoptosis, and up-regulate SDC-1(33, 34). In the present study we show that SDC-1 is increased in prostate tissue of mice fed an n-3 PUFA-enriched diet, and that, in human prostate cancer cells, the transcriptional pathway for n-3-PUFA regulation of SDC-1 expression involves the nuclear hormone receptor, peroxisome proliferator-activated receptor (PPAR) γ.
EXPERIMENTAL PROCEDURES
Materials—Human prostate cancer PC-3 and DU-145 cells were obtained from the American Type Culture Collection (Rockville, MD). Mouse anti-human syndecan 1 (B-B4) was purchased from Serotec (Oxford, UK), anti-heparan sulfate stub monoclonal antibody (3G10) was from Seikagaku America (Ijamsville, MD), and rabbit polyclonal anti-human syndecan 1 (H-174) was from Santa Cruz Biotechnology (Santa Cruz, CA). Troglitazone and GW259662 were from Cayman Chemical (Ann Arbor, MI), heparinase III was from Sigma, and chondroitin ABC lyase was from Seikagaku America. A Super Signal West Pico Kit was from Pierce, and FuGENE 6 transfection reagent was from Roche Applied Science. Luciferase and β-galactosidase assay kits were from Promega (Madison, WI). Dominant negative (d/n) PPARγ cDNA (L468/E471) was kindly supplied by Dr. V. Chatterjee, University of Cambridge. Preparation of fatty acid-bovine serum albumin (BSA) complexes and n-3 and n-6 PUFA-enriched low density lipoproteins (LDLs) have been previously described (33).
Animals—Male C57BL/6J mice were fed a high n-3 (n-6:n-3 ratio = 1:1) or a high n-6 (n-6:n-3 ratio = 40:1) diet for 8 weeks. Diet composition, body weights, fatty acid ratios in food, blood, and prostate tissues, and dissection of prostate lobes have been described (35). All animals were maintained in an isolated environment in barrier cages. Animal care was conducted in compliance with the state and federal Animal Welfare Acts and the standards and policies of the Department of Health and Human Services. The protocol was approved by our Institutional Animal Care and Use Committee.
Immunostaining of Mouse Prostate Tissue—Prostate glands were dissected from 8-week-old male mice fed a chow diet. Tissues were fixed in Carnoy fluid at 4 °C, dehydrated, embedded in paraffin, sectioned (5 μm), and deparaffinized. Immunostaining was conducted using a rabbit polyclonal antibody raised against a recombinant protein corresponding to amino acids 82-256 of human syndecan 1 (H-174) and the DAKO EnVion + system peroxidase (3,3′-diaminobenzidine) kit according to the manufacturer's instructions.
Cell Culture—PC-3 and DU-145 cells were maintained in Advanced Dulbecco's modified Eagle's medium supplemented with 1% fetal bovine serum (FBS) and Eagle's minimal essential medium with 10% FBS, respectively, penicillin/streptomycin, and l-glutamine at 37 °C in 5% CO2. In experiments measuring mRNA, cells were seeded in 6-well plates at a density of 5 × 105 cells/well in growth medium. After 6 h, the medium was changed to Dulbecco's modified Eagle's medium/F-12 with 0.5% FBS for 18 h and then supplemented with 100 μg/ml n-3 or n-6-enriched LDL, 30 μm DHA, 30 μm EPA, 30 μm LA, and 5 μm or 10 μm troglitazone for 8-24 h.
Fatty Acid Delivery to Cells—Cells were grown in 100-mm dishes until ∼50% confluent when the growth media was replaced with fresh media containing 0.5% FBS supplemented with LDL or BSA-fatty acid. FBS was used at low levels to minimize competition between its LDL and the n-6 or n-3 LDL added to the cultures. After 24 h, cell monolayers were washed twice with balanced salt solution (137 mm NaCl, 2.7 mm KCl, 1.45 mm KH2PO4, 20.3 mm Na2HPO4), and lipids were extracted with isopropanol for 24 h at 4 °C. Lipid extracts were phased into chloroform and saponified, and fatty acids were methylated and separated by gas-liquid chromatography (36). Phospholipids were separated from chloroform extracts by thin layer chromatography prior to fatty acid analysis.
Real-time PCR—RNA isolation and cDNA synthesis and real-time RT-PCR were as previously described (34). SDC-1 primers were 5′-ggagcaggacttcacctttg (forward) and 5′-ctcccagcacctctttcct (reverse). Versican primers were 5′-cccatgcgctacataaagtca-3′ (forward) and 5′-agaccatttgatgcggagaa-3′ (reverse). Perlecan primers were 5′-cgctggacacattcgtacct-3′ (forward) and 5′-accagggctcggaaataaac-3′ (reverse). Peptidylprolylisomerase B housekeeping gene primers were 5′-gcccaaagtcaccgtcaa (forward) and 5′-tccgaagagaccaaagatcac (reverse). Mouse SDC-1 primers were 5′-tggagaacaagacttcacctttg-3′ (forward) and 5′-ctcccagcacttccttcct-3′ (reverse). Mouse peptidylprolylisomerase B housekeeping gene primers were 5′-aacagcaagttccatcgtgtc-3′ (forward) and 5′-ctttcctcctgtgccatctc-3′ (reverse). Proteoglycan core protein data were normalized to the housekeeping control peptidylprolylisomerase B and are presented relative to control.
Enzyme Treatment and Western Blot Analysis—Cells were washed twice with ice-cold phosphate-buffered saline and lysed for 10 min on ice; debris was then removed by centrifugation. Proteins were precipitated by a 2.5-fold volume cold methanol at -20 °C for 2 h. The samples were again centrifuged at 14,000 rpm for 10 min. For membrane proteins, cells were collected in hypotonic buffer (10 mm Tris-HCl, 1 mm EDTA) with protease inhibitors and homogenized, and debris was removed by centrifugation at 8,000 × g. Membranes in the supernatant were pelleted at 100,000 × g, dissolved in lysis buffer, and precipitated with cold methanol as above. Pellets were washed with ice-cold acetone, dried, and resuspended in heparinase buffer (50 mm HEPES, 50 mm NaOAc, 150 mm NaCl, 5 mm CaCl2), and equivalent amounts of protein were treated with heparinase III (2 units/ml) and chondroitinase ABC (1 unit/ml) at 37 °C for 16 h. Proteins were separated by 3.5-15% SDS-PAGE and transferred onto polyvinylidene difluoride membrane. The membranes were blocked with TBST (10 mm Tris-base, 100 mm NaCl, 0.1% Tween 20, pH 7.5) containing 5% nonfat dry milk for 2 h at room temperature, washed with TBST for 5 min × 3, exposed to the primary antibodies in TBST containing 3% BSA at 4 °C overnight, washed three times with TBST, incubated with horseradish peroxidase-conjugated secondary antibody for 1 h, washed with TBST, and developed using the Super Signal West Pico Kit.
Wild-type and Mutant SDC-1 DR-1-luciferase Constructs—The one copy reporter construct DR-1 from the SDC-1 promoter (pGL3-DR-1 wt (37)) was generated by annealing the oligonucleotides 5′-cggttcttcccctttgctctctcggccgtttccgctacacccgagct-3′ and 5′-cgggtgtagcggaaacggccgagagagcaaaggggaagaaccggtac-3′ before ligation into KpnI- and SacI-digested pGL3-luciferase vector. The one copy mutant DR-1 reporter construct (pGL3-DR-1) was generated using the same method and the oligonucleotides 5′-cggttctAGTActGCcCTcactcggccgtttccgctacacccgagct-3′ and 5′-cgggtgtagcggaaacggccgagTgagGGCagTACTagaaccggtac-3′. Mutations are capitalized (37).
Transfection and Luciferase Assay—PC-3 cells were co-transfected with the PPRE-TK-luciferase reporter plasmid and a control plasmid containing the lacZ gene or with pcDNA3-d/n-PPARγ cDNA and pGL3-luc-DR-1 (wild type or mutant) using FuGENE 6 Transfection Reagent according to the manufacturer's instructions. Transfected cells were incubated with no or 100 μg/ml n-3 LDL, 30 μm DHA, or 10 μm troglitazone for 8 or 24 h. Luciferase activities were measured using a Luciferase Assay Kit in a TD-20e luminometer (Turner Designs Inc., Sunnyvale, CA), and β-galactosidase activity was measured using a β-Galactosidase assay kit in a SpectroMAX plate reader (Molecular Devices Corp., Sunnyvale, CA). Luciferase activities were normalized to β-galactosidase activity and are presented as the percentage of luciferase activity measured in the presence of stimulus, relative to the activity of control cells with no stimulation.
Nuclear Extract and Electrophoretic Gel Mobility Shift Assay—PC-3 cells were rinsed three times with phosphate-buffered saline, once with buffer A (20 mm HEPES (pH 7.9), 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride), scraped into buffer A containing 0.2% Nonidet P-40, and spun for 20 s at 15,000 rpm, 4 °C. Pellets were solubilized in 100 μl of buffer B (20 mm HEPES (pH 7.9), 1 mm EDTA, 1 mm EGTA, 420 mm NaCl, 1 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, 20% glycerol) and dialyzed against binding buffer (10 mm Tris, 50 mm KCl, 1 mm dithiothreitol, pH 7.5) overnight at 4 °C. DNA binding was performed by mixing 70,000 cpm of a 32P-end-labeled DNA fragment from SDC-1 promoter containing DR-1 (generated by annealing the oligonucleotides 5′-cggttcttcccctttgctctctcggccgtttccgctacacccgagct-3′ and 5′-cgggtgtagcggaaacggccgagagagcaaaggggaagaaccggtac-3′), 50 ng/μl poly(dI·dC) ·poly(dI·dC) (Sigma), 2.5% glycerol, 5 mm MgCl2, 0.05% Nonidet P-40, 20 μg of nuclear protein, 200 ng of unlabeled DNA, and/or 2 μl of PPARγ antibody (Cayman Chemical, Ann Arbor, MI) in a 20-μl reaction volume and incubated for 30 min at room temperature. Samples were loaded on a 5% polyacrylamide gel prerun for 60 min and run in TBE buffer (45 mm Tris base, 45 mm boric acid, 1 mm EDTA, pH 8.3) for 90 min at 200 V. Gels were dried and subjected to autoradiography.
Data Analysis—Data were analyzed by analysis of variance and Student's t test. The assays were carried out in triplicate, and the data are shown as means ± S.E.
RESULTS
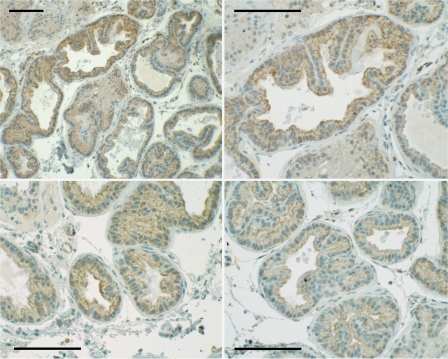
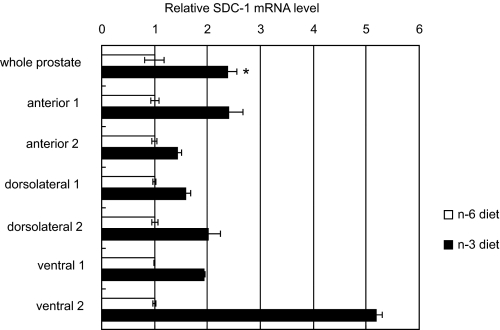
Syndecan 1 Expression in Mouse Prostate Tissue—Anterior, ventral, and dorsolateral prostate lobes were dissected from male mice, embedded in paraffin, and sectioned for immunostaining. An antibody directed against a sequence of the human SDC-1 core protein demonstrated heterogeneous cell surface staining of prostate epithelial cells that was particularly strong at the apical surface of many cells (Fig. 1). Prostate tissues from animals fed an n-3 or n-6 PUFA-enriched diet were similarly dissected and processed for mRNA analysis. Fig. 2 shows that SDC-1 mRNA in the combined prostate lobes was >2 fold higher in animals fed an n-3-enriched diet compared with those fed an n-6 PUFA-enriched diet. Individual lobes from two additional animals in each diet group were analyzed separately and demonstrated that higher SDC-1 expression was achieved in all prostate lobes of the n-3 fed animals.
FIGURE 1.
Immunostaining of SDC-1 in mouse prostate tissue. Mouse prostate glands were fixed, sectioned, and immunostained with antibody H-174 against a recombinant protein corresponding to amino acids 82-256 of the human SDC-1 core protein. Shown is the dorsolateral lobe at a magnification of 10× (upper left) or 20× (upper right, lower left, and lower right).
FIGURE 2.
SDC-1 mRNA in prostate tissues of mice fed n-3 or n-6-enriched diets. Grossly normal prostate glands were dissected from animals at 8 weeks of age, and RNA was isolated and measured by real-time RT-PCR. Shown are SDC-1mRNA in total prostate tissue from animals fed n-3 and n-6 PUFA-enriched diets (n = 3) and in isolated lobes of two additional animals fed n-3 and n-6 PUFA-enriched diets. *, p < 0.05.
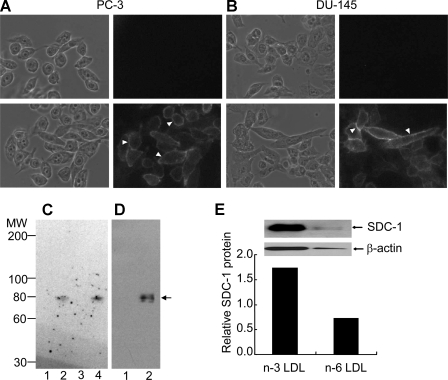
Syndecan-1 Expression in Human Prostate Cancer Cell Lines—To demonstrate the presence of SDC-1 in human prostate cancer cells, PC-3 cells (Fig. 3A) and DU-145 cells (Fig. 3B) were incubated with a mouse anti-human SDC-1 monoclonal antibody and visualized by a rhodamine-labeled second antibody. Strong immunostaining was observed around the periphery of the cells. A similar staining pattern was observed in a third prostate cancer cell line, LNCaP (not shown). To verify that SDC-1 was the predominant heparan sulfate proteoglycan present on human prostate cancer cells, whole cell lysates of PC-3 cells were either untreated or treated with glycosaminoglycan-degrading enzymes, and Western blotting was performed. Fig. 3C shows the Western analysis using monoclonal antibody 3G10, which recognizes the unsaturated hexuronic acid residue on a heparan sulfate core protein following heparan sulfate chain removal. An 80-kDa core protein was visible in lanes containing heparinase-treated samples. No other core protein bands were detected indicating that one major heparan sulfate species is associated with these cells. In addition, Fig. 3D also shows a Western analysis using a SDC-1 core protein-specific antibody that clearly identified a similarly migrating 80-kDa protein. We conclude that PC-3 cells express SDC-1 as their dominant heparan sulfate-containing proteoglycan. As shown in Fig. 3E, the SDC-1 core protein was markedly elevated in cells incubated with LDL enriched in n-3 PUFA compared with n-6 PUFA.
FIGURE 3.
Identification of SDC-1 in human prostate cancer cell lines. Phase contrast and rhodamine channel images of PC-3 (A) and DU-145 (B) cells. Cells were grown in 35-mm dishes, fixed, and stained with (bottom) or without (top) 0.25 μg/ml anti-SDC-1 followed by 25 μg/ml rhodamine-labeled goat anti-mouse IgG. Arrowheads indicate representative regions of cell-surface staining. C, Western blot analysis of PC-3 cell extracts after glycosaminoglycan-degrading enzyme treatments. Proteins were resolved on a 3.5-15% SDS-PAGE gel, transferred to polyvinylidene difluoride membranes, and immunoblotted with a mouse α-heparan sulfate stub monoclonal antibody. Lane 1, untreated; lane 2, heparinase-treated; lane 3, chondroitin ABC lyase-treated; lane 4, heparinase plus chondroitin ABC lyase-treated. Chondroitin ABC lyase degrades chondroitin sulfate and dermatan sulfate glycosaminoglycans but not heparan sulfate proteoglycan. D, Western analysis using a SDC-1 core protein-specific antibody. Lane 1, untreated; lane 2, heparinase-treated. The arrow denotes the 80-kDa core protein of SDC-1. E, Western analysis using the SDC-1 core protein-specific antibody of heparinase-treated membrane proteins of PC-3 cells incubated with 100 μg/ml n-3 PUFA- or n-6 PUFA-enriched LDL. Data are presented relative to a no LDL control.
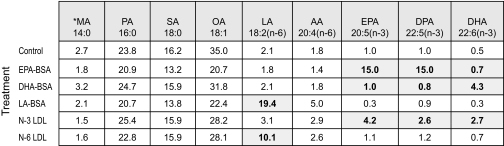
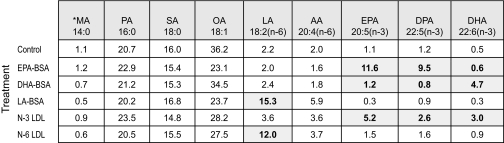
Delivery of PUFA to PC-3 Cells—To further examine in vitro regulation of SDC-1 expression by n-3 PUFA we modeled the two main physiological pathways for delivery of dietary fatty acids to cells: by LDL and by albumin. For this we used LDL isolated from vervet monkeys that had been fed diets enriched in fish oil. Our previous studies had shown that this LDL contained two major species of n-3 PUFA: EPA and DHA (33). As a control, n-6 PUFA-enriched LDL was isolated from vervet monkeys fed a diet supplemented with linoleic acid (LA (18:2, n-6)), which was the major fatty acid species in the n-6 LDL (33). Incubation of PC-3 cells with these LDL resulted in delivery of the respective fatty acids to the cells. Table 1 shows the fatty acid percent distribution in total cell lipids following 24-h incubation with either the PUFA-enriched LDL or BSA-bound EPA, DHA, or LA. In cells incubated with no fatty acid supplementation, the n-3 PUFA were ≤1% of total fatty acids. EPA-BSA addition resulted in an increase in EPA content to 15%. These cells effectively converted EPA to docosapentaenoic acid (DPA (22:5, n-3)), which also comprised 15% of total fatty acids. Metabolism of DPA to DHA, however, was not observed during this time period in the EPA-treated cells. DHA-BSA treatment resulted in increased DHA only. The n-3 LDL used for these studies did not contain a detectable level of DPA, but when the PC-3 cells were incubated with this LDL, it was observed that both EPA and DHA were delivered to the cells and that there was formation of DPA. Cells treated with LA-BSA or n-6 LDL showed an increase in LA to 19.4 and 10.1% of total fatty acids, respectively. Cell phospholipids were isolated from the total lipid extracts, and their fatty acid composition is shown in Table 2. The pattern of fatty acid distribution of the cell phospholipids mirrored that of the total cell lipids. Thus both LDL and PUFA-BSA were effective in delivering PUFA to the tumor cell phospholipids, and PC-3 cells were able to metabolize EPA to DPA but unable to metabolize the latter fatty acid further to DHA.
TABLE 1.
Percent fatty acid composition of PC-3 cell total lipids
* MA, myristic acid; PA, palmitic acid; SA, stearic acid; OA, oleic acid; LA, linoleic acid; AA, arachidonic acid; EPA, eicosapentaenoic acid; DPA, docosapentaenoic acid; DHA, docosahexaenoic acid. Shaded cells indicate fatty acids of particular interest.
TABLE 2.
Percent fatty acid composition of PC-3 cell phospholipids
* MA, myristic acid; PA, palmitic acid; SA, stearic acid; OA, oleic acid; LA, linoleic acid; AA, arachidonic acid; EPA, eicosapentaenoic acid; DPA, docosapentaenoic acid; DHA, docosahexaenoic acid. Shaded cells indicate fatty acids of particular interest.
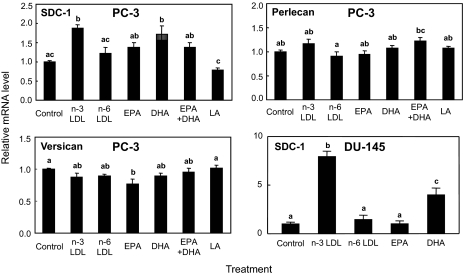
In Vitro Up-regulation of SDC-1 by n-3 PUFA—To demonstrate in vitro regulation of SDC-1 by n-3 PUFA, real-time RT-PCR was used to measure SDC-1 expression in PC-3 cells incubated with n-3 LDL or PUFA-BSA (Fig. 4). The first panel shows that n-3 LDL (100 μg/ml) and DHA-BSA (30 μm) were effective in elevating SDC-1 expression, whereas EPA (30 μm), LA (30 μm), or equimolar concentrations (15 μm each) of EPA plus DHA were ineffective. Higher concentrations of EPA and LA (up to 100 μm) were also ineffective (not shown) as was n-6 LDL at 100 (Fig. 4) or 500 μg/ml (not shown). The effect of n-3 LDL and DHA-BSA was specific for SDC-1 and not a general effect on all cell proteoglycans: neither n-3 LDL nor DHA-BSA stimulated the expression of versican or perlecan, two other proteoglycan species produced by these cells. In addition, n-3 LDL and DHA were also efficient in increasing the expression of SDC-1 in DU-145 prostate cancer cells, but as in PC-3 cells, n-6 LDL and EPA were ineffective.
FIGURE 4.
Effects of PUFA on SDC-1, versican, and perlecan expression. PC-3 or DU-145 cells were treated with 100 μg/ml n-3 PUFA- or n-6 PUFA-enriched LDL, 30 μm EPA-BSA, 30 μm DHA-BSA, 15 μm EPAplus 15 μm DHA, or 30 μm LA for 8 h. Total RNA was isolated and SDC-1 (top left, bottom right), versican (bottom left), and perlecan (top right) mRNA was determined by real-time RT-PCR. Data are means ± S.E. of three independent experiments. Values with different lowercase letters are significantly different (p < 0.05).
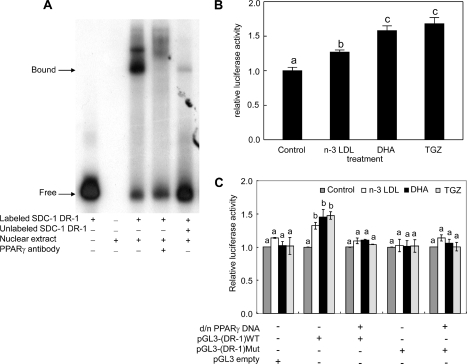
Regulation of SDC-1 by PPARγ—Our previous studies have implicated the nuclear receptor, PPARγ, in the control of SDC-1 expression in human breast cancer cells (34). To determine whether a similar pathway is involved in the n-3 PUFA up-regulation of SDC-1 in prostate cancer cells, we first used an electrophoretic gel mobility shift assay to demonstrate binding of a peroxisome proliferator-responsive element (PPRE) in the SDC-1 promoter to PPARγ (Fig. 5A). Nuclear extracts from PC-3 cells demonstrated a protein-DNA complex (Fig. 5A, third lane) that was competed by a 200-fold molar excess of the unlabeled oligonucleotide (Fig. 5A, fifth lane). The signal was reduced and supershifted in the presence of a PPARγ antibody supporting the involvement of PPARγ in the complex (Fig. 5A, fourth lane). To determine whether n-3 PUFA could elicit a PPAR response in the PC-3 cells, a reporter assay was used. PC-3 cells were transfected with a PPRE-TK-luciferase reporter plasmid and a control plasmid containing the lacZ gene. Luciferase activity was measured after a 24-h treatment of the cells with n-3 LDL, DHA-BSA, or the PPARγ agonist, troglitazone. As shown in Fig. 5B, all three treatments significantly increased luciferase activity indicating a PPAR response. To demonstrate a response specifically in the SDC-1 promoter, the luciferase reporter gene was placed under the control of either the wild-type (pGL3-(DR-1) wt) or mutant (pGL3-(DR-1)-mut) DR-1 element from the syndecan-1 promoter. PC-3 cells were co-transfected with wild-type or mutant SDC-1 DR-1-luciferase constructs and a d/n-PPARγ cDNA. No significant effect on luciferase activity in response to n-3 LDL, DHA, or troglitazone was observed in cells transfected with the empty pGL3-promoter vector (Fig. 5C). Whereas the pGL3-(DR-1) wt-luciferase reporter was activated by n-3 LDL, DHA, and tro-glitazone, co-transfection of the wild-type reporter with d/n-PPARγ cDNA eliminated the luciferase response. In addition, the transfected reporter gene containing a mutant SDC-1 DR-1 site failed to respond. These data establish the SDC-1 gene as a molecular target for n-3 PUFA and indicate that this targeting is mediated by PPARγ.
FIGURE 5.
Activation of PPARγand the human SDC-1 promoter by n-3 PUFA. A, electrophoretic gel mobility shift assay. Nuclear extracts from PC-3 cells were incubated with a [γ-32P]ATP-labeled oligonucleotide corresponding to the DR-1 element of the SDC-1 promoter. A specific supershifted complex was observed in the presence of anti-PPARγ antibody. Binding specificity was confirmed using a 200-fold molar excess of unlabeled corresponding oligonucleotide. B, PC-3 cells were co-transfected with a plasmid containing the luciferase gene driven by a PPAR response element (PPRE×3-tk-luciferase) and a control plasmid containing the lacZ gene. Following transfection, cells were treated with n-3 LDL (100 μg/ml), DHA (30 μm), or troglitazone (10 μm), and luciferase activity was measured at 24 h. Luciferase activity values were normalized to transfection efficiency monitored by co-transfected β-galactosidase expression. Values are mean ± S.E. of three independent experiments conducted in triplicate. C, PC-3 cells were co-transfected with a reporter construct containing the DR-1 from the SDC-1 promoter linked to a minimal promoter-luciferase reporter gene pGL3-(DR-1) wt or the same vector containing the mutant DR-1 (pGL3-(DR-1)mut) and a control plasmid containing the lacZ gene, with or without d/n-PPARγ cDNA. Following transfection, cells were treated with n-3 LDL (100 μg/ml), DHA (30 μm), or troglitazone (10 μm) for 24 h. Luciferase activity values are normalized to β-galactosidase activity and presented as the percentage of luciferase activity measured in the presence of stimulus, relative to the activity in control cells with no stimulus. Basal activity was reduced ∼2-fold by the d/n-PPARγ cDNA, but not changed by mutant DR-1. Values with different lowercase letters are significantly different (p < 0.05).
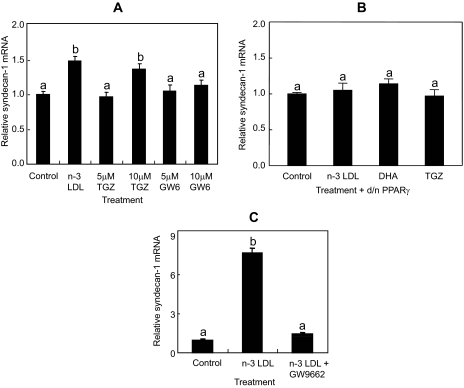
To confirm a specific involvement of PPARγ, as opposed to other PPAR isoforms, in n-3 PUFA up-regulation of SDC-1 in the PC-3 cells, SDC-1 expression was measured in response to PPAR isoform-specific agonists. Treatment with the PPAR agonist, troglitazone, as with n-3 LDL, resulted in increased expression of SDC-1, whereas treatment with the PPARδ agonist, GW610742, was ineffective (Fig. 6A). To further implicate PPARγ, cells were transfected with d/n-PPARγ cDNA. The up-regulation of SDC-1 by n-3 PUFA and troglitazone was blocked in cells transfected with d/n-PPARγ cDNA (Fig. 6B), thus connecting the n-3 LDL and DHA-induced PPAR response specifically to PPARγ. Finally, cells were treated with n-3 LDL in the presence of the PPARγ antagonist, GW259662. As shown in Fig. 6C, the PPARγ antagonist completely blocked the n-3 LDL stimulation of SDC-1 expression. The SDC-1 mRNA increase in response to n-3 LDL treatment peaks at 8 h (not shown), and the relatively higher response to n-3 LDL shown in this panel may be due to an 8-h treatment time compared with 24 h in panel A.
FIGURE 6.
Effect of PPAR agonists or antagonist on SDC-1 expression in PC-3 cells. A, cells were treated with 100 μg/ml n-3 LDL or 5 and 10 μm PPARγ agonist, troglitazone, or 5 and 10 μm PPARδ agonist, GW610742 for 24 h; B, cells were transfected with d/n-PPARγ cDNA and treated with 100 μg/ml n-3 LDL, 30 μm DHA, and 10 μm troglitazone for 8 h. C, cells were treated with 100 μg/ml n-3 LDL in the presence or absence of 15 μm PPARγ antagonist GW259662 for 8 h. Total RNA was isolated, and SDC-1mRNA was determined by real-time RT-PCR. Data are means ± S.E. in triplicate experiments. Values with different lowercase letters are significantly different (p < 0.05).
DISCUSSION
The novel finding of these studies is that SDC-1 is up-regulated by n-3 PUFA in prostate cells. This occurs both in vivo when mice are fed an n-3 PUFA-enriched diet and in vitro when human prostate cancer cells are exposed to n-3 PUFA. Although there is a growing body of evidence for a chemopreventive role for n-3 PUFA, there is no clear understanding of target genes and mechanisms that may bring about this role. Therefore the identification SDC-1 as a molecular target for n-3 PUFA is of significant interest. Due to the multifunctional properties of SDC-1, it is proposed that up-regulation of this proteoglycan would have profound effects on the interaction of the cells with their environment.
SDC-1 is the predominant cell surface proteoglycan on epithelial cells. Our studies with mouse prostate tissue identified a plasma membrane distribution of SDC-1 that exhibited a polarization to the apical surface of the glandular cells. Early studies by Rapraeger et al. (38) demonstrated the mobility of SDC-1 in the plasma membrane of cultured mouse mammary epithelial cells. Whereas an apical distribution was observed on actively growing cells, SDC-1 became sequestered at the basolateral surface of confluent monolayers. When cross-linked at the cell surface it associated with the actin cytoskeleton.
SDC-1 binds cells via its ectodomain to interstitial collagen (39), fibronectin (40), and throm-bospondin (41) and has been proposed to function as an anchor that links the cytoskeleton to the extracellular matrix. Thus staining of the basolateral surface of cells would be consistent with the matrix-binding properties of the proteoglycan. However, the distribution and expression of the SDC-1 in tissues has been shown to vary according to epithelial tissue (42). This suggests that discrete functions for SDC-1 may exist in different types of epithelia. Although the presence of SDC-1 in tissues is reported to be decreased on the cell surface adjacent to basement membrane (42), apical localization such as that observed in the mouse prostate epithelia (Fig. 1) was surprising. This staining pattern was particularly pronounced in the dorsolateral lobe, which, according to gene expression patterns, is equivalent to the human prostate peripheral zone (43). This zone is the site of origin of over 80% of human prostate carcinomas (44, 45). It is interesting to speculate that the observed distribution of SDC-1 may be associated with apical secretion from the cells, possibly of kallikreins such as the prostate specific antigen.
Our study is the first to show an increase in SDC-1 in tissues of animals fed an n-3-enriched diet, an effect that is consistent with a role for SDC-1 in tumor protection. Recently we have described the effects of PUFA-enriched diets in a mouse model with prostate-specific homozygous deletion of the tumor suppressor Pten (phosphatase and tensin homolog deleted on chromosome 10). Pten is the most frequently deleted gene in human prostate cancer. Using this animal model, in which prostate cancer has 100% penetrance, we have shown that a high n-3 PUFA diet inhibits tumor growth and increases survival, whereas a high n-6 PUFA diet has the opposite effect (35). Present data support our hypothesis that SDC-1 is a mediator of the tumor protective effects of n-3 PUFA. Future studies with our animal model will examine the relationship between SDC-1 expression and tumor progression. Despite a significant body of evidence that SDC-1 expression is inversely associated with tumor aggressiveness and poor prognosis in a variety of cancers (7-11, 46), the association of SDC-1 with prostate cancer has not been well studied. Kiviniemi et al. (19) reported an inverse relationship between SDC-1 and the Gleason grade of tumor. This was partially confirmed by Chen et al. (18) who found that SDC-1 expression was markedly reduced in locally invasive cancer compared with normal basal and secretory prostate epithelia. However, in high Gleason grade tumors, SDC-1 was associated with greater prostate specific antigen recurrence. In addition, prostate tissue microarray analysis showed a positive correlation between SDC-1 and tumor progression (20). One potential source of confusion is that all of these results were based on immunoreactivity to antibodies raised against epitopes on the SDC-1 core protein. Caution is needed in interpretation of such results, because the presence of heparanases in cancer tissue may degrade the heparan sulfate chains of SDC-1 and thus enhance the exposure of core protein epitopes to the antibodies. It is well known that in vitro degradation of glycosaminoglycan chains improves antibody recognition in Western analyses and immunoblotting, and this should be considered as a confounding factor in interpreting immunostaining data in tissues with high heparanase activity. Biochemical analyses are needed to clarify such data.
In all of the in vitro studies presented in this report we delivered PUFA to the cells by two routes that model the physiological delivery of dietary fatty acids to tumor cells: LDL and albumin. As shown in Table 1, both are similarly effective in delivering PUFA to the cells total lipid stores as well as specifically to cell phospholipids. One unexpected and intriguing finding was that of the two major n-3 PUFA present in n-3 LDL, DHA but not EPA was effective in stimulating the expression of SDC-1. DHA is synthesized from EPA by elongation and desaturation in a process defined by Sprecher (for review, see Ref. 47). Briefly, EPA (20:5, n-3) is converted to 22:5, n-3 by elongase (Elovl)-5, and then by Elovl-2 to 24:5, n-3. The next step requires desaturation of 24:5 by Δ6 desaturase to produce 24:6, n-3. This product then translocates from the endoplasmic reticulum to the peroxisome where the β oxidation pathway exerts an acyl chain shortening of C2 to produce DHA (22:6, n-3). The conversion of EPA to DHA is known to be inefficient in humans. In fact it has been shown that dietary supplementation with up to 4 g of EPA/day does not result in detectable levels of plasma DHA (48, 49). This low bioconversion is particularly evident in males (50). We observed that, when the PC-3 cells were treated with EPA, DPA was markedly increased by as early as 12 h (not shown) and by 24 h comprised 50% of the cell n-3 PUFA (Table 1). Further conversion to DHA was not achieved by this time indicating a deficit in the pathway after Elov-5. Similar results were obtained with DU-145 cells (not shown). Even over a longer time period (72 h, data not shown), we have observed little conversion to DHA in EPA-treated cells, and have at no time measured an increase in SDC-1 mRNA resulting from EPA treatment. It is not clear at present why EPA is not effective in activation of PPARγ or induction of SDC-1 expression. Studies are presently being undertaken to further clarify the metabolism of the long chain PUFA in relation to these effects in tumor cells.
An important finding from these studies is the establishment of a link in prostate cancer cells between n-3 LDL and DHA activation of PPARγ and subsequent induction of SDC-1. The reporter assays shown in Fig. 5 demonstrated a PPAR response to n-3 LDL and DHA that was inhibited in the presence of d/n-PPARγ cDNA. In addition, the PPARγ agonist troglitazone was effective in stimulating the expression of SDC-1, whereas a PPARγ antagonist, GW259662 completely blocked the effect of n-3 LDL on SDC-1 expression (Fig. 6). Importantly, we have for the first time demonstrated specific binding of PPARγ to a DR-1 element in the SDC-1 promoter. In hepatocytes, this DR-1 element in was shown to be a target gene for the farnesoid-X receptor (37). Available expression data show either absent or low levels of farnesoid-X receptor expression in PC-3 cells, whereas PPARγ is high (in the top 10% of genes expressed by these cells) (51, 52). Therefore PPARγ is more likely to be the relevant nuclear factor. This is supported by the fact that PPARγ can regulate expression of luciferase placed under the control of the SDC-1 DR-1. However, involvement of farnesoid-X receptor cannot be ruled out at this time. Similar to our observation in MCF-7 breast cancer cells (34), only DHA and not EPA was effective in stimulating the expression of SDC-1 in PC-3 cells. Although both EPA and LA bind PPARγ (53), both were shown to be poor activators of this PPAR isoform (54). Further studies are required to determine whether DHA activation of PPARγ is a direct effect or one mediated by an active metabolite of the fatty acid.
This work was supported, in whole or in part, by National Institutes of Health Grants P01CA106742 (to Y. Q. C., I. J. E., J. T. O., and J. M. C.), R01CA115958 (to I. J. E.), RO1CA114017 (to I. M. B.), HL-49373 (to L. L. R.), and R01CA107668 (to Y. Q. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SDC-1, syndecan 1; BPH, benign prostrate hyperplasia; BSA, bovine serum albumin; DHA, docosahexaenoic acid; d/n, dominant/negative; DPA, docosapentaenoic acid; EPA, eicosapentaenoic acid; LA, linoleic acid; LDL, low density lipoproteins; PPARγ, peroxisome proliferator-activated receptor γ; PPRE, PPAR-response elements; PUFA, polyunsaturated fatty acids; FBS, fetal bovine serum; RT, reverse transcription; wt, wild type.
Sun, H., Berquin, I. M., Owens, R. T., O'Flaherty, J. T., and Edwards, I. J. (2008) Cancer Res. 8, 2912-2919.
References
- 1.Bernfield, M., Gotte, M., Park, P. W., Reizes, O., Fitzgerald, M. L., Lincecum, J., and Zako, M. (1999) Annu. Rev. Biochem. 68 729-777 [DOI] [PubMed] [Google Scholar]
- 2.Liu, W., Litwack, E. D., Stanley, M. J., Langford, J. K., Lander, A. D., and Sanderson, R. D. (1998) J. Biol. Chem. 273 22825-22832 [DOI] [PubMed] [Google Scholar]
- 3.Woods, A., Oh, E. S., and Couchman, J. R. (1998) Matrix Biol. 17 477-483 [DOI] [PubMed] [Google Scholar]
- 4.Kaushal, G. P., Xiong, X., Athota, A. B., Rozypal, T. L., Sanderson, R. D., and Kelly, T. (1999) Br. J. Haematol. 104 365-373 [DOI] [PubMed] [Google Scholar]
- 5.Liebersbach, B. F., and Sanderson, R. D. (1994) J. Biol. Chem. 269 20013-20019 [PubMed] [Google Scholar]
- 6.Dhodapkar, M. V., Abe, E., Theus, A., Lacy, M., Langford, J. K., Barlogie, B., and Sanderson, R. D. (1998) Blood 91 2679-2688 [PubMed] [Google Scholar]
- 7.Numa, F., Hirabayashi, K., Kawasaki, K., Sakaguchi, Y., Sugino, N., Suehiro, Y., Suminami, Y., Hirakawa, H., Umayahara, K., Nawata, S., Ogata, H., and Kato, H. (2002) Int. J. Oncol. 20 39-43 [PubMed] [Google Scholar]
- 8.Matsumoto, A., Ono, M., Fujimoto, Y., Gallo, R. L., Bernfield, M., and Kohgo, Y. (1997) Int. J. Cancer 74 482-491 [DOI] [PubMed] [Google Scholar]
- 9.Inki, P., Joensuu, H., Grenman, R., Klemi, P., and Jalkanen, M. (1994) Br. J. Cancer 70 319-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anttonen, A., Kajanti, M., Heikkila, P., Jalkanen, M., and Joensuu, H. (1999) Br. J. Cancer 79 558-564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pulkkinen, J. O., Penttinen, M., Jalkanen, M., Klemi, P., and Grenman, R. (1997) Acta Otolaryngol. 117 312-315 [DOI] [PubMed] [Google Scholar]
- 12.Kumar-Singh, S., Jacobs, W., Dhaene, K., Weyn, B., Bogers, J., Weyler, J., and Van Marck, E. (1998) J. Pathol. 186 300-305 [DOI] [PubMed] [Google Scholar]
- 13.Nackaerts, K., Verbeken, E., Deneffe, G., Vanderschueren, B., Demedts, M., and David, G. (1997) Int. J. Cancer 74 335-345 [DOI] [PubMed] [Google Scholar]
- 14.Mennerich, D., Vogel, A., Klaman, I., Dahl, E., Lichtner, R. B., Rosenthal, A., Pohlenz, H. D., Thierauch, K. H., and Sommer, A. (2004) Eur. J. Cancer 40 1373-1382 [DOI] [PubMed] [Google Scholar]
- 15.Wiksten, J. P., Lundin, J., Nordling, S., Kokkola, A., and Haglund, C. (2000) Anticancer Res. 20 4905-4907 [PubMed] [Google Scholar]
- 16.Wiksten, J. P., Lundin, J., Nordling, S., Lundin, M., Kokkola, A., von Boguslawski, K., and Haglund, C. (2001) Int. J. Cancer 95 1-6 [DOI] [PubMed] [Google Scholar]
- 17.Stanley, M. J., Stanley, M. W., Sanderson, R. D., and Zera, R. (1999) Am. J. Clin. Pathol. 112 377-383 [DOI] [PubMed] [Google Scholar]
- 18.Chen, D., Adenekan, B., Chen, L., Vaughan, E. D., Gerald, W., Feng, Z., and Knudsen, B. S. (2004) Urology 63 402-407 [DOI] [PubMed] [Google Scholar]
- 19.Kiviniemi, J., Kallajoki, M., Kujala, I., Matikainen, M. T., Alanen, K., Jalkanen, M., and Salmivirta, M. (2004) APMIS 112 89-97 [DOI] [PubMed] [Google Scholar]
- 20.Zellweger, T., Ninck, C., Mirlacher, M., Annefeld, M., Glass, A. G., Gasser, T. C., Mihatsch, M. J., Gelmann, E. P., and Bubendorf, L. (2003) Prostate 55 20-29 [DOI] [PubMed] [Google Scholar]
- 21.Terry, P. D., Terry, J. B., and Rohan, T. E. (2004) J. Nutr. 134, (suppl.) 3412S-3420S [DOI] [PubMed] [Google Scholar]
- 22.Davis, B. C., and Kris-Etherton, P. M. (2003) Am. J. Clin. Nutr. 78, (suppl.) 640S-646S [DOI] [PubMed] [Google Scholar]
- 23.Leitzmann, M. F., Stampfer, M. J., Michaud, D. S., Augustsson, K., Colditz, G. C., Willett, W. C., and Giovannucci, E. L. (2004) Am. J. Clin. Nutr. 80 204-216 [DOI] [PubMed] [Google Scholar]
- 24.Yang, Y. J., Lee, S. H., Hong, S. J., and Chung, B. C. (1999) Clin. Biochem. 32 405-409 [DOI] [PubMed] [Google Scholar]
- 25.Norrish, A. E., Skeaff, C. M., Arribas, G. L., Sharpe, S. J., and Jackson, R. T. (1999) Br. J. Cancer 81 1238-1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mamalakis, G., Kafatos, A., Kalogeropoulos, N., Andrikopoulos, N., Daskalopulos, G., and Kranidis, A. (2002) Prostaglandins Leukot. Essent. Fatty Acids 66 467-477 [DOI] [PubMed] [Google Scholar]
- 27.Freeman, V. L., Meydani, M., Hur, K., and Flanigan, R. C. (2004) Cancer 101 2744-2754 [DOI] [PubMed] [Google Scholar]
- 28.Chung, B. H., Mitchell, S. H., Zhang, J. S., and Young, C. Y. (2001) Carcinogenesis 22 1201-1206 [DOI] [PubMed] [Google Scholar]
- 29.Rose, D. P., and Connolly, J. M. (1991) Prostate 18 243-254 [DOI] [PubMed] [Google Scholar]
- 30.Narayanan, N. K., Narayanan, B. A., and Reddy, B. S. (2005) Int. J. Oncol. 26 785-792 [PubMed] [Google Scholar]
- 31.Rose, D. P. (1997) Proc Soc. Exp. Biol. Med. 216 224-233 [DOI] [PubMed] [Google Scholar]
- 32.Larsson, S. C., Kumlin, M., Ingelman-Sundberg, M., and Wolk, A. (2004) Am. J. Clin. Nutr. 79 935-945 [DOI] [PubMed] [Google Scholar]
- 33.Edwards, I. J., Berquin, I. M., Sun, H., O'Flaherty, J. T., Daniel, L. W., Thomas, M. J., Rudel, L. L., Wykle, R. L., and Chen, Y. Q. (2004) Clin. Cancer Res. 10 8275-8283 [DOI] [PubMed] [Google Scholar]
- 34.Sun, H., Berquin, I. M., and Edwards, I. J. (2005) Cancer Res. 65 4442-4447 [DOI] [PubMed] [Google Scholar]
- 35.Berquin, I. M., Min, Y., Wu, R., Wu, J., Perry, D., Cline, J. M., Thomas, M. J., Thornburg, T., Kulik, G., Smith, A., Edwards, I. J., D'Agostino, R., Zhang, H., Wu, H., Kang, J. X., and Chen, Y. Q. (2007) J. Clin. Invest. 117 1866-1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parks, J. S., and Gebre, A. K. (1991) J. Lipid Res. 32 305-315 [PubMed] [Google Scholar]
- 37.Anisfeld, A. M., Kast-Woelbern, H. R., Meyer, M. E., Jones, S. A., Zhang, Y., Williams, K. J., Willson, T., and Edwards, P. A. (2003) J. Biol. Chem. 278 20420-20428 [DOI] [PubMed] [Google Scholar]
- 38.Rapraeger, A., Jalkanen, M., and Bernfield, M. (1986) J. Cell Biol. 103 2683-2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koda, J. E., Rapraeger, A., and Bernfield, M. (1985) J. Biol. Chem. 260 8157-8162 [PubMed] [Google Scholar]
- 40.Saunders, S., and Bernfield, M. (1988) J. Cell Biol. 106 423-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun, X., Mosher, D. F., and Rapraeger, A. (1989) J. Biol. Chem. 264 2885-2889 [PubMed] [Google Scholar]
- 42.Hayashi, K., Hayashi, M., Jalkanen, M., Firestone, J. H., Trelstad, R. L., and Bernfield, M. (1987) J. Histochem. Cytochem. 35 1079-1088 [DOI] [PubMed] [Google Scholar]
- 43.Berquin, I. M., Min, Y., Wu, R., Wu, H., and Chen, Y. Q. (2005) J. Biol. Chem. 280 36442-36451 [DOI] [PubMed] [Google Scholar]
- 44.McNeal, J. E. (1969) Cancer 23 24-34 [DOI] [PubMed] [Google Scholar]
- 45.Sakr, W. A., and Grignon, D. J. (1998) Anal. Quant. Cytol. Histol. 20 417-423 [PubMed] [Google Scholar]
- 46.Fujiya, M., Watari, J., Ashida, T., Honda, M., Tanabe, H., Fujiki, T., Saitoh, Y., and Kohgo, Y. (2001) Jpn. J. Cancer Res. 92 1074-1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sprecher, H. (2002) Prostaglandins Leukot. Essent. Fatty Acids 67 79-83 [DOI] [PubMed] [Google Scholar]
- 48.Park, Y., and Harris, W. S. (2003) J. Lipid Res. 44 455-463 [DOI] [PubMed] [Google Scholar]
- 49.Arterburn, L. M., Hall, E. B., and Oken, H. (2006) Am. J. Clin. Nutr. 83 (suppl.) 1467S-1476S [DOI] [PubMed] [Google Scholar]
- 50.Williams, C. M., and Burdge, G. (2006) Proc. Nutr. Soc. 65 42-50 [DOI] [PubMed] [Google Scholar]
- 51.Hughes-Fulford, M., Li, C. F., Boonyaratanakornkit, J., and Sayyah, S. (2006) Cancer Res. 66 1427-1433 [DOI] [PubMed] [Google Scholar]
- 52.Zhao, H., Kim, Y., Wang, P., Lapointe, J., Tibshirani, R., Pollack, J. R., and Brooks, J. D. (2005) Prostate 63 187-197 [DOI] [PubMed] [Google Scholar]
- 53.Xu, H. E., Lambert, M. H., Montana, V. G., Parks, D. J., Blanchard, S. G., Brown, P. J., Sternbach, D. D., Lehmann, J. M., Wisely, G. B., Willson, T. M., Kliewer, S. A., and Milburn, M. V. (1999) Mol. Cell 3 397-403 [DOI] [PubMed] [Google Scholar]
- 54.Forman, B. M., Chen, J., and Evans, R. M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 4312-4317 [DOI] [PMC free article] [PubMed] [Google Scholar]