Abstract
Heparanase is an endo-β-d-glucuronidase that degrades heparan sulfate in the extracellular matrix and on the cell surface. Human proheparanase is produced as a latent protein of 543 amino acids whose activation involves excision of an internal linker segment (Ser110–Gln157), yielding the active heterodimer composed of 8- and 50-kDa subunits. Applying cathepsin L knock-out tissues and cultured fibroblasts, as well as cathepsin L gene silencing and overexpression strategies, we demonstrate, for the first time, that removal of the linker peptide and conversion of proheparanase into its active 8 + 50-kDa form is brought about predominantly by cathepsin L. Excision of a 10-amino acid peptide located at the C terminus of the linker segment between two functional cathepsin L cleavage sites (Y156Q and Y146Q) was critical for activation of proheparanase. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry demonstrates that the entire linker segment is susceptible to multiple endocleavages by cathepsin L, generating small peptides. Mass spectrometry demonstrated further that an active 8-kDa subunit can be generated by several alternative adjacent endocleavages, yielding the precise 8-kDa subunit and/or slightly elongated forms. Altogether, the mode of action presented here demonstrates that processing and activation of proheparanase can be brought about solely by cathepsin L. The critical involvement of cathepsin L in proheparanase processing and activation offers new strategies for inhibiting the prometastatic, proangiogenic, and proinflammatory activities of heparanase.
Heparanase is an endo-β-d-glucuronidase that degrades heparan sulfate side chains of heparan sulfate proteoglycans (1–4). Enzymatic cleavage of heparan sulfate proteoglycans by heparanase contributes to disassembly of the extracellular matrix (ECM)2 and basement membranes, resulting in accelerated cell invasion (1–4). Heparanase activity has been traditionally correlated with cell invasion associated with cancer metastasis, a consequence of structural modifications that loosen the ECM barrier (5, 6). This notion gained further support by employing siRNA and ribosome technologies, clearly demonstrating heparanase-mediated heparan sulfate cleavage and ECM remodeling as a critical prerequisite for cancer metastasis, angiogenesis, and inflammation (7). Heparanase up-regulation has been documented in an increasing number of human carcinomas (3, 8). In many cases, heparanase induction correlates with increased tumor metastasis, tumor vascular density, and shorter postoperative survival of cancer patients, providing strong clinical support for the pro-metastatic and pro-angiogenic functions of the enzyme (3, 9, 10).
Given the ability of heparanase to affect cell and tissue function in a variety of normal and pathophysiological processes, its activity must be kept under tight regulation. Control of heparanase post-translational modifications (4, 11, 12), including proteolytic processing and activation (13–16), represent important regulatory mechanisms. The human heparanase cDNA has an open reading frame that encodes for 543 amino acids yet the latent proheparanase is 65-kDa in size due to glycosylation (1, 11). Proheparanase undergoes proteolytic processing involving the removal of an internal segment of 48 amino acids residing between Ser110 (site 1) and Gln157 (site 2), yielding an active heterodimer composed of 8- and 50-kDa subunits (13–16). Until now, the mode of processing and the fate of the internal linker segment (Ser110–Gln157) were only partially elucidated (13). Mutagenesis at site 1 and its flanking sequences failed to identify critical residues for proteolytic cleavage, whereas processing at site 2 required a bulky hydrophobic amino acid (Tyr156) at position P2 of the cleavage site (13). The critical involvement of Tyr156 in accurate cleavage at site 2 and the inhibitory effect of a cell permeable inhibitor of cathepsin L implicate a cathepsin L-like proteolytic activity in the processing of proheparanase (13).
Cathepsin L, a papain-like cysteine proteinase, is synthesized as a preproenzyme (17). The prepeptide is removed in the endoplasmic reticulum and the pro-enzyme undergoes autoactivation in the acidic environment of late endosomes and/or lysosomes. The mature form is stored within lysosomes where it functions as an endopeptidase (17). In addition to its role in terminal protein degradation inside lysosomes, cathepsin L is implicated in multiple physiological and pathological processes (18), primarily by virtue of its involvement in pro-enzyme processing and protein maturation (19). For example, in the thymic cortex, cathepsin L plays a key role in cleaving the CLIP peptide thereby allowing for antigen presentation in the context of major histocompatibility complex class II molecules (20). Cathepsin L within secretory vesicles is responsible for the generation of several peptide neurotransmitters and hormones, including enkephalin, β-endorphin, and ACTH (21–23). Recently, a cathepsin L isoform that lacks a signal peptide was reported to function in processing of the CDP/Cux transcription factor and thereby in cell cycle regulation (24). Moreover, cathepsin L and other cysteine proteases have been implicated in tumor development and progression through degradation of protein constituents of the plasma membrane and the ECM (25–28). It was recently shown that deletion of cathepsin L in the RIP1-Tag2 (RT2) mouse model of pancreatic islet cell tumorigenesis resulted in a profound reduction in tumor growth (average decrease of 88%), and a significant impairment in tumor invasion (25).
The present study was undertaken to characterize the mechanism of proheparanase processing. Applying cathepsin L knock-out fibroblasts and cathepsin L gene silencing and overexpression strategies, we demonstrate that conversion of proheparanase into its active form is brought about almost exclusively by cathepsin L. Furthermore, the precise mode of action of cathepsin L in proheparanase processing was elucidated in a cell-free system.
MATERIALS AND METHODS
Cell Culture—Human breast carcinoma MDA-MB-435 and MCF-7 cells (8, 30) and human choriocarcinoma JAR (31) cells were grown in Dulbecco's modified Eagle's medium (4.5 g of glucose/liter (Biological Industries, Beit-Haemek, Israel) supplemented with 10% fetal calf serum, glutamine, penicillin, and streptomycin. Cultures of bovine corneal endothelial cells were established from steer eyes and maintained in Dulbecco's modified Eagle's medium (1 g of glucose/liter) supplemented with 5% newborn calf serum and 10% fetal calf serum, as described (6, 30). Primary cell cultures of fibroblasts derived from cathepsin L knock-out RIP1-Tag2 (RT2) tumors (cat L-/- fibroblasts) or from wild-type RT2 tumors (WT fibroblasts) (25) were grown in high glucose Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics.
Tissue Samples—The generation of cathepsin L knock-out mice has been previously described (32). Liver and spleen, as well as serum, were collected from wild-type and cathepsin L knock-out mice. Tissues were homogenized in lysis buffer (1% Brij 35, 150 mm NaCl, 50 mm Tris-HCl, pH 7.5) supplemented with a mixture of protease inhibitors (Sigma) (13) and the supernatant fraction was subjected to heparanase enzymatic activity, as described below.
Antibodies—Polyclonal rabbit anti-human hepararanase antibody (pAb 1453) was raised against the full-length 65-kDa proheparanase and affinity purified, as described (33). Monoclonal anti-heparanase antibody (monoclonal antibody 01385-126), recognizing both the 50-kDa subunit and the 65-kDa proheparanase was kindly provided by Dr. P. Kussie (ImClone Systems Inc., New York, NY). Polyclonal antibody 810 (pAb 810) and anti-CKLE were raised in rabbit against synthetic peptides residing in the 8-kDa subunit (96GTKTDFLIFDPKK108) or the linker segment (127CKYGSIPPDVEEKLRLE143), respectively. Monoclonal antibody (CPLH-2D4) directed against procathepsin L was purchased from Santa Cruz Biotechnology (catalog number sc-32801, Santa Cruz, CA). Monoclonal antibodies directed against mature cathepsin L (clone 33/2, ascites fluid, catalog number C2970) and against α-tubulin (clone B-5-1-2, catalog number T6074) were purchased from Sigma.
Preparation of Dishes Coated with ECM—Bovine corneal endothelial cells (second to fifth passage) were plated into 35-mm tissue culture dishes at an initial density of 2 × 105 cells/ml and cultured as described above, except that 4% dextran T-40 was included in the growth medium. Na 352SO4 (25 mCi/ml) (Amersham Biosciences, Buckinghamshire, UK) was added on days 2 and 5 after seeding and the cultures were incubated with the label without medium change. On day 12, the subendothelial ECM was exposed by dissolving the cell layer with PBS containing 0.5% Triton X-100 and 20 mm NH4OH, followed by four washes with PBS (6, 30). The ECM remained intact, free of cellular debris and firmly attached to the entire area of the tissue culture dish.
Heparanase Activity—Cells were lysed by three cycles of freezing and thawing in heparanase reaction solution (0.15 NaCl, 20 mm phosphate citrate buffer, pH 5.8, 1 mm dithiothreitol, 1 mm CaCl2). Cell lysates were incubated (24 h, 37 °C, pH 5.8) with 35S-labeled ECM. The medium was centrifuged and the supernatant containing sulfate-labeled degradation fragments was analyzed by gel filtration on Sepharose CL-6B column (0.9 × 30 cm). Fractions (0.2 ml) were eluted with PBS and their radioactivity was counted in a β-scintillation counter (6). Degradation fragments of heparan sulfate side chains are eluted from Sepharose 6B at 0.5 < Kav < 0.8 (peak II, fractions 20–35). Nearly intact heparan sulfate proteoglycans are eluted just after the V0 (Kav < 0.2, peak I) (6, 30). Each experiment was performed at least three times and the variation in elution positions (Kav values) did not exceed ±15%.
SDS-PAGE and Western Blot Analysis—Cells (1 × 106) were lysed in buffer containing 1% Brij 35, 150 mm NaCl, 50 mm Tris-HCl, pH 7.5, or in buffer containing 1% Nonidet P-40, 10 mm EDTA in PBS, both supplemented with a mixture of protease inhibitors (Sigma) (13). Cells were incubated with the lysis buffer for 15–30 min on ice, cell debris were removed by centrifugation and the protein concentration in the supernatants was determined using the Bradford protein assay (Bio-Rad). Samples of up to 100 μg of total protein were subjected to SDS-PAGE (10% acrylamide) under reducing conditions and proteins were transferred to a polyvinylidene difluoride membrane (Millipore Corporation, Billerica, MA). The membrane was probed with the appropriate antibody, followed by horseradish peroxidase-conjugated secondary antibody and a chemiluminescent substrate (West-one, iNtron Biotechnology, Gyeonggido, Korea) (13). For detection of heparanase, samples containing up to 1 mg of total protein were first added to a mixture of 30 μl of heparin-agarose beads and 30 μl of concanavalin A-agarose beads (Sigma) and incubated with rotation for 2 h at room temperature, followed by overnight incubation at 4 °C. Beads were then washed twice with saline, boiled (7 min) in sample buffer, and the supernatants were subjected to SDS-PAGE and immunoblotting, as described above.
Generation of Heparanase Deletion Mutant—Deletion mutant of human heparanase cDNA was constructed according to site-specific mutagenesis by the overlap extension approach (13). Two fragments were amplified in two separate PCR using the full-length heparanase cDNA as a template. PCR 1 amplified the fragment that contains the deletion mutation together with the upstream sequence, utilizing a reverse primer containing the deletion mutation and a common forward primer containing the wild-type heparanase sequence (1UERI-hpa). PCR 2 amplified the DNA fragment that contains the deletion mutation together with downstream sequences, utilizing the forward primer containing the deletion mutation and a common reverse primer containing the wild-type heparanase sequence (4LHpa). The two PCR purified fragments (JETquick gel extraction spin kit, Genomed, Lohne, Germany) were mixed, denatured, annealed, and extended, and the product was amplified in a third PCR, using the 1UERIhpa and 4Lhpa primers. The PCR 3 product was subcloned into pcDNA3-Hepa (containing the full-length heparanase) digested with EcoRI and AflII using the TaKaRa DNA ligation kit II (Takara Bio Inc., Shiga, Japan). The external primers are: 1UERIHpa, 5′-CTTCAGCATCTTAGCCGTCTTT-3′ and 4LhHpa, 5′-GCAGCCAGGTGAATTCCCAAGAT-3′. The reverse and forward primers for the mutation are: 5′-GGGCCATTCCAACCGTAA-3′ and 5′-TTACGGTTGGAATGGCCC-3′, respectively. All PCR were performed using the TaKaRa Ex Taq™ polymerase (TaKaRa Bio Inc.) and the following cycling conditions: 95 °C for 3 min followed by 32 cycles of 96 °C for 18 s, 60 °C for 90 s, then an elongation step of 72 °C for 70 s (13).
Transfection—JAR cells (70–80% confluence) were transfected (48 h, 37 °C) with pcDNA vectors containing the full-length heparanase cDNA or mutated heparanase cDNA, using FuGENE transfection reagent (Roche Applied Science) according to the manufacturer's instructions. Transfected cells were selected with 400–600 μg/ml G-418 (Invitrogen) and stable populations of heparanase expressing cells were obtained (13).
siRNA Transfection—Anti-human cathepsin L siRNA was designed according to Zheng et al. (34) from the human cathepsin L cDNA sequence, 91AAGTGGAAGGCGATGCACAAC111, and was synthesized by Dharmacon RNA Technologies (Lafayette, CO). MDA-MB-435 breast carcinoma or JAR choriocarcinoma cells were seeded into 6-well plates and grown in 2.5 ml of Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. After 24 h in culture, MDA-MB-435 (20–30% confluent) cells were transfected with 25 μl of 20 μm stock solution of siRNA duplexes (0.2 μm final concentration) using the GeneSilencer siRNA transfection reagent kit, according to the manufacturer's protocol (Genlantis, San Diego, CA). Forty-eight h later, the cells were re-transfected with another dose of siRNA and incubated for an additional 48 h. JAR cells were transfected with cathepsin L siRNA as described above and 24 h later were retransfected with siRNA plus pcDNA containing the full-length heparanase or empty vector. The cells were harvested by a rubber policeman, solubilized in lysis buffer containing protease inhibitors, and subjected to Western blot analysis and heparanase activity assay, as described above.
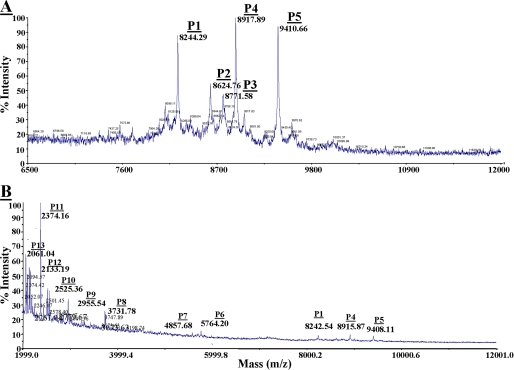
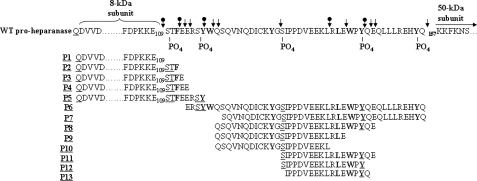
Mass Spectrometry—Recombinant, full-length proheparanase, or linker-truncated 50 + 8-kDa linear heparanase (hepa-3GS), generated by replacing the linker segment with a spacer of three glycine-serine pairs (15), were incubated in 100 mm Tris buffer, pH 6.8, supplemented with 8 mm EDTA and 4 mm DTT, for 80 min at 37 °C, with or without human liver-derived cathepsin L (Sigma). The reaction mixtures were purified on high performance liquid chromatography reverse-phase C18 column and subjected to matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass analysis. Masses of peptides (P1–P13) determined by MALDI-TOF corresponded to the calculated masses (±20 δ) of peptides derived from the linker segment and adjacent portions of the 8-kDa subunit, taking into account phosphorylation modifications on particular tyrosines, serines, and threonines. The peptides (P1–P13), their corresponding sequences and calculated masses (see Fig. 6) (including additional 80 δ for each phosphate group) were: P1, QD1... E110, 8246 δ; P2, QD1... ESTF112, 8581.9 δ, + 80 δ (1PO4) = 8661.9 δ; P3, QD1... ESTFE113, 8711 δ, + 80 δ (1PO4) = 8791 δ; P4, QD1... ESTFEE114, 8840 δ, + 80 δ (1PO4) = 8920 δ; P5, QD1... ESTFEERSY117, 9246.6 δ, + 2X80 δ (2PO4) = 9406.6 δ; P6, E114... Q157, 5478 δ, +4X80 δ (4PO4) = 5798 δ; P7, S120... Q157, 4628 δ + 3X80 δ (3PO4) = 4868 δ; P8, Q119... E148, 3575 δ, + 2X80 δ (2PO4) = 3734 δ; P9, Q119... E143, 2871 δ, + 80 δ (1PO4) = 2951 δ; P10, Q120.... L140, 2472.8 δ + 80 δ (1PO4) = 2553 δ; P11, S131... E148, 2210 δ + 2X80 δ (2PO4) = 2370 δ; P12, S131... Y146, 1953 δ, + 2X80 δ (2PO4) = 2113 δ; and P13, I122... E148, 1994 δ + 80 δ (1PO4) = 2042 δ.
FIGURE 6.
MALDI-TOF analysis of peptides generated by cathepsin L from recombinant proheparanase. Recombinant proheparanase was incubated (1 μg) with human cathepsin L (0.5 μg/ml) and subjected to MALDI-TOF analysis (A and B). Thirteen masses of peptides (P1–P13) corresponding to sequences with calculated masses similar to the measured masses (MALDI-TOF) ±20 daltons (see “Materials and Methods”) were detected. A, P1 (8244.29 daltons) corresponds to the exact size of the 8-kDa subunit (36QD1VV to Glu109). P2, P3, P4, and P5 are elongated forms of the 8-kDa subunit plus 110STF112, 110STFE113, 110STFEE114, and 110STFERSY117, respectively. B, P1, P4, and P5 were described in A. Peptides P6–P13 are all derived from the linker segment (see Fig. 7).
RESULTS
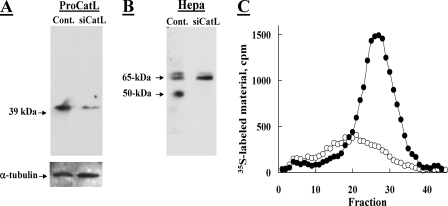
Knockdown of Cathepsin L Markedly Suppresses Proheparanase Processing—To investigate the role of cathepsin L in proheparanase processing, we utilized siRNA mediated silencing of cathepsin L (34) in JAR human choriocarcinoma cells capable of proheparanase processing, but devoid of heparanase gene expression and enzymatic activity (13). Cells were first transfected with anti-cathepsin L siRNA, or were mock transfected. Twenty-four h later the cells were transiently co-transfected for an additional 24 h with a pcDNA-Hepa vector that encodes for the full-length 65-kDa heparanase. The marked decrease (∼75%) in the level of procathepsin L in the siRNA-transfected cells (Fig. 1A) was accompanied by an almost complete inhibition of proheparanase processing into the 50-kDa heparanase subunit (Fig. 1B) and by a profound decrease in heparanase enzymatic activity, compared with mock transfected cells (Fig. 1C). The lack of detectable 50-kDa subunit, representing active heparanase, in cathepsin L siRNA-transfected JAR cells (Fig. 1B), indicates that in this experimental system (i.e. cells devoid of endogenous heparanase activity) proteolytic enzymes other than cathepsin L do not play a significant role in heparanase processing and activation.
FIGURE 1.
Knockdown of procathepsin L inhibits processing of exogenous proheparanase by JAR choriocarcinoma cells. Human choriocarcinoma JAR cells, devoid of endogenous heparanase, were subjected to two sequential transfections with 2 μm anti-procathepsin L siRNA at a 48-h interval (siCat L), or were mock transfected (Control). 72 h after the first transfection, cells were transiently transfected with a pcDNA plasmid encoding the full-length heparanase. 24 h later the cells were lysed and subjected to Western blot analysis of procathepsin L (A) and heparanase (B), as described under “Materials and Methods.” C, heparanase activity. Lysates of siCat L (○) and mock (•) transfected cells (both also transfected with the full-length heparanase) were lysed and incubated (7 h, 37 °C, pH 6.0) with sulfate-labeled ECM. Sulfate-labeled degradation fragments released into the incubation medium were analyzed by gel filtration on Sepharose 6B, as described under “Materials and Methods.”
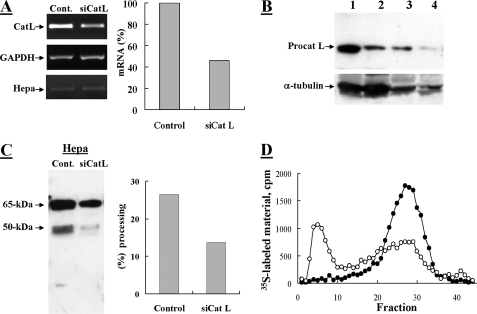
Next, the effect of siRNA-mediated cathepsin L silencing on proheparanase processing was examined in MDA-MB-435 human breast carcinoma cells expressing high levels of endogenous heparanase (30). RT-PCR analysis of anti-cathepsin L siRNA-transfected cells showed a 54% knockdown of procathepsin L mRNA compared with mock transfected cells (Fig. 2A). Heparanase mRNA levels remained unchanged, excluding the possibility of an off-target effect of cathepsin L siRNA on heparanase mRNA. At the protein level, procathepsin L was reduced by 71% in cathepsin L-siRNA-transfected versus mock transfected cells as revealed by Western blot analysis (Fig. 2B) and densitometric analysis (not shown). Knockdown of cathepsin L resulted in a marked decrease (65%) in the level of the processed form of heparanase (represented by the 50-kDa subunit) as determined by Western blot and densitometric analysis (Fig. 2C). Consequently, heparanase enzymatic activity was markedly reduced (65%) in lysates obtained from cells transfected with anti-cathepsin L siRNA as compared with mock transfected cells (Fig. 2D). These results demonstrate that cathepsin L mediates the processing of proheparanase in MDA-MB-435 cells, although to a lesser extent than in JAR cells. This difference may be due to the long half-life of the already processed active heparanase (35) that masks the actual decreased level of processing obtained during the short term treatment with cathepsin L siRNA, incomplete depletion of cathepsin L, and/or the presence of additional cathepsin L-like proteases in MDA-MB-435 versus JAR cells. Altogether, these results clearly demonstrate that cathepsin L mediates the processing of endogenous and acquired proheparanase in MDA-MB-435 and JAR carcinoma cells, respectively.
FIGURE 2.
Knockdown of procathepsin L in MDA-MB-435 cells inhibits processing of proheparanase. MDA-MB-435 cells were subjected to two sequential transfections with 2 μm anti-procathepsin L siRNA at a 48-h interval (siCat L), or were mock transfected and treated with reagents alone (Control). A, semi-quantitative RT-PCR analysis of cathepsin L (Cat L), heparanase (Hepa), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Densitometric analysis (right) revealed a 54% decrease in the mRNA level of siCat L-treated cells versus control cells. Cells were lysed and subjected to Western blot analysis of: B, procathepsin L (lanes 1 (Control) and 2 (siCatL): 100 μg of cell lysate; lanes 3 (Control) and 4 (siCatL): 25 μg and cell lysate); and C, heparanase (pAb 1453), as described under “Materials and Methods.” Proheparanase processing, indicated by the generation of a 50-kDa subunit, was decreased (2.2-fold) in siCat L-transfected cells compared with mock transfected cells, as revealed by densitometry analysis (right panel). D, heparanase activity. siCat L (○) and mock (•) transfected cell lysates were incubated with 35S-labeled ECM and analyzed for heparanase enzymatic activity, as described in the legend to Fig. 1.
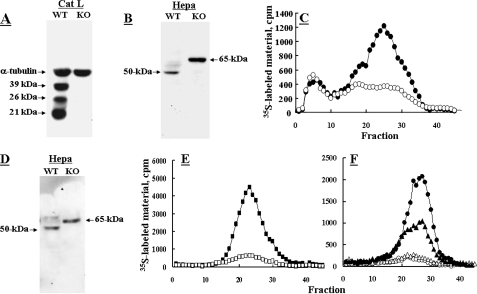
Cathepsin L Knock-out Fibroblasts Are Incapable of Proheparanase Processing—Fibroblasts derived from cathepsin L knock-out RT2 pancreatic islet tumors (25) were tested for their ability to process proheparanase in comparison with fibroblasts from wild-type (WT) RT2 tumors (Fig. 3). WT fibroblasts expressed high levels of properly processed highly active heparanase, as revealed by Western blot analysis (Fig. 3B) and heparanase enzymatic activity (Fig. 3C). In contrast, cathepsin L knock-out fibroblasts (Fig. 3A) expressed high levels of the full-length 65-kDa pro-enzyme (Fig. 3B) and almost no heparanase enzymatic activity (Fig. 3C). Next, WT and cathepsin L knock-out fibroblasts were stably transfected with pcDNA-Hepa. Although in cathepsin L knockout fibroblasts the overexpressed heparanase accumulated as a 65-kDa latent proheparanase, in the WT fibroblasts proheparanase underwent proper processing into a highly active enzyme (Fig. 3, D and E). In subsequent studies, heparanase enzymatic activity was examined in tissue extracts derived from WT and cathepsin L knock-out mice. As demonstrated in Fig. 3F, samples (i.e. liver, spleen) derived from cathepsin L knock-out mice exhibited very little or no detectable heparanase activity as opposed to high activity levels in corresponding samples derived from wild-type mice. Collectively, these results emphasize the critical role of cathepsin L as the principal and likely sole protease responsible for proheparanase processing and activation.
FIGURE 3.
Cathepsin L knock-out (KO) fibroblasts and tissues are unable to process proheparanase. Fibroblasts derived from either cathepsin L knock-out RT2 tumors (○) or wild-type RT2 tumors (•) were lysed (1% Nonidet P-40, 10 mm EDTA in PBS supplemented with protease inhibitors) and subjected for Western blot analysis of: A, cathepsin L (39-, 26-, and 21-kDa forms) and α-tubulin; and B, heparanase, as described under “Materials and Methods.” C, cell lysates were also analyzed for heparanase activity assay, as described under “Materials and Methods.” D and E, cathepsin L knock-out fibroblasts (□) and wild-type fibroblasts (▪) were also stably transfected with heparanase and subjected to Western blotting for heparanase (D) and determination of heparanase enzymatic activity (E). F, tissues (liver, ○, •; spleen Δ, ▴) derived from cathepsin L knock-out (•, ▴) or WT (○, Δ) mice were homogenized in lysis buffer and the supernatant fraction (500 μg) was analyzed (37 °C, 18 h, pH 6.0) for heparanase enzymatic activity, as described in the legend to Fig. 1.
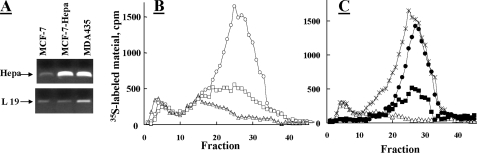
Overexpression of Cathepsin L Augments Processing and Activation of Proheparanase in MCF-7 Cells—Next, we investigated the effect of cathepsin L overexpression on the ability of cells to process and activate the 65-kDa proheparanase protein. For this purpose, we used MCF-7 human breast carcinoma cells which, unlike MDA-MB-435 cells, express low levels of both active cathepsin L (36, 37) and heparanase (30). Semi-quantitative RT-PCR demonstrated that MCF-7 cells stably transfected with pcDNA-Hepa express high levels of proheparanase mRNA (Fig. 4A) and protein (not shown), comparable with those found in WT MDA-MB-435 cells. Nevertheless, little or no significant increase in heparanase activity was detected in heparanase-transfected MCF-7 cells (Fig. 4B) despite their high heparanase mRNA expression level (Fig. 4A). These results indicate that increased expression of heparanase is not sufficient to confer a comparable increase in heparanase enzymatic activity; rather that it is processing of the proheparanase protein that is limiting.
FIGURE 4.
Overexpression of cathepsin L in MCF-7 cells augments processing of proheparanase. Parental MDA-MB-435 (○), MCF-7 (Δ), and MCF-7 cells stably transfected with heparanase (MCF-7-Hepa, □) were analyzed for: A, heparanase (Hepa) mRNA levels (RT-PCR); B, heparanase enzymatic activity (37 °C, 5 h, pH 6.0); and C, heparanase activity in MCF-7 cells overexpressing cathepsin L. MCF-7 cells were mock transfected (Δ), transiently double transfected with a plasmid encoding the full-length heparanase and empty plasmid (▪), or transiently double transfected with plasmids encoding heparanase and cathepsin L (•). Cell lysates were analyzed for heparanase activity (37 °C, 5 h, pH 6.0) in comparison with WT MDA-MB-435 cells (×), as described in the legend to Fig. 1.
We next tested whether the high level of cathepsin L activity in MDA-MB-435 cells (36, 37) is responsible for a more effective processing and generation of active heparanase in these cells than in MCF-7 cells. For this purpose, MCF-7 cells were transiently doubly transfected with vectors encoding for heparanase and cathepsin L, and compared with cells doubly transfected with heparanase and control empty vector. As depicted in Fig. 4C, transfection with both heparanase and cathepsin L resulted in a marked increase in heparanase enzymatic activity as compared with MCF-7 cells transfected with heparanase alone. Notably, this activity was similar in magnitude to that observed with MDA-MB-435 cells (Fig. 4C). These results clearly demonstrate that the limiting factor in heparanase-transfected MCF-7 cells is cathepsin L, emphasizing again its critical involvement in proheparanase processing.
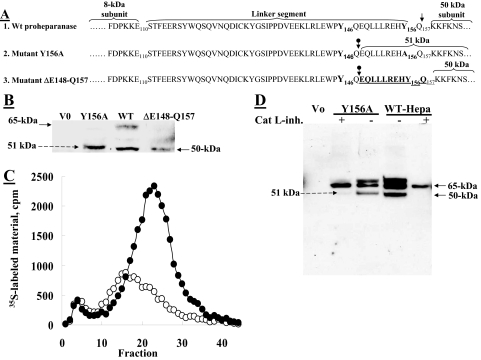
A 10-Amino Acid Peptide at the C Terminus of the Linker Segment Is Critical for Proheparanase Processing and Activation—We have previously demonstrated that substitution of Tyr156 with alanine (Y156A) altered the correct processing of proheparanase, resulting in generation of a 51-kDa inactive polypeptide instead of the characteristic 50-kDa subunit (13). We speculated that a point mutation (Y156A) in the potential cathepsin L motif Y156Q at the C terminus of the linker peptide will expose the next potential cleavage site (i.e. Y146Q), along the linker segment (internal cleavage site) resulting in a 10-amino acid peptide (Glu148–Gln157), which remains attached to the 50-kDa subunit and thereby blocks access to the active site of the enzyme, as predicted by the three-dimensional model of heparanase (13). To validate this mode of action, we generated a deletion mutant ΔGlu148–Gln157 (Fig. 5A) that lacks the 10-amino acid peptide (Glu148–Gln157) at the C terminus of the linker that precedes the 50-kDa subunit, and consequently eliminates the cathepsin L cleavage site (Y156Q) but still contains the remaining 38 amino acid portion of the linker (between Ser110–Gln147), including the internal Y146Q-cathepsin L cleavage motif. To investigate the processing and enzymatic activity of this deletion mutant versus the point-mutated (Y156A) proheparanase, we transfected JAR cells with vectors encoding either the wild-type proheparanase, the deletion mutant ΔGlu148–Gln157, or the Y156A mutant (Fig. 5A). Western blot analysis revealed that unlike the Y156A mutant, which was improperly processed, yielding a 51-kDa subunit (Fig. 5B) with very low heparanase activity (Fig. 5C, ○), similar to that of mock transfected cells (V0), the deletion mutant ΔGlu148–Gln157 was properly processed, yielding a 50-kDa subunit (Fig. 5B) and a highly active enzyme (Fig. 5C, •). Thus, whereas the Y156A point mutant resulted in an improperly processed (elongated 51-kDa subunit) and inactive heparanase due to the presence of a 10-amino acid peptide that blocks access to the active site, the ΔGlu148–Gln157-mutated proheparanase (lacking the 10 amino acid peptide at the C terminus of the linker segment) underwent proteolytic cleavage at an internal cleavage site (Y146Q), yielding a properly processed 50-kDa subunit and active heparanase. Therefore, these results indicate that activation of proheparanase requires proteolytic excision of the 10-amino acid peptide at the C terminus of the linker segment. Moreover, these results demonstrate that the Y146Q motif serves as a functional cathepsin L endocleavage site located within the linker segment upstream to the Y156Q site, yet could be identified only when the latter motif is mutated (Fig. 5B).
FIGURE 5.
A 10-amino acid peptide at the C terminus of the linker segment is critical for proheparanase activation. A, schematic presentation of WT proheparanase undergoing processing at Y156Q (↓), yielding the accurate 50-kDa subunit (1). Point-mutated (Y156A) proheparanase, yielding a 51-kDa polypeptide composed of the 50-kDa subunit conjugated to a 10-amino acid peptide at the C terminus of the linker (2), due to cleavage inside the linker segment at the cathepsin L motif Y146Q (circle with down arrow). Deletion mutant, ΔGlu148–Gln157, spanning the 10-amino acid peptide at the C terminus of the linker segment, including the Y156Q cleavage site, which undergoes processing at Y146Q, generating the proper 50-kDa subunit (3). B, JAR cells were stably transfected with empty plasmid (V0), pcDNA encoding the full-length heparanase cDNA (WT), pcDNA encoding the point mutated heparanase cDNA (Y156A), or pcDNA encoding the deletion mutant (ΔGlu148–Gln157). Cell lysates were subjected to Western blot analysis using anti-heparanase pAb 1453. C, JAR cells transfected with the Y156A (○) or ΔGlu148–Gln157 (•) heparanase mutants were assayed for heparanase enzymatic activity (7 h, pH 6.0, 37 °C) on sulfate labeled ECM. D, JAR cells were either mock transfected (Vo) or transiently transfected with wild-type heparanase (WT) or the Y156A point-mutated heparanase. The cells were grown (24 h) in the absence or presence of 0.72 μm cathepsin L inhibitor I (catalog number 219421, Calbiochem), extracted (1% Nonidet P-40, 10 mm EDTA in PBS supplemented with protease inhibitors), and subjected to Western blot analysis of heparanase, applying pAb 1435.
To further validate the functionality of these two sites (Y156Q; Y146Q) as cathepsin L cleavage motifs, JAR cells transfected with vectors encoding either the WT proheparanase or the Y156A mutant were grown in the presence or absence of the cell permeable cathepsin L inhibitor I (Calbiochem, Merck, Darmstadt, Germany) and subjected to Western blot analysis (Fig. 5D). In the absence of the cathepsin L inhibitor the WT proheparanase was properly processed, whereas the Y156A mutant yielded the 51-kDa subunit (Fig. 5D), as expected. In contrast, in the presence of the inhibitor, processing was abolished at both sites resulting in accumulation of the 65-kDa proenzyme (Fig. 5D); further verifying that these sites are indeed cathepsin L cleavage motifs.
Because the intact linker segment could not be detected by pAb CKLE antibodies that specifically recognize the linker segment (not shown), we assumed that other endocleavage sites (apart from the Y146Q site) exist along the linker segment. Accumulation of the 65-kDa proheparanase in the presence of cathepsin L inhibitor (Fig. 5D) further suggest susceptibility of the linker peptide to multiple endocleavages by cathepsin L, functioning as the primary protease responsible for removing the entire linker segment (see below).
MALDI-TOF Analysis of Proheparanase Cleavage Peptides Generated by Cathepsin L—We have previously demonstrated that recombinant latent proheparanase is properly processed by cathepsin L in a cell-free system, yielding an active enzyme composed of the 8- and 50-kDa subunits (13). To verify the accurate size of the 8-kDa heparanase subunit generated by cathepsin L and the fate of the linker segment, a reaction mixture consisting of recombinant proheparanase incubated with cathepsin L was subjected to MALDI-TOF mass spectrometry (MS) analysis. Incubation of the full-length 65-kDa proheparanase with cathepsin L yielded several peptides of different masses, none of which was detected upon incubation of proheparanase in reaction buffer alone (not shown), indicating that these are cleavage products of proheparanase generated by cathepsin L. Five main masses (P1–P5) ranging from ∼ 8 to ∼ 9 kDa were revealed by this analysis (Fig. 6A). The determined mass of 8244.29 daltons (P1) is in agreement with the expected calculated mass of the full-length 8-kDa subunit (Gln36–Glu109) including Gln36 that resides after cleavage of the signal peptide at the consensus site Gln34–Ala–Gln36 (16). This result indicates that cathepsin L yields the precise 8-kDa subunit. Other masses were 8624.76 (P2), 8771.58 (P3), 8917.89 (P4), and 9410.66 (P5) daltons (Fig. 6A), in agreement with masses that correspond to Gln36–Phe112, Gln36–Glu113, Gln36–Glu114, and Gln36–Tyr117, respectively (see “Materials and Methods”). These are species of the 8-kDa subunit with extended C termini, likely generated by several endocleavages that take place upstream to site 1 within the N terminus of the linker segment. The occurrence of multiple adjacent endocleavages upstream to site 1 is in agreement with site-directed mutagenesis indicating tolerance for point mutation substitutions in this site and its flanking regions, yielding an active enzyme (13). Thus, it appears that abolishing the exact endocleavage at site 1 can be compensated by adjacent upstream endocleavages, yielding a slightly elongated 8-kDa subunit that is still functional.
MS analysis revealed that in the presence of cathepsin L, shorter heparanase peptides (∼2 to ∼5 kDa) were generated, in addition to the principal peptides (P1–P5) corresponding to the 8-kDa subunit (see above). Attenuated processing of recombinant proheparanase yielded up to 8 peptides (Fig. 6B) with masses of 5764.2, 4857.68, 3731.78, 2955.54, 2525.36, 2374.16, 2133.19, and 2061.04 daltons (Fig. 6B) corresponding to Glu114–Gln157, Ser120–Gln157, Gln119–Glu148, Gln119–Glu143, Gln119–Leu140, Ser131–Glu148, Ser131–Tyr146, and Ile122–Gln148, respectively (see “Materials and Methods”), all derived from the linker segment. Notably, when recombinant single chain heparanase in which the linker segment was replaced by 3 glycine-serine residues (hepa-3GS) (15) was incubated with cathepsin L, none of these peptides were detected by mass spectrometry (not shown). Altogether, the MS analysis suggests that cathepsin L acts at multiple endocleavage sites along the linker segment, thereby removing it in small fragments rather than as an intact peptide. This observation is supported by the observation that the linker segment could not be detected by an antibody (α-CKEL) that specifically recognizes the linker peptide in the context of the intact proenzyme (not shown). Notably, determination of a particular MS peptide sequence took into account the occurrence of phosphorylation modification (additional 80 δ) on particular tyrosine or serine residues along the linker (Fig. 7), consistent with our observation that the linker segment is preferentially phosphorylated on serine and tyrosine residues.3
FIGURE 7.
Multiple endocleavage sites along the linker segment. The primary sequence of the linker segment is shown, flanked by the 8-kDa subunit at the N terminus, and the 50-kDa subunit at the C terminus. Sequences of peptides 1 to 13 established by mass spectrometry (Fig. 7) were overlapped, highlighting nine typical cathepsin L endocleavage sites (↓) taking place at the first or second amino acid upstream to a bulky amino acid (bold), and five atypical cathepsin L endocleavage sites (circle with down arrow) (C terminus of Ser109, Phe111, Tyr117, Leu130, and Tyr146). PO4 indicates the predicted phosphorylated sites. Phosphorylation of the underlined amino acids in peptides 1–13 was taken into account in calculating their molecular mass.
Proposed Mode of Action—The 13 peptides identified by the MS analysis were aligned on the basis of overlapping sequences (Fig. 7). Interestingly, five putative phosphorylation sites (Ser110, Ser116 or Tyr117, Tyr129 or Ser131, Tyr146 and Tyr156) are in accordance with the overlap among the different peptides, supporting their existence (Fig. 7). This alignment highlighted seven internal cleavage sites with characteristic cathepsin L cleavage motifs, all containing an aromatic or hydrophobic amino acid at positions P2 or P3 of the cleavage site (Fig. 7). Altogether, the mass spectrometry data suggest that the entire linker segment can be removed solely by cathepsin L through multiple endocleavages.
DISCUSSION
Cathepsin L is a characteristic lysosomal cysteine proteinase of the papain superfamily of peptidases (18) that functions primarily as an endoprotease in the lysosome. In addition, the enzyme has been implicated in multiple physiological and pathological processes (18). Applying gene silencing, knock-out, and overexpression approaches, we demonstrate that alterations in the endogenous cellular levels of cathepsin L markedly affect the processing of proheparanase in carcinoma cells and immortalized fibroblasts. Of particular significance is the lack of detectable processing and activation of the latent 65-kDa enzyme in JAR cells co-transfected with the full-length latent enzyme and anti-cathepsin L siRNA (Fig. 1), and in tissues and cultured fibroblasts derived from cathepsin L knock-out mice (Fig. 3, E and F), suggesting that proheparanase cannot be properly processed and activated by proteolytic enzymes other than cathepsin L.
Unlike JAR cells, MDA-MB-435 breast carcinoma cells express high levels of endogenous heparanase. The lack of complete inhibition of heparanase processing and enzymatic activity in MDA-MB-435 cells transfected with anti-cathepsin L siRNA (Fig. 2) may be attributed to the exceedingly long half-life (30 h) of the endogenous active enzyme in these cells (35). Alternatively, the presence of additional cathepsin L-like proteases in MDA-MB-435 cells compared with JAR cells could contribute to this difference. The critical role of cathepsin L in proheparanase processing in breast carcinoma cells is further supported by the finding that overexpression of cathepsin L markedly increased the processing and activation of proheparanase when overexpressed in co-transfected MCF-7 cells, which otherwise express low endogenous levels of the proenzyme (Fig. 4B). By contrast, MCF-7 cells overexpressing the 65-kDa proenzyme alone, exhibited a very low heparanase enzymatic activity. Interestingly, MCF-7 cells exhibit a low cathepsin L activity primarily due to their high content of endogenous cathepsin L inhibitors (36, 37). This balance may be altered upon cathepsin L overexpression. In contrast, MDA-MB-435 cells express relatively low levels of endogenous cathepsin L inhibitors (36, 37), and hence proheparanase is readily processed and activated by these cells. Altogether, our results indicate that cathepsin L is the principal and most likely the sole enzyme responsible for processing and activation of proheparanase.
Applying deletion and point mutations, we show that a 10-amino acid peptide at the C terminus of the linker segment, located between the two functional cathepsin L cleavage sites Y156Q and Y146Q, abolished heparanase activity and that its removal by cathepsin L is critical for proheparanase activation. These results are consistent with the predicted three-dimensional model of heparanase, demonstrating that a 10-amino acid peptide at the C terminus of the linker is sufficient for blocking the accessibility of the substrate (heparan sulfate) to the active site of heparanase (13). This peptide can therefore be applied as a prototype to design peptides and peptidomimetics that inhibit heparanase enzymatic activity.
According to mass spectrometry analysis (Fig. 7), processing of recombinant proheparanase with cathepsin L generated the precise 8-kDa subunit (P1) plus four slightly elongated forms (P2, P3, P4, and P5), suggesting the occurrence of five endocleavage sites targeted by cathepsin L upstream of site 1 (Glu109) at Phe111, Glu112, Glu113, and Tyr117, respectively (Figs. 6 and 7). The feasibility of multiple adjacent endocleavages upstream of site 1 is in agreement with site-directed mutagenesis indicating tolerance for point mutations substitutions in this site and its flanking region, yielding an active enzyme (13). Thus, it appears that abolishing the primary endocleavage at site 1 can be compensated by adjacent upstream endocleavages, yielding a slightly elongated 8-kDa subunit that is still functional. Moreover, mass spectrometry analysis (Fig. 6C) revealed up to 8 peptides (P6–P13) all originating from the linker segment, indicating that cathepsin L attacks at multiple endocleavage sites along the linker segment (Figs. 6 and 7), namely, Trp118, Gln119, Gly130, Leu140, Glu143, Tyr146, Gln147, Glu148, and Tyr156, suggesting that the linker peptide is not removed as an intact segment. In support of this hypothesis is the observation that an intact linker segment could not be detected by Western blot analysis (not shown).
Among the 11 endocleavage sites revealed by the MS analysis (Figs. 6 and 7), nine are typical recognition motifs of cathepsin L that preferentially include bulky (aromatic or hydrophobic) amino acids at the P2 or P3 position of the cleavage sites (38, 39). Y156Q, Y146Q (aromatic acid (tyrosine) at P2 of the cleavage), Y146Q/E (tyrosine at P3 of the cleavage), L143E (hydrophobic acid (leucine) at P2 of the cleavage), Y129G (tyrosine at P2), W118Q (tryptophan at P2) or YWQ (tyrosine at P3) and F111E (phenylalanine at P2) or F111EE (phenylalanine at P3). A similar cleavage pattern was noted for processing of proenkephalin by cathepsin L at particular mono or dibasic amino acids where an aromatic acid is located at P2 or P3 of the cleavage site (21). The other cleavages at Glu109-(site 1), Phe111-, Tyr117-, Leu140-, and Tyr146 do not exhibit typical cathepsin L motifs. Although atypical cleavages by cathepsin L have been reported (39), an alternative explanation is that the Glu109, Phe111, Tyr117, Leu140, and Tyr146 may be regarded as the C terminus of peptides generated by endocleavage at an adjacent upstream typical motif (Fig. 7), which is subsequently blunted by the very weak carboxypeptidase activity of cathepsin L itself (40), in a stepwise manner. Thus, the 8-kDa subunit and four other peptides derived from the linker (e.g. P2, P5, P10, and P12) may be generated by either direct endocleavage at atypical sites, or by endocleavage at cathepsin L typical sites followed by a carboxypeptidase activity at the peptide C terminus. The mass spectrometry analysis also demonstrated that the linker segment is preferentially phosphorylated on serine and tyrosine residues, suggesting their possible regulatory role in heparanase processing through an effect on the accessibility of particular cathepsin L cleavage sites, as reported for HSP-70 (41).
The following observations have enabled us to determine herein the detailed mechanism of proheparanase processing. Applying MDA-MB-435 and MCF-7 breast carcinoma cells we have demonstrated that proheparanase processing is mediated by cathepsin L. Moreover, applying JAR cells devoid of endogenous heparanase, and cathespin L knock-out fibroblasts and tissues, we showed that proheparanase processing is brought about by cathepsin L as the primary and possibly sole protease. Applying site-directed mutagenesis we have demonstrated that proper proheparanase processing and activation requires proteolytic removal of a 10-amino acid peptide located between two functional cathepsin L cleavage sites, Y156Q, at the C terminus of the linker segment and Y146Q, inside the linker segment, emphasizing the key role of cathepsin L in proheparanase processing as it removes the most critical part of the linker segment and exposes the heparanase active site. Moreover, mass spectrometry analysis revealed that the entire linker segment is susceptible to multiple cleavages by cathepsin L and that the 8-kDa subunit can be generated by several alternative adjacent endocleavages, yielding the precise 8-kDa subunit or slightly elongated, but still active forms. Altogether, these results indicate that proper processing and activation of proheparanase can be brought about solely by cathepsin L.
Clearly, a better understanding of the mechanism of proheparanase processing requires elucidation of heparanase and cathepsin L cellular trafficking to more accurately identify the cellular site of processing. We have noticed that despite incomplete cathepsin L gene silencing in JAR cells (Fig. 1A), processing of proheparanase was fully abrogated (Fig. 1, B and C), suggesting that high levels of cathepsin L are needed for proheparanase processing, as previously demonstrated for other physiological functions of cathepsin L (42, 43). Overexpression of cathepsin L and other lysosomal proteases affects their sorting from the lysosomes and subsequent targeting into specific granules or secretory vesicles, where they perform a particular physiological function. For example, in transformed fibroblasts, up-regulated procathepsin L is targeted into dense core multivesicular bodies, also known as secretory lysosomes. These dense bodies are generated by fusion of vesicles of endosomal origin with either lysosomes or the plasma membrane where they are secreted as single vesicles termed exosomes (29). Our preliminary results suggest that heparanase and cathepsin L are most probably co-localized in multivesicular bodies (not shown).
The involvement of cathepsin L in tumor progression through proteolytic degradation of structural constituents of the ECM is well documented (25–27). Our results indicate that the tumor promoting effect of cathepsin L may be due, at least in part, to its role in proheparanase processing. Elucidation of heparanase trafficking and secretion may offer new tools for suppressing the proangiogenic and prometastatic properties of heparanase.
Acknowledgments
We acknowledge Drs. Thomas Reinheckel and Christoph Peters, University of Freiburg, Germany, for providing the original breeding pairs of cathepsin L knock-out mice.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1-CA106456 (to I. V.) and R01-CA125162-01 (to J. A. J.) from NCI. This work was also supported by Israel Science Foundation Grant 549/06, the Israel Cancer Research Fund, the Rappaport Family Institute Fund, the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum (DKFZ), and Israel's Ministry of Science and Technology (MOST). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ECM, extracellular matrix; siRNA, small interfering RNA; ACTH, adrenocorticotropic hormone; pAb, polyclonal antibody; PBS, phosphate-buffered saline; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; RT, reverse transcriptase; WT, wild type; MS, mass spectrometry; δ, daltons.
G. Abboud-Jarrous, R. Atzmon, T. Peretz, C. Palermo, B. B. Gadea, J. A. Joyce, and I. Vlodavsky, unpublished results.
References
- 1.Parish, C. R., Freeman, C., and Hulett, M. D. (2001) Biochim. Biophys. Acta 1471 M99-M108 [DOI] [PubMed] [Google Scholar]
- 2.Edovitsky, E., Elkin, M., Zcharia, E., Peretz, T., and Vlodavsky, I. (2004) J. Natl. Cancer Inst. 96 1219-1230 [DOI] [PubMed] [Google Scholar]
- 3.Ilan, N., Elkin, M., and Vlodavsky, I. (2006) Int. J. Biochem. Cell Biol. 38 2018-2039 [DOI] [PubMed] [Google Scholar]
- 4.Vreys, V., and David, G. (2007) J. Cell. Mol. Med. 11 427-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakajima, M., Irimura, T., and Nicolson, G. L. (1988) J. Cell. Biochem. 36 157-167 [DOI] [PubMed] [Google Scholar]
- 6.Vlodavsky, I., Fuks, Z., Bar-Ner, M., Ariav, Y., and Schirrmacher, V. (1983) Cancer Res. 43 2704-2711 [PubMed] [Google Scholar]
- 7.Edovitsky, E., Lerner, I., Zcharia, E., Peretz, T., Vlodavsky, I., and Elkin, M. (2006) Blood 107 3609-3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, I., Maly, B., Simon, I., Meirovitz, A., Pikarsky, E., Zcharia, E., Peretz, T., Vlodavsky, I., and Elkin, M. (2007) Clin. Cancer Res. 13 4069-4077 [DOI] [PubMed] [Google Scholar]
- 9.Koliopanos, A., Friess, H., Kleeff, J., Shi, X., Liao, Q., Pecker, I., Vlodavsky, I., Zimmermann, A., and Buchler, M. W. (2001) Cancer Res. 61 4655-4659 [PubMed] [Google Scholar]
- 10.Gohji, K., Okamoto, M., Kitazawa, S., Toyoshima, M., Dong, J., Katsuoka, Y., and Nakajima, M. (2001) J. Urol. 166 1286-1290 [PubMed] [Google Scholar]
- 11.Simizu, S., Ishida, K., Wierzba, M. K., and Osada, H. (2004) J. Biol. Chem. 279 2697-2703 [DOI] [PubMed] [Google Scholar]
- 12.Simizu, S., Suzuki, T., Muroi, M., Lai, N. S., Takagi, S., Dohmae, N., and Osada, H. (2007) Cancer Res. 67 7841-7849 [DOI] [PubMed] [Google Scholar]
- 13.Abboud-Jarrous, G., Rangini-Guetta, Z., Aingorn, H., Atzmon, R., Elgavish, S., Peretz, T., and Vlodavsky, I. (2005) J. Biol. Chem. 280 13568-13575 [DOI] [PubMed] [Google Scholar]
- 14.McKenzie, E., Young, K., Hircock, M., Bennett, J., Bhaman, M., Felix, R., Turner, P., Stamps, A., McMillan, D., Saville, G., Ng, S., Mason, S., Snell, D., Schofield, D., Gong, H., Townsend, R., Gallagher, J., Page, M., Parekh, R., and Stubberfield, C. (2003) Biochem. J. 373 423-435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nardella, C., Lahm, A., Pallaoro, M., Brunetti, M., Vannini, A., and Steinkuhler, C. (2004) Biochemistry 43 1862-1873 [DOI] [PubMed] [Google Scholar]
- 16.Fairbanks, M. B., Mildner, A. M., Leone, J. W., Cavey, G. S., Mathews, W. R., Drong, R. F., Slightom, J. L., Bienkowski, M. J., Smith, C. W., Bannow, C. A., and Heinrikson, R. L. (1999) J. Biol. Chem. 274 29587-29590 [DOI] [PubMed] [Google Scholar]
- 17.Nomura, T., and Fujisawa, Y. (1997) Biochem. Biophys. Res. Commun. 230 143-146 [DOI] [PubMed] [Google Scholar]
- 18.Turk, B., Turk, D., and Turk, V. (2000) Biochim. Biophys. Acta 1477 98-111 [DOI] [PubMed] [Google Scholar]
- 19.Britton, C., and Murray, L. (2004) J. Cell Sci. 117 5133-5143 [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa, T., Roth, W., Wong, P., Nelson, A., Farr, A., Deussing, J., Villadangos, J. A., Ploegh, H., Peters, C., and Rudensky, A. Y. (1998) Science 280 450-453 [DOI] [PubMed] [Google Scholar]
- 21.Yasothornsrikul, S., Aaron, W., Toneff, T., and Hook, V. Y. (1999) Biochemistry 38 7421-7430 [DOI] [PubMed] [Google Scholar]
- 22.Hook, V. Y. (2006) Cell. Mol. Neurobiol. 26 449-469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasothornsrikul, S., Greenbaum, D., Medzihradszky, K. F., Toneff, T., Bundey, R., Miller, R., Schilling, B., Petermann, I., Dehnert, J., Logvinova, A., Goldsmith, P., Neveu, J. M., Lane, W. S., Gibson, B., Reinheckel, T., Peters, C., Bogyo, M., and Hook, V. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 9590-9595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goulet, B., Baruch, A., Moon, N. S., Poirier, M., Sansregret, L. L., Erickson, A., Bogyo, M., and Nepveu, A. (2004) Mol. Cell 14 207-219 [DOI] [PubMed] [Google Scholar]
- 25.Gocheva, V., Zeng, W., Ke, D., Klimstra, D., Reinheckel, T., Peters, C., Hanahan, D., and Joyce, J. A. (2006) Genes Dev. 20 543-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joyce, J. A., Baruch, A., Chehade, K., Meyer-Morse, N., Giraudo, E., Tsai, F. Y., Greenbaum, D. C., Hager, J. H., Bogyo, M., and Hanahan, D. (2004) Cancer Cell 5 443-453 [DOI] [PubMed] [Google Scholar]
- 27.Frohlich, E., Mohrle, M., and Klessen, C. (2004) Arch. Dermatol. Res. 295 411-421 [DOI] [PubMed] [Google Scholar]
- 28.Palermo, C., and Joyce, J. A. (2008) Trends Pharmacol. Sci. 29 22-28 [DOI] [PubMed] [Google Scholar]
- 29.Keller, S., Sanderson, M. P., Stoeck, A., and Altevogt, P. (2006) Immunol. Lett. 107 102-108 [DOI] [PubMed] [Google Scholar]
- 30.Vlodavsky, I., Friedmann, Y., Elkin, M., Aingorn, H., Atzmon, R., Ishai-Michaeli, R., Bitan, M., Pappo, O., Peretz, T., Michal, I., Spector, L., and Pecker, I. (1999) Nat. Med. 5 793-802 [DOI] [PubMed] [Google Scholar]
- 31.Goshen, R., Hochberg, A. A., Korner, G., Levy, E., Ishai-Michaeli, R., Elkin, M., de Groot, N., and Vlodavsky, I. (1996) Mol. Hum. Reprod. 2 679-684 [DOI] [PubMed] [Google Scholar]
- 32.Roth, W., Deussing, J., Botchkarev, V. A., Pauly-Evers, M., Saftig, P., Hafner, A., Schmidt, P., Schmahl, W., Scherer, J., Anton-Lamprecht, I., Von Figura, K., Paus, R., and Peters, C. (2000) FASEB J. 14 2075-2086 [DOI] [PubMed] [Google Scholar]
- 33.Zetser, A., Bashenko, Y., Miao, H. Q., Vlodavsky, I., and Ilan, N. (2003) Cancer Res. 63 7733-7741 [PubMed] [Google Scholar]
- 34.Zheng, X., Chou, P. M., Mirkin, B. L., and Rebbaa, A. (2004) Cancer Res. 64 1773-1780 [DOI] [PubMed] [Google Scholar]
- 35.Gingis-Velitski, S., Zetser, A., Kaplan, V., Ben-Zaken, O., Cohen, E., Levy-Adam, F., Bashenko, Y., Flugelman, M. Y., Vlodavsky, I., and Ilan, N. (2004) J. Biol. Chem. 279 44084-44092 [DOI] [PubMed] [Google Scholar]
- 36.Zajc, I., Sever, N., Bervar, A., and Lah, T. T. (2002) Cancer Lett. 187 185-190 [DOI] [PubMed] [Google Scholar]
- 37.Xing, R., Addington, A. K., and Mason, R. W. (1998) Biochem. J. 332 499-505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lecaille, F., Authie, E., Moreau, T., Serveau, C., Gauthier, F., and Lalmanach, G. (2001) Eur. J. Biochem. 268 2733-2741 [DOI] [PubMed] [Google Scholar]
- 39.Loser, R., Schilling, K., Dimmig, E., and Gutschow, M. (2005) J. Med. Chem. 48 7688-7707 [DOI] [PubMed] [Google Scholar]
- 40.Judice, W. A., Puzer, L., Cotrin, S. S., Carmona, A. K., Coombs, G. H., Juliano, L., and Juliano, M. A. (2004) Eur. J. Biochem. 271 1046-1053 [DOI] [PubMed] [Google Scholar]
- 41.Laumas, S., Abdel-Ghany, M., Leister, K., Resnick, R., Kandrach, A., and Racker, E. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 3021-3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn, K., Yeyeodu, S., Collette, J., Madden, V., Arthur, J., Li, L., and Erickson, A. H. (2002) Traffic 3 147-159 [DOI] [PubMed] [Google Scholar]
- 43.Collette, J., Bocock, J. P., Ahn, K., Chapman, R. L., Godbold, G., Yeyeodu, S., and Erickson, A. H. (2004) Int. Rev. Cytol. 241 1-51 [DOI] [PubMed] [Google Scholar]