Abstract
Inh3 (inhibitor-3) is a potent inhibitor of protein phosphatase-1 that selectively associates with PP1γ1 and PP1α but not the PP1β isoform. We demonstrate that Inh3 is a novel substrate for caspase-3 and is degraded in vivo during apoptosis induced by actinomycin D. Inh3 was not degraded in apoptotic MCF-7 cells, which lack caspase-3. These experiments establish that Inh3 is a novel physiological substrate of caspase-3. Electroporation of the caspase-3-resistant Inh3-D49A mutant into HL-60 cells resulted in a significant attenuation of apoptosis induced by actinomycin D. These results show that Inh3 degradation contributes to the apoptotic process. Immunofluorescence based examination of the subcellular localizations of Inh3 and PP1γ1 revealed a major relocalization of the cellular pool of PP1γ1 from the nucleolus to the nucleus and then to the cytoplasm during actinomycin D-induced apoptosis. A similar redistribution of PP1α from the nucleus to the cytoplasm occurred. These results are consistent with an unexpected discovery that significant fractions of the cellular pools of PP1γ1 and PP1α are associated with Inh3 in HL-60 cells. Thus, Inh3 is a major factor in the cellular economy of PP1γ1 and PP1α subunits. The unscheduled relocalization of this large a pool of PP1 subunits and their release from a potent inhibitor could deregulate a diverse range of essential cellular processes and signaling pathways. We discuss the significance of these findings in relation to working hypotheses whereby Inh3 destruction could contribute to the apoptotic process.
Serine/threonine protein phosphatase-1 activity is involved in the regulation of a remarkably diverse range of cellular functions (1–3). Protein phosphatase-1 activity is provided by as many as 100 enzyme forms in which the catalytic subunit, PP12, is associated with one or more different subunits (1). These noncatalytic subunits function as targeting proteins, which serve to specify the substrates that are acted on by PP1. The PP1 catalytic subunit is a 37-kDa protein and is ubiquitously expressed in mammalian cells as three closely related isoforms, PP1α, PP1β/δ, and PP1γ1 (4). The structural basis for the interaction of PP1 with a large number of partners is due to a hydrophobic pocket, distal to the active site, that recognizes a sequence motif of the type RVXF (5–7). Spatial restriction by the targeting subunits is the primary mechanism for the endowment of specificity to the PP1 catalytic subunit, which by itself is relatively nonspecific (8). Each PP1-targeting subunit complex can be considered to be a Ser/Thr phosphatase with its own specificity (1, 6). Gene deletion of PP1-binding proteins in yeast has been shown to generate specific phenotypes associated with loss of dephosphorylation of specific PP1 substrates (9, 10). A corollary of the targeting hypothesis is that no free active PP1 is present in the cell, since this could lead to unregulated and nonspecific dephosphorylation of phospho-Ser/Thr-containing proteins.
There are also potent inhibitors of the PP1 catalytic subunit with IC50 values in the nanomolar range (1). These were originally identified as heat-stable and trypsin-sensitive proteins (11), and they now are recognized as regulatory/targeting subunits (1, 12–14). The best known of these inhibitor proteins are Inh1 (inhibitor-1) (15, 16), its neuronal homologue DARPP-32 (17), and Inh2 (inhibitor-2) (12). This group also includes Inh3 (inhibitor-3), which inhibits protein phosphatase-1 activity with an IC50 in the nanomolar range (18).
Human Inh3 is a small protein of 126 residues that has a calculated molecular mass of 14 kDa but exhibits anomalous behaviors on SDS-PAGE with an apparent molecular mass of 23 kDa and on gel filtration chromatography with an Mr of 55,000 (18). Inh3 exhibits specificity for its interactions with PP1α and PP1γ1, since it does not co-immunoprecipitate with PP1β (19). Inh3 loses its inhibitory functions after in vitro phosphorylation with several protein kinases that include protein kinase A, protein kinase C, and casein kinase 2.3 The cellular functions of Inh3 are unknown, but it is a nuclear protein that co-localizes with PP1γ1 in the nucleoli and PP1α in the centrosome and also is present in the mitotic apparatus (19).
The apoptotic pathways are complex, interconnected signaling systems that respond to death signals that bind to death receptors (extrinsic pathways) and to intracellular stress (intrinsic pathway) (20, 21). Binding of apoptotic signals, such as FasL and TRAIL, to transmembrane receptors leads to activation of the initiator caspases, caspase-8 and caspase-10 (20). The intrinsic pathway is triggered by intracellular stress (e.g. DNA damage, genotoxic chemicals, ionizing radiation, cytokine withdrawal, or mitochondrial damage). These signals lead to the release of cytochrome c from the mitochondria (22), which triggers the assembly of the apoptosome, a signaling platform that activates caspase-9 (20, 23). The initiator caspases in turn activate the effector or executioner caspases (caspase-3, -6, and -7). Caspase-3 is the major executioner caspase, since it degrades the majority of the caspase substrates (24, 25).
The release of cytochrome c is regulated by the interactions of proapoptotic and prosurvival proteins that are members of the Bcl-2 family of proteins; these play a key role in cellular decisions between survival and apoptosis in response to genotoxic stress and cytokine withdrawal. In unperturbed cells, Bcl-2, Bcl-XL, Bcl-w, Mcl-1, and A1 restrain Bax and Bak, which promote the release of cytochrome c. Cell damage is sensed by the proapoptotic BH3-only proteins, Bim, Bid, Puma, BAD (Bcl-2 antagonist of cell death), and Noxa, which bind to the prosurvival Bcl-2 proteins via their BH3 domains (26–28).
Several members of the Bcl-2 group of proteins and the BH3-only proteins are regulated by phosphorylation/dephosphorylation (29). This includes Bcl-2, whose phosphorylation enhances its antiapoptotic functions (30). One of the BH3-only proteins, BAD, plays a role in growth factor (cytokine) regulation of cell survival/apoptosis, and its proapoptotic functions are inhibited by several protein kinases that include Akt, Rsk, PAK, p70S6K, and protein kinase A. Dephosphorylation of BAD allows it to initiate a proapoptotic response by interaction with members of the Bcl-2 proteins (31–33). BAD phosphorylation is maintained by growth factor signaling, and withdrawal of growth factors leads to dephosphorylation of BAD and the activation of the intrinsic mitochondrial pathway (28, 34). Several protein phosphatases have been implicated in BAD dephosphorylation, including PP1α (35–37), PP2A (38), PP2B (39), and PP2C (40). Caspase-9 activation also has been reported to be dependent on PP1α activity (41). Electroporation of PP1α catalytic subunit into cells has been shown to trigger apoptosis (42, 43), whereas electroporation of Inh2 delayed apoptosis (42).
In this study, we provide the first evidence that Inh3 is a novel substrate for caspase-3 in vitro and in vivo. Inh3 is rapidly degraded by caspase-3 during act-D-induced apoptosis. Electroporation of a caspase-3-resistant Inh3 mutant into cultured HL-60 cells attenuated the progression of the apoptotic response, suggesting that Inh3 degradation actively participates in apoptosis.
EXPERIMENTAL PROCEDURES
Materials—Actinomycin D, camptothecin, etoposide, cycloheximide, okadaic acid, and Hoechst 33258 were obtained from Sigma. Recombinant human caspase-3 and benzyloxycarbonyl-Asp-Glu-Val-Asp-fluoromethyl ketone (Z-DEVD-fmk) as the methylated form Z-D(OMe)-E(OMe)-V-D(OMe)-fmk, were purchased from Calbiochem/EMD Biosciences. Nickel-nitrilotriacetic acid-agarose was obtained from Qiagen. TALON metal affinity resin was obtained from Clontech. Antibodies were obtained from the following sources: anti-phospho-Chk2 (Thr68), anti-phospho-ERK, anti-ERK, and anti-PARP from Cell Signaling; isoform-specific anti-PP1α (C-19), isoform-specific anti-PP1β (N-19), isoform-specific anti-PP1γ (C-19), mouse monoclonal anti-PP1 (E-9), anti-caspase-3 (H-277), anti-BAD (C-7), anti-phospho-BAD (Ser112)-R, and anti-GAPDH (6C5) from Santa Cruz Biotechnology; anti-polyhistidine from Sigma. A rabbit polyclonal antibody to full-length Inh3 was prepared as previously described (19). The Inh3-specific rabbit polyclonal antibody recognizing an epitope corresponding to amino acids 69–83 was kindly provided by Dr. S. Vijayaraghavan (Kent State University). Control rabbit IgG (sc-2027) was obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Rabbit muscle phosphorylase b and phosphorylase kinase were generous gifts from Dr. G. Carlson (University of Kansas).
Cell Culture and Treatments—HL-60 (human promyelocytic leukemia) and MOLT-4 (human T cell lymphoblastic leukemia) cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum. MCF-7 (human breast carcinoma) cells were cultured in minimum essential medium supplemented with 10% fetal bovine serum. All cells were cultured at 37 °C in a humidified atmosphere of 5% CO2. Suspension cells were only treated when the viability exceeded 95%.
Plasmid Construction and Mutagenesis—The construct used for N-terminally His6-tagged Inh3 expression was obtained by cloning the Inh3 coding sequence (18) into the SmaI site of the expression vector pQE-32 (Qiagen). The oligonucleotide primer pair used was 5′-ATG GCC GAG GCA GGG GCT-3′ (forward) and 5′-TTA GTG CTG CAT TGG CCC TGG-3′ (reverse). The ATG initiating codon and TAA stop codon are underlined. The mutation was generated with the QuikChange XL site-directed mutagenesis kit (Stratagene). For the Inh3(D49A) mutation, the oligonucleotide primer pair used was 5′-GG ACA AGT GAC ACT GTG GCC AAT GAA CAC ATG GGC CG-3′ and 5′-CG GCC CAT GTG TTC ATT GGC CAC AGT GTC ACT TGT CC-3′. Mutated codons are underlined. All constructs were verified by sequence analysis.
Expression and Purification of Inh3—His6-Inh3 and His6-Inh3(D49A) were expressed in Escherichia coli strain BL21 Star. Briefly, 2 liters of culture were grown in Terrific Broth supplemented with 50 μg/ml ampicillin to an optical density of 0.6 at A600, and protein expression was induced overnight at 26 °C by the addition of 0.1 mm isopropyl β-d-thiogalactoside. The cells were harvested by centrifugation and resuspended in 100 ml of ice-cold lysis buffer (50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole, 0.1 mm phenylmethylsulfonyl fluoride, 1 mm benzamide, pH 8.0). The cells were disrupted by passage through a French Press at 1500 p.s.i. Insoluble material was removed by centrifugation, and the supernatant was heated at 80 °C for 15 min and then centrifuged at 10,000 rpm for 30 min. The supernatant was applied to a nickel-nitrilotriacetic acid-agarose affinity column (Qiagen). After extensive washes with 50 mm imidazole in 50 mm NaH2PO4, 300 mm NaCl, pH 8.0, the protein was eluted with 150 mm imidazole in the same buffer.
Inh3 Immunodepletion and Western Blotting—Cell lysates were prepared by suspending the cells in lysis buffer (1% Non-idet P-40, 1.5 mm MgCl2, 10 mm KCl, 1 mm dithiothreitol, 10 mm HEPES, pH 7.9) in the presence of a protease inhibitor mixture (Sigma). The amounts of total protein were quantified using the Bio-Rad protein assay according to the manufacturer's instructions. Cell lysates (2.5 mg/ml) were precleared by incubation with 30 μl/ml protein A-agarose beads (Santa Cruz Biotechnology) for 1 h at 4 °C. After brief centrifugation, the supernatant (500 μl) was incubated with 20 μg of anti-Inh3 antibody overnight at 4 °C. Protein A-agarose beads (60 μl) were added to the sample for an additional 2 h at 4 °C. After centrifugation, the supernatant was collected, mixed with 3× SDS-PAGE sample buffer, boiled for 5 min, separated on SDS-polyacrylamide gels, and blotted onto polyvinylidene difluoride membranes (Bio-Rad). The membranes were blocked with 5% nonfat milk in TBST (50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 0.05% Tween 20) at room temperature for 1 h. They were then washed with phosphate-buffered saline, pH 7.4 (PBS) and incubated with the primary antibody in 5% bovine serum albumin in PBS overnight at 4 °C. After washing with TBST three times for 5 min each, the membranes were incubated with the secondary antibody at room temperature for 1 h. They were then washed three times with TBST and visualized by enhanced chemiluminescence (Pierce). Inh3 depletion was checked by Western blot using anti-Inh3 antibody. Densitometric analysis was performed using the Alpha Innotech ChemiImager 5500 and its software.
In Vitro Cleavage of Inh3 by Caspase-3—Wild-type or mutant Inh3 (0.5 μg) was treated with 200 units of recombinant active caspase-3 (Calbiochem/EMD Biosciences) in a buffer containing 50 mm HEPES (pH 7.4), 100 mm NaCl, 10% glycerol, and 10 mm dithiothreitol for various periods of time at 37 °C. The samples were then analyzed by Western blotting using specific antibodies.
Assessment of Apoptotic Cell Number—Determination of the apoptotic state of cells was determined by examination of Hoechst 33258 dye-stained cells by immunofluorescence microscopy using a Zeiss Axioplan 2 fluorescence microscope with a ×20 objective lens (42). Hoechst 33258 dye (5 μg/ml from 1 mg/ml stock in H2O) was added, and the cells were incubated for 10 min at 37 °C under 5% CO2. Normal cells were considered to have Hoechst-stained smooth nuclei, whereas nuclear staining cells having condensed, fragmented chromatin were judged to be apoptotic cells. A minimum of 300 cells were counted for each determination.
DNA Fragmentation—DNA was extracted using a High Pure PCR template preparation kit (Roche Applied Science). DNA samples were electrophoretically separated on 1.5% agarose gels. After electrophoresis, gels were visualized by staining with ethidium bromide under UV light.
Electroporation of Purified Inh3 into HL-60 Cells—Cells (5 × 106 cells/ml) were transferred to an electrocuvette (BTX) with a 0.4-cm electrode gap and mixed with purified Inh3 (20 μg) in Opti-MEM medium (Invitrogen) in a volume of 600 μl. Electroporation was performed with a BTX ECM 830 square wave electroporation system (Harvard Apparatus). Conditions for electroporation were optimized by monitoring the electroporation with His-tagged Inh3 followed by Western blotting. The pulse used was 280 V for 20 ms. After the pulse, the cells were transferred to RPMI 1640 medium supplemented with 10% fetal bovine serum. Cell viability of the electroporated cells was more than 90% after counting by trypan blue exclusion assay.
Overlay Assays—Purified Inh3 (0.5 μg) or the same amount of caspase-3-treated Inh3 was separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% nonfat milk for 2 h at room temperature and then incubated with purified PP1α (2 μg/ml) overnight at 4 °C. The membrane was washed extensively with PBS and probed with anti-PP1 antibody.
In Vitro Immunoprecipitation—Purified Inh3 (0.5 μg) and PP1α (0.5 μg) were incubated in PBS for 1 h at 4 °C. PP1α antibody (0.2 μg) was added to the mixture and kept for 1 h at 4 °C. Protein G-agarose beads (20 μl) were added to the mixture for an additional 1 h at 4 °C. After washing with PBS three times, bound proteins were eluted and subjected to Western blotting with anti-Inh3 antibody.
In Vitro Binding Assay—Cells electroporated with His-Inh3(D49A) or mock-electroporated cells were treated with act-D (4 μm) for 5 h. Cells were washed three times with PBS and harvested in lysis buffer in the presence of a protease inhibitor mixture. After centrifugation, the supernatants (about 500 μg of proteins) were added to 100 μl of TALON metal affinity resin (Clontech) and incubated for 1 h on a shaker. The beads were washed three times with 1 ml of 50 mm imidazole in TBS. Two hundred microliters of 290 mm imidazole in TBS were added to the beads and incubated for 10 min with shaking to release bound polyhistidine proteins. The eluted supernatants were collected by centrifugation and were subjected to immunoblotting.
TALON beads were saturated with His-Inh3 and washed three times with 50 mm imidazole in TBS. The beads bound ∼15 mg of His-Inh3/ml of resin. For pull-down assays of free PP1 from cell lysates, 500 μg of protein were added to 100 μl of His-Inh3-TALON beads and incubated for 1 h on a shaker. The beads were washed three times with 1 ml of 50 mm imidazole in TBS. The bound proteins were extracted with loading buffer and subjected to SDS-PAGE and immunoblotted with antibody against PP1.
Protein Phosphatase-1 Activity Assay—32P-Labeled rabbit muscle phosphorylase a was prepared by phosphorylation of phosphorylase b with phosphorylase kinase. [γ-32P]ATP was purchased from MP Biomedicals, Inc. The assay was performed as previously described (18). Recombinant PP1α was expressed in E. coli and purified as previously described (44).
Immunofluorescence Microscopy—HL-60 cells were washed with PBS and cytocentrifuged onto microscope slides. Slides were submerged in methanol at -20 °C for 20 min, followed by suspension in 80% ice-cold ethanol overnight at -20 °C. The cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min on ice, blocked with 2% bovine serum albumin in PBS for 1 h, and incubated with 8 μg/ml anti-Inh3 rabbit polyclonal antibody and 8 μg/ml anti-PP1α goat polyclonal antibody or 8 μg/ml anti-PP1γ goat polyclonal antibody followed by 15 μg/ml fluorescein isothiocyanate-labeled goat anti-rabbit IgG (H + L) secondary antibody (Jackson ImmunoResearch) for Inh3 and 15 μg/ml Rhodamine Red-X-labeled donkey anti-goat IgG (H + L) secondary antibody (Jackson ImmunoResearch) for PP1α or PP1γ. DNA staining was performed in 1 μg/ml 4′,6-diamidino-2-phenylindole for 10 min at room temperature. Localization of Inh3 or PP1α was determined by using a Zeiss Axioplan 2 fluorescence microscope with a ×40 objective lens. The images were captured using a Zeiss AxioCamMR3 camera and AxioVision release 4.6 software.
RESULTS
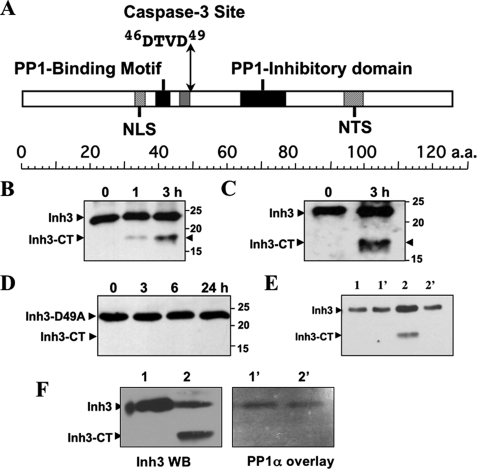
Inh3 Harbors a Caspase-3 Cleavage Site and Is a Substrate for Caspase-3 in Vitro—Caspases have a specificity for a 4-amino acid sequence, numbered P1–P4, with cleavage taking place after the invariant P4 Asp residue (24). Caspase-3 has a specificity for the sequence DEVD, but it also cleaves Ras GTPase-activating protein (45) and MEKK1 (mitogen-activated protein kinase/ERK kinase kinase 1) (46) at DTVD sequences. Inh3 harbors a putative caspase-3 cleavage site, 46DTVD49 (Fig. 1A), located between the two PP1 interaction sites, which consist of a RVXF motif (18) and an adjacent inhibitory domain, both of which are required for inhibition of PP1.4 Cleavage of Inh3 at this site would be expected to lead to separation of the two PP1 binding domains and the loss of high affinity binding to PP1.
FIGURE 1.
Inhibitor-3 is cleaved by caspase-3 in vitro at a DTVD cleavage site. A, domain map of human Inh3. The diagram shows the location of the putative caspase-3 cleavage site (46DTVD49), the nuclear localization signal (NLS), the nucleolar targeting signal (NTS) (19), the PP1 binding motif (39KKVEW43) (18), and the inhibitory domain that lies between residues 64 and 77. B, in vitro cleavage of Inh3 by caspase-3. Purified recombinant His6-Inh3 was incubated with recombinant human caspase-3 for the indicated times and Western blotted with a rabbit polyclonal antibody against Inh3 (see “Experimental Procedures”). Inh3-CT is the 17.5-kDa cleavage product. C, purified recombinant Inh3 was treated with caspase-3 as in B, except that a peptide-specific antibody to amino acids 69–83 of Inh3 was used. D, mutation of the caspase-3 site in Inh3 renders it resistant to cleavage. Recombinant Inh3(D49A) was treated with caspase-3 as in B, and the digests were analyzed by Western blotting with a polyclonal antibody against Inh3. One representative experiment of three is shown in A–D. E, the Inh3-CT cleavage product does not immunoprecipitate with PP1. Purified Inh3 or Inh3 predigested with caspase-3 was incubated with PP1α for 1 h at 4 °C, immunoprecipitated with PP1α antibody, and Western blotted with anti-Inh3 antibody (see “Experimental Procedures”). Lane 1, Inh3 input; lane 1′, immunoprecipitate of PP1α plus Inh3; lane 2, caspase-3-cleaved Inh3 input; lane 2′, immunoprecipitate of PP1α plus caspase-3-cleaved Inh3. F, overlay assay. Left, Western blot of Inh3 and caspase-3-treated Inh3 with Inh3 antibody. Lane 1, Inh3; lane 2, caspase-3-cleaved Inh3. Right, overlay blot of Inh3 and caspase-3-treated Inh3 with PP1. Lanes 1′ and 2′ correspond to lanes 1 and 2 in the left panel. The membranes were blocked with 5% nonfat milk proteins and then incubated with purified PP1α. The membrane was washed and then probed with anti-PP1 antibody (see “Experimental Procedures”). Essentially identical results were obtained in two independent experiments for the data shown in E and F. a.a., amino acids; WB, Western blot.
Inh3 was shown to be a substrate for caspase-3 in vitro by incubation of purified Inh3 with active recombinant caspase-3. Inh3 has a molecular mass of 14 kDa but migrates anomalously on SDS-PAGE with an apparent size of 23 kDa, which may be due to its high content of charged amino acid residues (18). Western blotting with a polyclonal antibody against Inh3 revealed the formation of a cleavage product of 17.5 kDa (Fig. 1B). Similar results were obtained with a polyclonal antibody against residues 69–83 of Inh3 (Fig. 1C). Cleavage at the DTVD sequence of Inh3 by caspase-3 would be expected to result in fragments of 5.5 and 17.5 kDa, assuming that these behaved in the same manner as full-length Inh3. The 5.5-kDa fragment was not detected, possibly because it is not recognized by the antibodies used.
The invariant Asp residue (Asp49) that is essential for caspase-3 substrate recognition (24) was mutated to alanine. Purified Inh3(D49A) was resistant to caspase-3 degradation (Fig. 1D), providing evidence that Inh3 is cleaved at the 46DTVD49 site.
The 17.5-kDa Inh3-CT fragment produced by caspase-3 degradation was tested for its ability to bind to PP1 by in vitro immunoprecipitation with anti-PP1α and by Western blot with Inh3 antibody (Fig. 1E). Lanes 2 and 2′ show the immunoprecipitation of a mixture of caspase-3-cleaved Inh3 and PP1α. The Inh3-CT fragment was not co-immunoprecipitated with PP1α (Fig. 1E, lanes 2 and 2′). The inability of Inh3-CT to interact with PP1 was also demonstrated by overlay blotting with PP1α (Fig. 1F). The left panel shows a Western blot with Inh3 antibody of a digest of Inh3 with caspase-3, similar to the experiment of Fig. 1B. Lane 1 shows Inh3, and lane 2 shows caspase-3-treated Inh3. The right-hand panel shows an overlay blot of the same two solutions for PP1 binding. The results confirm that the Inh3-CT fragment does not bind to PP1α. We were not able to detect the putative 5.5-kDa fragment in this assay, although it might have been expected to retain an interaction with PP1; this could be due to technical reasons because of a weak affinity or because of the small size of the fragment, which may have been poorly retained by the membrane.
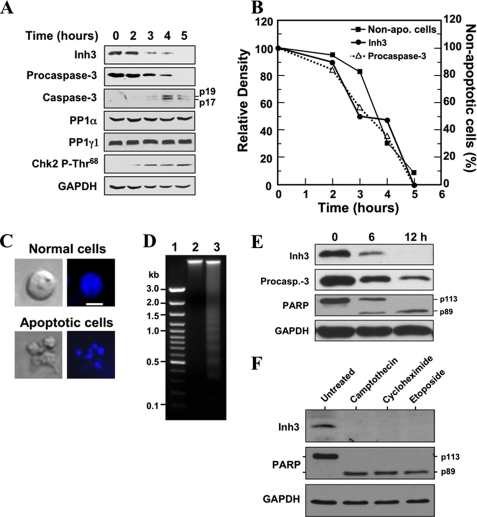
Inh3 Is Degraded in Vivo during Actinomycin D-induced Apoptosis— The effects of act-D-induced apoptosis on Inh3 levels were examined in order to address the question of whether it is an in vivo target of caspase-3. This was performed in the HL-60 cell line, which is a promyelocytic leukemia cell line that has been extensively studied as a model for hematopoietic differentiation (47) and apoptosis induced with act-D (48). Inh3 was degraded in a time-dependent manner in act-D-treated HL-60 cells and was completely degraded by 5 h (Fig. 2A). The activation of caspase-3 was monitored by the disappearance of procaspase-3 and the characteristic appearance of the 19- and 17-kDa polypeptides (Fig. 2A). The conversion of procaspase-3 to active caspase-3, which possesses polypeptides of 17 and 12 kDa, occurs by autoproteolysis; the 19-kDa polypeptide is further processed to the 17-kDa subunit (24). It is noteworthy that we did not observe the Inh3-CT fragment in Western blots of apoptotic cells with anti-Inh3, indicating that subsequent degradation of this cleavage product might be very rapid in vivo. This would not be surprising, since Inh3 is highly sensitive to proteolysis and also contains sequences in the C terminus that resemble PEST sequences (18).
FIGURE 2.
Inh3 is degraded during act-D-induced apoptosis in HL-60 cells in parallel with the activation of caspase-3. A, HL-60 cells were incubated with 4 μm act-D for the indicated times; cell lysates were prepared and subjected to Western blot analysis using the indicated antibodies. GAPDH was used as a protein loading control in the Western blots. B, the levels of Inh3 (solid circles) and procaspase-3 (open triangles) in the blots in A were quantified via densitometry and plotted against time. The percentage of apoptotic cells and nonapoptotic cells at each time point was determined by Hoechst staining (see “Experimental Procedures”). The data were plotted as percentage of nonapoptotic cells (solid squares). C, Hoechst 33258 staining of cells. The apoptotic cells exhibited irregular Hoechst nuclear staining with multiple bright specks of chromatin fragmentation and condensation. Normal cells were considered to have Hoechst-stained smooth nuclear regions. The images on the left are those obtained by light microscopy, and the images on the right were obtained by fluorescence microscopy. The white bar in the top right image is the scale for 10 μm. D, DNA fragmentation pattern of act-D-treated cells. HL-60 cells were untreated (lane 2) or treated with act-D for 5 h (lane 3). DNA was extracted and analyzed on 1.5% agarose gel electrophoresis after ethidium bromide staining. Lane 1, marker DNA. E, Inh3 is degraded in parallel with PARP during act-D-induced apoptosis. MOLT-4 cells were treated with 4 μm act-D for the indicated times. The cells were lysed and Western blotted for Inh3, caspase-3, PARP, and GAPDH. F, Inh3 degradation occurs during apoptosis induced by camptothecin, cycloheximide, or etoposide. HL-60 cells were treated with 10 μm camptothecin, 250 μm cycloheximide, or 250 μm etoposide for 5 h. The cells were then lysed and Western blotted for Inh3, PARP, and GAPDH. One representative experiment of three is shown.
The lysates were also Western blotted for PP1α and PP1γ1. These did not show appreciable changes during apoptosis (Fig. 2A). Western blots for Chk2-phosphothreonine 68 with a phosphospecific antibody were also performed as an index of its activation (Fig. 2A). The levels of Chk2-phosphothreonine 68 increased from 2 to 5 h, consistent with its activation by ATM (ataxia-telangiectasia mutated), as would be expected by activation of the intrinsic apoptotic signaling pathways (49–51).
The relative levels of Inh3 and procaspase-3 in the blots in Fig. 2A were determined by densitometry and plotted against time. These displayed a nearly co-incident time course of degradation (Fig. 2B). The time course of induction of apoptosis was determined by counting the number of apoptotic cells by fluorescence microscopy after staining with Hoechst 33258 dye (see “Experimental Procedures”). The results were plotted as the percentage of nonapoptotic cells (Fig. 2B). This time course lagged slightly behind that for caspase-3 activation (disappearance of procaspase-3) and Inh3 degradation (Fig. 2B). The appearance of the apoptotic cells (irregular Hoechst nuclear staining with multiple bright specks of chromatin fragmentation and condensation) is shown in Fig. 2C. The apoptotic response was also confirmed by examination of the chromatin by agarose gel electrophoresis (Fig. 2D). Ethidium bromide staining revealed the classical ladder of DNA separated by oligonucleosome-sized intervals (52). Thus, the data show that the degradation of Inh3 is a novel apoptotic response, since it is temporally associated with the activation of caspase-3 and the progression of apoptosis in the cell population.
The apoptotic response of Inh3 was also examined in act-D-treated MOLT-4 human T lymphoblastic leukemia cells. Western blots were performed for Inh3, procaspase-3, and poly-(ADP-ribose) polymerase (PARP) (Fig. 2E). PARP is an important regulator of DNA damage repair and is a well established substrate of caspase-3. It is cleaved from a 113-kDa protein to a 89-kDa protein during apoptosis (53, 54). The time course for the degradation of Inh3 in MOLT-4 cells is similar to that for the decrease in procaspase-3 and the appearance of the cleaved form of PARP (Fig. 2E). It was noted that Inh3 and PARP degradation were slower in MOLT-4 cells than we had found in HL-60 cells, consistent with previous reports that MOLT-4 cells are less sensitive to act-D than are HL-60 cells (55).
The coupling between procaspase-3 activation and Inh3 degradation during apoptosis observed above (Fig. 2B) is consistent with the view that Inh3 is cleaved in vivo by activated caspase-3. It would be expected that Inh3 degradation should be observed under other apoptotic stimuli that result in caspase-3 activation. Inh3 levels were examined in HL-60 cells that were treated with other chemical agents that have been extensively used to induce apoptosis. These were camptothecin and etoposide, which are topoisomerase poisons that cause DNA damage, and cycloheximide (56, 57). All three agents induced the cleavage of Inh3 as well as PARP (Fig. 2F).
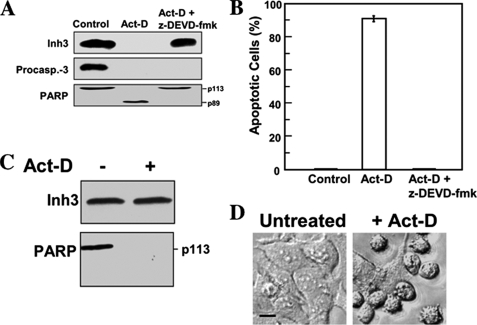
Cleavage of Inh3 in Vivo during Apoptosis Is Mediated by Caspase-3—As Inh3 is highly sensitive to proteolysis (18), it could be degraded by other caspases or non-caspase proteases that are activated during apoptosis (e.g. the calpains) (58). In order to obtain further evidence that caspase-3 is the agent for the degradation of Inh3 in vivo during apoptosis, the effects of the caspase-3 inhibitor Z-DEVD-fmk (59) were examined. The cleavages of Inh3 and PARP were completely blocked by pretreatment of HL-60 cells with Z-DEVD-fmk (100 μm) for 1 h prior to treatment with act-D (Fig. 3A). In parallel, Z-DEVD-fmk blocked act-D-induced apoptosis (Fig. 3B).
FIGURE 3.
Inh3 is not degraded during act-D-induced apoptosis in the presence of the caspase inhibitor, Z-DEVD-fmk, or in a caspase-3-deficient cell line. A, HL-60 cells were incubated with or without caspase inhibitor (Z-DEVD-fmk; 100 μm) for 1 h and then treated with 4 μm act-D for an additional 5 h to induce apoptosis. Cell lysates were subjected to Western blot analysis using antibodies against Inh3, caspase-3, and PARP. B, the percentage of apoptotic cells was determined by the Hoechst assay (see “Experimental Procedures”). The error bar indicates the mean ± S.D. (n = 3). C, MCF-7 cells, which lack caspase-3, were treated with 16 μm act-D for 24 h. Rounded and detached cells were collected, lysed, and Western blotted for Inh3 and PARP. D, phase-contrast microscopy of MCF-7 cells treated as in C, showing the morphological differences between untreated and act-D-treated cells. Phase images were obtained using a Zeiss Axioplan 2 fluorescence microscope with a ×20 objective (see “Experimental Procedures”). Scale bar (black horizontal bar in the left image), 10 μm.
Although Z-DEVD-fmk is often used as a selective inhibitor of caspase-3, it also inhibits caspase-7 and caspase-8, albeit with lower efficiencies (59). As a further test of the role of caspase-3 in Inh3 degradation, the effects of apoptosis on Inh3 levels were examined in a caspase-3-deficient cell line, MCF-7. The MCF-7 human breast carcinoma cell line is caspase-3-deficient because of the deletion of exon 3 in the caspase-3 gene (60). MCF-7 cells were treated with act-D for 24 h and analyzed by Western blotting for Inh3. Inh3 was completely resistant (Fig. 3C). These experiments provide an unequivocal demonstration that caspase-3 is required for the degradation of Inh3 during apoptosis and a positive identification of caspase-3 as the in vivo agent for the degradation of Inh3. It should be noted that MCF-7 cells nevertheless undergo apoptosis after treatment with act-D. In contrast to Inh3, PARP was still degraded (Fig. 3C), and examination of cell morphology (cell shrinkage, a rounded morphology, and increased detachment) confirmed that apoptosis had occurred (Fig. 3D). This is consistent with the evidence for caspase-3-independent mechanisms for PARP degradation (61) and caspase-7 activation (62) in MCF-7 cells during apoptosis.
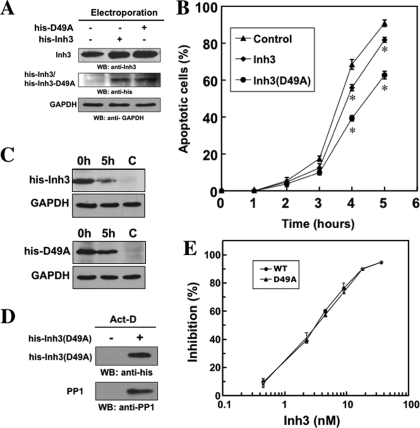
Stabilization of Inh3 against Caspase-3 Cleavage Attenuates act-D-induced Apoptosis—The finding that Inh3 is an in vivo target for caspase-3 suggests that the consequent release of PP1α and PP1γ1 from inhibition could be a contributory element in the induction of apoptosis, bearing in mind that they are implicated in the regulation of many cellular processes. Irrespective of mechanism, it has been shown that PP1 activity is proapoptotic via the electroporation of the free PP1 catalytic subunit into HL-60 cells (42). In order to assess whether Inh3 degradation by caspase-3 contributes to the apoptotic response, recombinant His-tagged wild-type His-Inh3 and His-Inh3(D49A) were introduced into HL-60 cells by protein electroporation (see “Experimental Procedures”). The efficiency of protein delivery using electroporation was determined by Western blot analysis (Fig. 4A). Western blotting with Inh3 antibody indicated that the amounts of total Inh3 after electroporation had increased (Fig. 4A, top). Densitometry of the blots showed that the amounts of His-Inh3 and His-Inh3(D49A) that were introduced by electroporation were 0.5 and 0.8 times the levels of endogenous Inh3, respectively. Confirmation of the introduction of the His-tagged proteins was made by Western blotting with anti-polyhistidine antibody (Fig. 4A, middle).
FIGURE 4.
Introduction of the caspase-3 resistant Inh3(D49A) mutant into HL-60 cells by electroporation attenuates act-D-induced apoptosis. Purified recombinant His-Inh3(D49A) or His-Inh3 was introduced into HL-60 cells by electroporation (see “Experimental Procedures”). A, the uptake of electroporated His-Inh3 or His-Inh3(D49A) was monitored by Western blot analysis with an anti-Inh3 antibody. The relative densities of the blots were determined to be 1:1.5:1.8 for the control cells, cells electroporated with His-Inh3, and cells electroporated with His-Inh3(D49A), respectively. The center panel shows the same membrane from the upper panel after stripping and Western blotting (WB) with anti-polyhistidine antibody. GAPDH was used as the loading control. B, mock-electroporated cells (solid triangles) and cells electroporated with His-Inh3 (solid diamonds) or His-Inh3(D49A) (solid circles) were treated with act-D (4 μm) for 1–5 h. The percentage of apoptotic cells was determined by Hoechst staining (see “Experimental Procedures”). Data are presented as mean ± S.D. from three independent experiments conducted in parallel (*, p < 0.005 comparing control and Inh3(D49A) or Inh3). C, HL-60 cells electroporated with His-Inh3 or His-Inh3(D49A), as indicated, were untreated (0 h) or treated with act-D (4 μm) for 5 h. Cell lysates were analyzed by immunoblotting using anti-polyhistidine antibody. C, mock-electroporated cells. D, pull-down assay for binding of PP1 to His-Inh3(D49A). His-Inh3(D49A) was electroporated into HL-60 cells. After act-D treatment for 5 h, cell lysates were pulled down with TALON metal affinity resin and subjected to immunoblotting using anti-polyhistidine (top) and anti-PP1 (bottom) (see “Experimental Procedures”). Left lane, apoptotic mock-electroporated cells; right lane, apoptotic cells electroporated with His-Inh3(D49A). The results shown were representative of three independent experiments. E, the D49A mutation of Inh3 does not affect its ability to inhibit PP1 activity. Purified recombinant His-tagged Inh3 (solid circles) and Inh3(D49A) (solid triangles) were assayed for the inhibition of PP1α activity measured using 32P-labeled muscle phosphorylase a as the substrate. Data are shown as the mean ± S.D. (n = 3) (see “Experimental Procedures”).
The effects of the introduction of His-tagged Inh3 and its D49A mutant into HL-60 cells on the time course of the induction of apoptosis by act-D were determined (Fig. 4B). The introduction of wild type Inh3 slightly reduced the apoptotic response; the reduction from the control was statistically significant. The introduction of Inh3(D49A) resulted in a significant reduction of the fraction of apoptotic cells from 90 to 60% by the 5 h time point (Fig. 4B). These results provide evidence for a dominant negative effect of Inh3(D49A) on the progression of apoptosis. The findings support the view that Inh3 degradation contributes to the apoptotic response in HL-60 cells. The effects observed are even more striking when it is considered that the ratio of introduced Inh3(D49A) to endogenous Inh3 was about 1:1.
Three additional control experiments were performed. Fig. 4C shows Western blots using anti-polyhistidine antibody of the lysates of HL-60 cells into which His-Inh3 and His-Inh3(D49A) had been electroporated after 5 h of treatment with act-D. The results show that His-Inh3(D49A) is more stable in apoptotic HL-60 cells than His-Inh3. HL-60 cells into which His-Inh3(D49A) had been introduced by electroporation were subjected to pull-downs of the His-Inh3(D49A) with TALON metal affinity beads (see “Experimental Procedures”) after act-D treatment. Western blots for His-Inh3(D49A) with anti-polyhistidine antibody and for PP1 with mouse monoclonal anti-PP1 antibody were performed (Fig. 4D). No PP1 was pulled down by the metal affinity beads in the control cells, which had not been electroporated with Inh3(D49A). In the apoptotic cells into which Inh3(D49A) had been electroporated, Inh3(D49A) was pulled down, showing that it is not degraded after apoptosis and that it is associated with PP1. The next issue that was addressed was whether the Inh3(D49A) mutant retained the ability to inhibit PP1 activity. The IC50 values of recombinant His-tagged Inh3 and His-tagged Inh3(D49A) for the inhibition of PP1 activity were determined (18). Both inhibited PP1 activity in an indistinguishable manner with an IC50 of ∼3 nm (Fig. 4E).
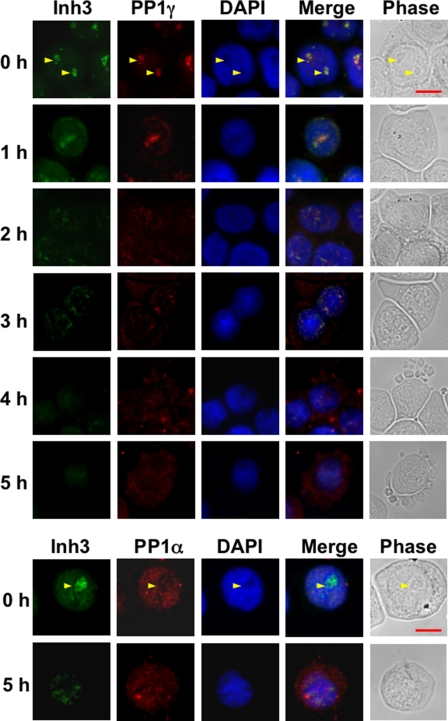
The Subcellular Localization of PP1γ1 and PP1α Is Altered during Apoptosis—Studies of the spatiotemporal distribution of PP1γ1 by time lapse imaging of PP1γ1 have shown that it is dynamically relocalized during the mammalian cell cycle and that it is implicated in nucleolar function and the regulation of chromosome segregation and cytokinesis (63). We have previously shown that PP1γ1 is largely present in the nucleoli in HEK 293 cells during anaphase and is dependent on Inh3 for its nucleolar localization (19) (i.e. there is reasonable evidence that the bulk of PP1γ1 in nucleoli is likely to be associated with Inh3).
The behavior of Inh3 and PP1γ1 during the 5-h time course for the induction of apoptosis in HL-60 cells by act-D is shown in Fig. 5 (top). Inh3 fluorescence in HL-60 cells (0 h panel) is present in the nucleus but not in the cytosol and is concentrated in the nucleoli, as has been previously reported for HeLa cells (19). Inh3 fluorescence was lost from the nucleoli and was evenly distributed to the nuclear compartment by 1 h. The nucleoli are known to be rapidly dispersed during apoptosis, and this has been demonstrated in HL-60 cells (64). This took place within 1 h in our experimental system, as shown by staining of the cells for RNA with pyronin Y (data not shown). The dispersal of Inh3 fluorescence to the nuclear compartment by the 1 h time point is consistent with the loss of the nucleoli. From 2 to 5 h, Inh3 fluorescence in the nucleus faded, consistent with its degradation during apoptosis. (Since we find that the Inh3-CT fragment does not accumulate during apoptosis (Fig. 2), we attribute the observed fluorescence to be due to intact Inh3.) The remaining Inh3 did not leave the nuclear compartment, although the nucleoli had been disrupted. In addition to a nuclear localization, it was noticed that Inh3 fluorescence retained an association with regions of denser fluorescence in the nuclei long after the nucleoli are disrupted, together with some retention of co-localization with PP1γ1. This is evident in the images for the 3-h period. In studies of other nucleolar constituents during apoptosis, it has been noticed that these may retain some associations with granular structures after loss of the visible nucleoli (65).
FIGURE 5.
Subcellular localization of Inh3, PP1α, and PP1γ1 in normal and apoptotic HL-60 cells. HL-60 cells were untreated or treated with 4 μm act-D for the times indicated. Cells were cytocentrifuged, fixed, permeabilized, and immunostained as described under “Experimental Procedures.” Top, cells were double-stained for Inh3 (green fluorescence) and for PP1γ1(red fluorescence). Bottom, HL-60 cells were untreated or treated with 4 μm act-D for 5 h. Cells were cytocentrifuged, fixed, permeabilized, and immunostained for Inh3 (green fluorescence) and for PP1α (red fluorescence). Fluorescence images were obtained using an Axioplan 2 fluorescence microscope with a ×40 objective. The yellow arrows in the 0 h rows show the nucleoli. Scale bar (shown by the red horizontal bar in the phase image in the upper right panel), 10 μm. DAPI, 4′,6-diamidino-2-phenylindole.
PP1γ1 (Fig. 5, top) is also concentrated in the nucleoli as previously reported for HeLa cells (19) and is released to the nuclear compartment by 1 h, similar to the behavior of Inh3 fluorescence. From 2 h onward, PP1γ1 fluorescence is increasingly visible in the cytoplasm and parallels the loss of Inh3 nuclear fluorescence. Taken together with the changes in Inh3 fluorescence, the data are consistent with the view that the degradation of Inh3 is accompanied by the relocalization of free PP1γ1 from the nucleoli to the nuclear and cytosolic compartments.
We also examined PP1α localization in HL-60 cells (Fig. 5, bottom) at 0 and 5 h after treatment with act-D. PP1α exhibits a nuclear localization and is present in the centrosomes, consistent with a previous report (63). In apoptotic cells, PP1α was redistributed from the nucleus to the cytoplasm at 5 h. However, this redistribution cannot be as closely linked to Inh3 degradation as in the case of PP1γ1, which is co-localized largely to the nucleoli, since the nuclear fluorescence for Inh3 is much weaker than in the nucleoli. Nevertheless, the data show that PP1α loses its nuclear localization and is translocated to the cytosol during apoptosis.
These studies show that there is a major relocalization of the nucleolar/nuclear pools of PP1γ1 and PP1α in apoptotic HL-60 cells that coincides with the degradation of Inh3. This relocation is an impressive apoptotic event that has not been previously described and, taken on its own, could reflect a mechanism that would result in the disruption of cellular phosphorylation/dephosphorylation systems. This would be the consequence of the release of free active PP1 subunits as well as their intracellular redistribution.
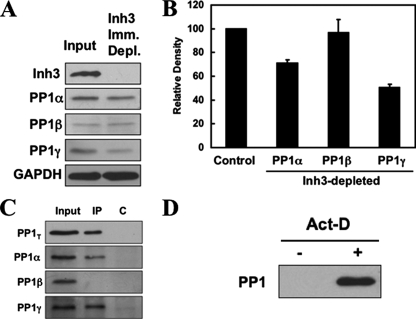
Determination of the Fractions of PP1α and PP1γ1 Associated with Inh3 in HL-60 Cells—Although relocalization of PP1γ1 and PP1α is coincident with the destruction of Inh3, it does not establish a causal relationship. However, if all of the PP1γ1 and PP1α that are relocalized arose through their release from Inh3, this would imply that the Inh3-PP1 complexes represent a sizable fraction of the cellular pools of these PP1 subunits.
The fractional amounts of PP1α and PP1γ1 that are associated with Inh3 in HL-60 cell lysates were estimated by determining how much of the two remained in lysates immunodepleted of Inh3. This was performed by immunoprecipitation with amounts of Inh3 antibody that would immunodeplete Inh3 in a single immunoprecipitation. The results of a typical experiment are shown in Fig. 6A, which shows the Western blots for Inh3, as well as the PP1 isoforms in the immunodepleted supernatants. Inh3 was completely depleted as shown by Western blots for Inh3 (Fig. 6A, top). The levels of PP1α and PP1γ1 were reduced, but those of PP1β were unchanged, consistent with our previous findings that it does not interact with Inh3 (19). The amounts of PP1α, PP1β, and PP1γ1 remaining after immunodepletion of Inh3 from cell lysates were then determined by densitometry from a similar experiment performed in triplicate (Fig. 6B). This showed that 51 and 71%, respectively, of PP1γ1 and PP1α remained in the immunodepleted supernatants (i.e. at least 49% of PP1γ1 and 29% of PP1α had been removed, whereas PP1β was unchanged). This analysis of the fractions of PP1γ1 and PP1α associated with Inh3 was surprising, but it is consistent with their subcellular relocalization during apoptosis.
FIGURE 6.
Inh3 is associated with significant fractions of the PP1α and PP1γ1 isoforms in HL-60 cells. HL-60 cell lysates were immunoprecipitated with a polyclonal antibody against Inh3 using amounts of antibody that were predetermined to be sufficient to immunodeplete the lysates (see “Experimental Procedures”). A, the Inh3-immunodepleted supernatants were Western blotted with antibodies against Inh3 and with isoform-specific antibodies against PP1α, PP1β, or PP1γ1, as indicated. GAPDH was used as a loading control. B, the amounts of PP1α, PP1β, and PP1γ1 remaining in the Inh3-depleted supernatants relative to the input (Control) were determined by densitometric analysis (see “Experimental Procedures”) and shown as a bar diagram. The data are presented as mean ± S.D. (n = 3). C, the immunoprecipitates (IP) from the experiment shown in A were immunoblotted with a non-isoform-specific antibody against PP1 (shown as PP1T) or with isoform-specific antibodies against PP1α, PP1β, or PP1γ1. The lanes shown are the input, the immunoprecipitate (IP), and the immunoprecipitation performed with normal rabbit IgG (lane C). D, pull-down assays of untreated and act-D-treated HL-60 lysates for free PP1. HL-60 cells were treated with 4 μm act-D for 5 h. Cell lysates were pulled down using TALON metal affinity beads that were presaturated with His-Inh3. The bound proteins were extracted with loading buffer and subjected to SDS-PAGE and immunoblotted with antibody against PP1 (non-isoform-specific).
Western blots with PP1 antibodies of the immunoprecipitates with Inh3 antibody under the conditions used in these experiments are shown in Fig. 6C. The results confirm that PP1α and PP1γ1 were immunoprecipitated and that PP1β did not interact with Inh3.
In order to demonstrate that free PP1 is generated during apoptosis, we performed an experiment using His-tagged Inh3 attached to TALON metal affinity beads. The beads were incubated with amounts of His-Inh3 sufficient to saturate the beads (see “Experimental Procedures”). These were then used for pull-down assays of nonapoptotic and apoptotic HL-60 cells (treatment with act-D for 5 h). No PP1 was bound to the Inh3-saturated beads in nonapoptotic cell lysates (Fig. 6D, left lane). This result provides an experimental test of the concept that there is little or no free PP1 in cells (3, 6). This contrasts with the results for the apoptotic HL-60 cell lysates, where PP1 was present in pull-down with the His-Inh3 beads, proving that free PP1 is present in the cell lysates from apoptotic cells (Fig. 6D, right lane). This supports the view that apoptotic degradation of Inh3 results in the release of free PP1.
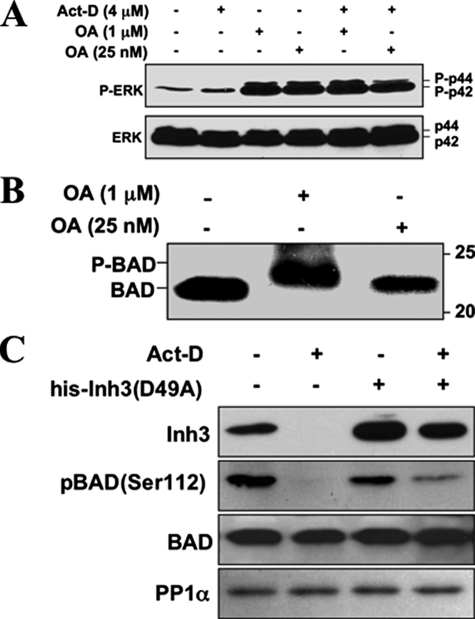
Involvement of Inh3 and PP1 Activity in BAD Phosphorylation/Dephosphorylation in Vivo—The findings that the Inh3(D49A) mutant attenuates the progression of apoptotic response in act-D-induced apoptosis in HL-60 cells supports the hypothesis that release of PP1 activity plays a role in the apoptotic response. Protein phosphatase-1 activity has been implicated in the dephosphorylation of a number of apoptosis-related proteins (66). In particular, PP1α activity has been implicated in the dephosphorylation of BAD (35–37, 42).
Experiments were first performed to confirm that PP1 activity was involved in BAD dephosphorylation in HL-60 cells. This was done by examining the effects of okadaic acid on BAD dephosphorylation. Okadaic acid (OA) has a 100-fold greater inhibitory effect on PP2A than PP1 in vitro (67). HL-60 cells were treated with 25 nm OA or a 40-fold higher concentration of 1 μm OA, conditions where PP2A would be selectively inhibited or where both PP2A and PP1 activity would be inhibited, respectively. As a control, levels of phospho-ERK, a known substrate of PP2A for which dephosphorylation is inhibited by low levels of OA (68), were examined (Fig. 7A). Treatment with 25 nm OA was sufficient to block the dephosphorylation of ERK as shown by the increased levels of phospho-ERK (as the p42 and p44 forms) in HL-60 cells in both untreated cells and in cells treated with act-D for 5 h (Fig. 7A). This confirmed that the lower concentration of OA (25 nm) would inhibit PP2A in our system.
FIGURE 7.
PP1 activity is involved in the dephosphorylation of BAD, and BAD dephosphorylation during act-D-induced apoptosis is inhibited by electroporation of Inh3(D49A). A, HL-60 cells were treated with 1 μm or 25 nm OA for 5 h, in the absence and presence of act-D (4 μm), as indicated. Cell lysates were Western blotted with a phosphospecific antibody for ERK (top), and with an antibody against ERK (bottom). B, HL-60 cells were treated with 1 μm or 25 nm OA for 5 h. Cells were then analyzed for BAD by Western blotting. The positions of phosphorylated (P-BAD) and nonphosphorylated BAD are indicated. Data for A and B are representative of three experiments. C, HL-60 cells were electroporated with or without purified His-Inh3(D49A) prior to treatment with act-D for 5 h or without act-D, as indicated. Cell lysates were Western blotted for Inh3, BAD-phosphoserine 112, BAD, and PP1α. The data are representative of three independent experiments.
The effects of 25 nm and 1 μm OA on BAD phosphorylation were examined in HL-60 cells. The lower concentration of 25 nm OA had no effect on BAD migration, but 1 μm OA caused a mobility shift of BAD to the hyperphosphorylated form (Fig. 7B). The lower concentration of 25 nm OA that is only sufficient to inhibit PP2A was unable to induce a mobility shift, supporting the proposal that PP1 activity is involved in the dephosphorylation of BAD.
We then examined the effects of electroporation of the Inh3(D49A) mutant into HL-60 cells on the levels of BAD-phosphoserine 112. HL-60 cells were treated with act-D for 5 h to induce apoptosis. Cells were Western blotted for Inh3, BAD-phosphoserine 112 with a phosphospecific antibody, BAD protein, and PP1α (Fig. 7C). BAD-phosphoserine 112 was dephosphorylated after induction of apoptosis. When His-tagged Inh3(D49A) was electroporated into HL-60 cells before treatment with act-D, BAD-phosphoserine 112 (Fig. 7C) was partially protected from dephosphorylation. This is consistent with an in vivo role for protein phosphatase-1 activity in the dephosphorylation of BAD, subsequent to the release of active PP1 by the degradation of Inh3.
DISCUSSION
This study provides evidence that Inh3 is a novel substrate for caspase-3. Inh3 is cleaved at the sequence 46DTVD49, a previously reported recognition sequence for caspase-3 (45, 46). More significantly, our data establish that Inh3 is a novel in vivo cellular substrate for caspase-3 and is degraded in a temporal manner consistent with the activation of caspase-3 and the induction of apoptosis. The findings that Inh3 is stable in caspase-3-deficient MCF-7 cells shows that Inh3 cleavage during apoptosis is specifically mediated by caspase-3. Inh3 is a nuclear protein, whereas procaspase-3 is cytosolic, so that it is important to note that it has been shown that caspase-3 is translocated to the nucleus after its activation (69).
Almost 400 mammalian caspase-3 substrates have been listed (70); however, not all of these have been shown to contribute to the apoptotic process (24, 25). A key experiment we have performed shows that caspase-3 cleavage of Inh3 is indeed relevant to the apoptotic response in HL-60 cells; viz. the electroporation of the Inh3(D49A) caspase-3-resistant mutant resulted in a significant (∼30%) attenuation of the progression of apoptosis induced by act-D. Future investigation of whether the caspase-3-mediated degradation of Inh3 holds for apoptosis triggered by the extrinsic pathways involving death receptors or by withdrawal of trophic factors should lead to further insights into the status of Inh3 as a factor in apoptosis. Given the preeminent role of caspase-3 as an executioner caspase (70), this would not be surprising.
The caspase-3 site is strategically located between the two interaction domains of Inh3, the KKVEW motif and the inhibitory domain (Fig. 1A). Severance of the connection between the two domains, purely on thermodynamic principles, would convert the system to one in which the two fragments would have much weaker affinities for PP1 than the interconnected domains, as has been shown for DARPP-32 (71).
We have also shown that a major translocation of PP1γ1 and PP1α occurs during apoptosis (Fig. 5). Nucleolar PP1γ1 and Inh3 were released to the nuclear compartment within 1 h, consistent with the dissolution of the nucleoli, and PP1γ1 fluorescence was released to the cytosol during the 2–5 h period in parallel with the loss of nuclear Inh3 fluorescence. PP1α is also released from the nuclear compartment to the cytosol during the same time period. These observations are consistent with the view that degradation of Inh3 may be accompanied by release of free PP1 catalytic subunits, whose potential deleterious effects would be enhanced by their loss of cellular localization. The amounts of PP1α and PP1γ1 that are released may be of sufficient magnitude to have an impact on cellular phosphoproteins, based on evidence that a significant portion of their cellular pools is associated with Inh3 (Fig. 6B). In addition, the use of metal chelate beads saturated with His-tagged Inh3 for pull-down assays of untreated and act-D-treated cells provided a direct demonstration for the release of free PP1 catalytic subunits in apoptotic cells5 (Fig. 6D).
The simplest hypothesis for the impact of Inh3 degradation on apoptosis is that this results in the release and translocation of free PP1α and PP1γ1 subunits, whose activities could potentially cause disruptions of the phosphorylation states of proteins involved in essential cellular processes or even generate proapoptotic responses by modification of apoptosis-signaling proteins.
Our previous findings that Inh3 and PP1γ1 are localized to the nucleolus leads to the thought that the Inh3-PP1γ1 complex may be involved in nucleolar processes (19). However, taken together with the large fraction of PP1γ1 that is involved, consideration must be given to another aspect of nucleolar function that has emerged during the past decade, viz. that it plays roles in control of the cell cycle, mitosis, and stress responses (72, 73). The trafficking of key nuclear proteins to the nucleolus is an emerging paradigm for a cellular strategy that allows the controlled sequestration and release of proteins from the nucleoli as a regulatory mechanism (72, 73). Thus, the question is raised as to whether nucleolar localization of Inh3-PP1γ1 represents a mechanism for sequestration of PP1γ1 until an appropriately scheduled release takes place.
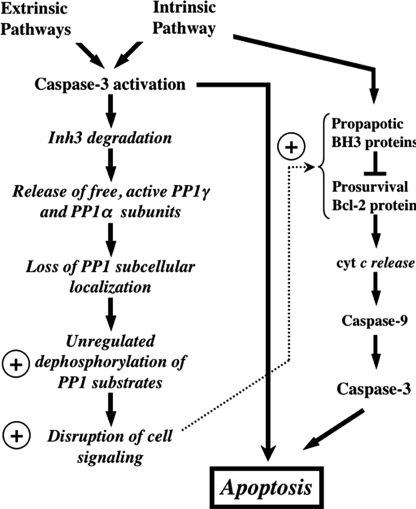
The potential significance of caspase-3-mediated degradation of Inh3 is shown diagrammatically in Fig. 8. The left-hand column shows the activation of caspase-3, which is triggered by elements of both the intrinsic and extrinsic pathways. Although we have only demonstrated that Inh3 is degraded by caspase-3 activation during apoptosis initiated through the intrinsic pathway, it seems reasonable that this might occur independently of the manner of caspase-3 activation. Below this we indicate the potential consequences of Inh3 degradation. These include the loss of constraints on PP1 localization, the deinhibition of PP1 activity, and the relocalization of PP1 to different cellular compartments.
FIGURE 8.
Overview of the potential impact of caspase-3 degradation of Inh3 on cell functions. This diagram shows the potential impact of caspase-3 on Inh3 degradation within a simplified scheme of the apoptotic pathways. Since caspase-3 is a primary executioner caspase, Inh3 degradation is expected to accompany caspase-3-activated apoptotic signaling pathways whether these are generated through the intrinsic or extrinsic pathways. Inh3 is a potent PP1 inhibitor and controls a significant fraction of the cellular pool of PP1γ1 and PP1α. In addition, it also controls the subcellular localization of PP1γ1 to the nucleolus. Degradation of Inh3 leads to release of active PP1 catalytic subunits, which are no longer restrained to their normal subcellular compartments. As a consequence, unregulated dephosphorylation of PP1 substrates by the active PP1 catalytic subunits takes place, leading to disruption of cellular processes. The circled plus sign is used here to indicate a proapoptotic effect. Among the PP1 substrates are a subset of those that are involved in apoptotic signaling pathways. Their unregulated dephosphorylation might also affect apoptotic signaling. On the right side of the diagram is a simplified outline of the intrinsic pathway to indicate the interactions of the Bcl-2 and BH3 family of proteins, of which several, including Bcl-2 and BAD, have been reported to be regulated by protein phosphatase-1 activity. Unregulated dephosphorylation of these proteins could produce a proapoptotic signaling effect. This would provide a potential feedback mechanism that is shown by the dotted arrow.
In the right-hand column, the prosurvival Bcl-2 protein family and the proapoptotic BH3 proteins with which they interact are shown (Fig. 8). Several members of this group of proteins, including BAD, have been reported to be substrates of PP1α, and their dephosphorylation elicits a proapoptotic signal (31–33). Dephosphorylation of BAD, for example, by deinhibition of PP1 as a consequence of Inh3 degradation, could provide a potential route for cross-talk between the extrinsic and intrinsic pathways as well as a potential route for a feedback amplification of caspase-3 activation.
In summary, the studies reported here provide evidence that Inh3 is a physiological substrate for caspase-3 and is degraded during apoptosis. Moreover, Inh3 degradation itself may contribute to the process of apoptosis. Inh3 is associated with a significant fraction of PP1γ1 and PP1α, suggesting that it may play a significant role in the overall cellular economy or distribution of PP1 subunits. These novel findings raise a number of questions regarding the cellular functions of Inh3 that may reward further investigation.
Acknowledgments
We thank Dr. Marietta Lee for critical discussions of this work.
This work was supported, in whole or in part, by National Institutes of Health Grant 18512. This work was also supported by grants from Philip Morris USA Inc. and Philip Morris International and by a gift from Col. Melvin Freeman toward the acquisition of the fluorescence microscope used in these studies. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PP1, protein phosphatase-1 catalytic subunit; act-D, actinomycin D; BH3, Bcl-2 homology-3 domain; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ERK, extracellular signal-regulated kinase; PARP, poly(ADP ribose) polymerase; PBS, phosphate-buffered saline; TBS, Tris-buffered saline; Z, benzyloxycarbonyl; fmk, fluoromethyl ketone; OA, okadaic acid.
Z. Q. Qi and E. Y. C. Lee, unpublished observations.
L. Zhang and E. Y. C. Lee, manuscript in preparation.
A report published since the submission of this paper has demonstrated that Inh3 forms a ternary complex with PP1 and Sds22 in vitro and in COS1 cell lysates (74). The loss of Inh3 might be expected to result in release of a PP1-Sds22 complex (rather than free PP1). However, it may be noted that although the existence of this ternary complex has been demonstrated, a rigorous evaluation of the fraction of the cellular pool of Inh3 that is associated with a ternary Inh3-PP1-Sds22 complex remains to be performed.
References
- 1.Ceulemans, H., and Bollen, M. (2004) Physiol. Rev. 84 1-39 [DOI] [PubMed] [Google Scholar]
- 2.Cohen, P. (1989) Annu. Rev. Biochem. 58 453-508 [DOI] [PubMed] [Google Scholar]
- 3.Lee, E. Y., Zhang, L., Zhao, S., Wei, Q., Zhang, J., Qi, Z. Q., and Belmonte, E. R. (1999) Front. Biosci. 4 D270-D285 [DOI] [PubMed] [Google Scholar]
- 4.Shima, H., Hatano, Y., Chun, Y. S., Sugimura, T., Zhang, Z., Lee, E. Y., and Nagao, M. (1993) Biochem. Biophys. Res. Commun. 192 1289-1296 [DOI] [PubMed] [Google Scholar]
- 5.Egloff, M. P., Johnson, D. F., Moorhead, G., Cohen, P. T., Cohen, P., and Barford, D. (1997) EMBO J. 16 1876-1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao, S., and Lee, E. Y. (1997) J. Biol. Chem. 272 28368-28372 [DOI] [PubMed] [Google Scholar]
- 7.Barford, D., Das, A. K., and Egloff, M. P. (1998) Annu. Rev. Biophys. Biomol. Struct. 27 133-164 [DOI] [PubMed] [Google Scholar]
- 8.Egloff, M. P., Cohen, P. T., Reinemer, P., and Barford, D. (1995) J. Mol. Biol. 254 942-959 [DOI] [PubMed] [Google Scholar]
- 9.Tu, J., Song, W., and Carlson, M. (1996) Mol. Cell. Biol. 16 4199-4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alms, G. R., Sanz, P., Carlson, M., and Haystead, T. A. (1999) EMBO J. 18 4157-4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandt, H., Lee, E. Y., and Killilea, S. D. (1975) Biochem. Biophys. Res. Commun. 63 950-956 [DOI] [PubMed] [Google Scholar]
- 12.Li, M., Satinover, D. L., and Brautigan, D. L. (2007) Biochemistry 46 2380-2389 [DOI] [PubMed] [Google Scholar]
- 13.Mi, J., Guo, C., Brautigan, D. L., and Larner, J. M. (2007) Cancer Res. 67 1082-1089 [DOI] [PubMed] [Google Scholar]
- 14.Satinover, D. L., Brautigan, D. L., and Stukenberg, P. T. (2006) Cell Cycle 5 2268-2274 [DOI] [PubMed] [Google Scholar]
- 15.Shenolikar, S. (1994) Annu. Rev. Cell Biol. 10 55-86 [DOI] [PubMed] [Google Scholar]
- 16.Connor, J. H., Quan, H., Oliver, C., and Shenolikar, S. (1998) Methods Mol. Biol. 93 41-58 [DOI] [PubMed] [Google Scholar]
- 17.Svenningsson, P., Nishi, A., Fisone, G., Girault, J. A., Nairn, A. C., and Greengard, P. (2004) Annu. Rev. Pharmacol. Toxicol. 44 269-296 [DOI] [PubMed] [Google Scholar]
- 18.Zhang, J., Zhang, L., Zhao, S., and Lee, E. Y. (1998) Biochemistry 37 16728-16734 [DOI] [PubMed] [Google Scholar]
- 19.Huang, H. S., Pozarowski, P., Gao, Y., Darzynkiewicz, Z., and Lee, E. Y. (2005) Arch. Biochem. Biophys. 443 33-44 [DOI] [PubMed] [Google Scholar]
- 20.Riedl, S. J., and Salvesen, G. S. (2007) Nat. Rev. Mol. Cell. Biol. 8 405-413 [DOI] [PubMed] [Google Scholar]
- 21.Boatright, K. M., and Salvesen, G. S. (2003) Curr. Opin. Cell Biol. 15 725-731 [DOI] [PubMed] [Google Scholar]
- 22.Adams, J. M. (2003) Genes Dev. 17 2481-2495 [DOI] [PubMed] [Google Scholar]
- 23.Bao, Q., and Shi, Y. (2007) Cell Death Differ. 14 56-65 [DOI] [PubMed] [Google Scholar]
- 24.Earnshaw, W. C., Martins, L. M., and Kaufmann, S. H. (1999) Annu. Rev. Biochem. 68 383-424 [DOI] [PubMed] [Google Scholar]
- 25.Fischer, U., Janicke, R. U., and Schulze-Osthoff, K. (2003) Cell Death Differ. 10 76-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cory, S., Huang, D. C., and Adams, J. M. (2003) Oncogene 22 8590-8607 [DOI] [PubMed] [Google Scholar]
- 27.Galonek, H. L., and Hardwick, J. M. (2006) Nat. Cell Biol. 8 1317-1319 [DOI] [PubMed] [Google Scholar]
- 28.Adams, J. M., and Cory, S. (2007) Oncogene 26 1324-1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puthalakath, H., and Strasser, A. (2002) Cell Death Differ. 9 505-512 [DOI] [PubMed] [Google Scholar]
- 30.Deng, X., Gao, F., Flagg, T., and May, W. S., Jr. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 153-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zha, J., Harada, H., Yang, E., Jockel, J., and Korsmeyer, S. J. (1996) Cell 87 619-628 [DOI] [PubMed] [Google Scholar]
- 32.Yang, E., Zha, J., Jockel, J., Boise, L. H., Thompson, C. B., and Korsmeyer, S. J. (1995) Cell 80 285-291 [DOI] [PubMed] [Google Scholar]
- 33.Datta, S. R., Katsov, A., Hu, L., Petros, A., Fesik, S. W., Yaffe, M. B., and Greenberg, M. E. (2000) Mol. Cell 6 41-51 [PubMed] [Google Scholar]
- 34.Willis, S. N., Fletcher, J. I., Kaufmann, T., van Delft, M. F., Chen, L., Czabotar, P. E., Ierino, H., Lee, E. F., Fairlie, W. D., Bouillet, P., Strasser, A., Kluck, R. M., Adams, J. M., and Huang, D. C. (2007) Science 315 856-859 [DOI] [PubMed] [Google Scholar]
- 35.Ayllon, V., Martinez, A. C., Garcia, A., Cayla, X., and Rebollo, A. (2000) EMBO J. 19 2237-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayllon, V., Cayla, X., Garcia, A., Fleischer, A., and Rebollo, A. (2002) Eur. J. Immunol. 32 1847-1855 [DOI] [PubMed] [Google Scholar]
- 37.Ayllon, V., Cayla, X., Garcia, A., Roncal, F., Fernandez, R., Albar, J. P., Martinez, C., and Rebollo, A. (2001) J. Immunol. 166 7345-7352 [DOI] [PubMed] [Google Scholar]
- 38.Chiang, C. W., Harris, G., Ellig, C., Masters, S. C., Subramanian, R., Shenolikar, S., Wadzinski, B. E., and Yang, E. (2001) Blood 97 1289-1297 [DOI] [PubMed] [Google Scholar]
- 39.Wang, H. G., Pathan, N., Ethell, I. M., Krajewski, S., Yamaguchi, Y., Shibasaki, F., McKeon, F., Bobo, T., Franke, T. F., and Reed, J. C. (1999) Science 284 339-343 [DOI] [PubMed] [Google Scholar]
- 40.Tamura, S., Toriumi, S., Saito, J., Awano, K., Kudo, T. A., and Kobayashi, T. (2006) Cancer Sci. 97 563-567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dessauge, F., Cayla, X., Albar, J. P., Fleischer, A., Ghadiri, A., Duhamel, M., and Rebollo, A. (2006) J. Immunol. 177 2441-2451 [DOI] [PubMed] [Google Scholar]
- 42.Wang, R. H., Liu, C. W., Avramis, V. I., and Berndt, N. (2001) Oncogene 20 6111-6122 [DOI] [PubMed] [Google Scholar]
- 43.Berndt, N., Dohadwala, M., and Liu, C. W. (1997) Curr. Biol. 7 375-386 [DOI] [PubMed] [Google Scholar]
- 44.Zhang, A. J., Bai, G., Deans-Zirattu, S., Browner, M. F., and Lee, E. Y. (1992) J. Biol. Chem. 267 1484-1490 [PubMed] [Google Scholar]
- 45.Wen, L. P., Madani, K., Martin, G. A., and Rosen, G. D. (1998) Cell Death Differ. 5 729-734 [DOI] [PubMed] [Google Scholar]
- 46.Deak, J. C., Cross, J. V., Lewis, M., Qian, Y., Parrott, L. A., Distelhorst, C. W., and Templeton, D. J. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5595-5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsiftsoglou, A. S., Pappas, I. S., and Vizirianakis, I. S. (2003) Pharmacol. Ther. 100 257-290 [DOI] [PubMed] [Google Scholar]
- 48.Martin, S. J., Lennon, S. V., Bonham, A. M., and Cotter, T. G. (1990) J. Immunol. 145 1859-1867 [PubMed] [Google Scholar]
- 49.Kastan, M. B., and Bartek, J. (2004) Nature 432 316-323 [DOI] [PubMed] [Google Scholar]
- 50.Bartek, J., Lukas, C., and Lukas, J. (2004) Nat. Rev. Mol. Cell. Biol. 5 792-804 [DOI] [PubMed] [Google Scholar]
- 51.Melchionna, R., Chen, X. B., Blasina, A., and McGowan, C. H. (2000) Nat. Cell Biol. 2 762-765 [DOI] [PubMed] [Google Scholar]
- 52.Wyllie, A. H. (1980) Nature 284 555-556 [DOI] [PubMed] [Google Scholar]
- 53.Hong, S. J., Dawson, T. M., and Dawson, V. L. (2004) Trends Pharmacol. Sci. 25 259-264 [DOI] [PubMed] [Google Scholar]
- 54.Nicholson, D. W., Ali, A., Thornberry, N. A., Vaillancourt, J. P., Ding, C. K., Gallant, M., Gareau, Y., Griffin, P. R., Labelle, M., Lazebnik, Y. A., Munday, N. A., Raju, S. M., Smulson, M. E., Yamin, T. T., Yu, V. L., and Miller, D. K. (1995) Nature 376 37-43 [DOI] [PubMed] [Google Scholar]
- 55.Martin, S. J., Bonham, A. M., and Cotter, T. G. (1990) Biochem. Soc. Trans. 18 634-636 [DOI] [PubMed] [Google Scholar]
- 56.Roos, W. P., and Kaina, B. (2006) Trends Mol. Med. 12 440-450 [DOI] [PubMed] [Google Scholar]
- 57.Li, T. K., and Liu, L. F. (2001) Annu. Rev. Pharmacol. Toxicol. 41 53-77 [DOI] [PubMed] [Google Scholar]
- 58.Lu, Q., and Mellgren, R. L. (1996) Arch. Biochem. Biophys. 334 175-181 [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Calvo, M., Peterson, E. P., Leiting, B., Ruel, R., Nicholson, D. W., and Thornberry, N. A. (1998) J. Biol. Chem. 273 32608-32613 [DOI] [PubMed] [Google Scholar]
- 60.Janicke, R. U., Sprengart, M. L., Wati, M. R., and Porter, A. G. (1998) J. Biol. Chem. 273 9357-9360 [DOI] [PubMed] [Google Scholar]
- 61.Ferguson, H. A., Marietta, P. M., and Van Den Berg, C. L. (2003) J. Biol. Chem. 278 45793-45800 [DOI] [PubMed] [Google Scholar]
- 62.Germain, M., Affar, E. B., D'Amours, D., Dixit, V. M., Salvesen, G. S., and Poirier, G. G. (1999) J. Biol. Chem. 274 28379-28384 [DOI] [PubMed] [Google Scholar]
- 63.Trinkle-Mulcahy, L., Sleeman, J. E., and Lamond, A. I. (2001) J. Cell Sci. 114 4219-4228 [DOI] [PubMed] [Google Scholar]
- 64.Martelli, A. M., Robuffo, I., Bortul, R., Ochs, R. L., Luchetti, F., Cocco, L., Zweyer, M., Bareggi, R., and Falcieri, E. (2000) J. Cell. Biochem. 78 264-277 [PubMed] [Google Scholar]
- 65.Martelli, A. M., Zweyer, M., Ochs, R. L., Tazzari, P. L., Tabellini, G., Narducci, P., and Bortul, R. (2001) J. Cell. Biochem. 82 634-646 [DOI] [PubMed] [Google Scholar]
- 66.Garcia, A., Cayla, X., Guergnon, J., Dessauge, F., Hospital, V., Rebollo, M. P., Fleischer, A., and Rebollo, A. (2003) Biochimie (Paris) 85 721-726 [DOI] [PubMed] [Google Scholar]
- 67.Cohen, P., Holmes, C. F., and Tsukitani, Y. (1990) Trends Biochem. Sci. 15 98-102 [DOI] [PubMed] [Google Scholar]
- 68.Sontag, E., Nunbhakdi-Craig, V., Sontag, J. M., Diaz-Arrastia, R., Ogris, E., Dayal, S., Lentz, S. R., Arning, E., and Bottiglieri, T. (2007) J. Neurosci. 27 2751-2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kamada, S., Kikkawa, U., Tsujimoto, Y., and Hunter, T. (2005) J. Biol. Chem. 280 857-860 [DOI] [PubMed] [Google Scholar]
- 70.Luthi, A. U., and Martin, S. J. (2007) Cell Death Differ. 14 641-650 [DOI] [PubMed] [Google Scholar]
- 71.Desdouits, F., Cheetham, J. J., Huang, H. B., Kwon, Y. G., da Cruz e Silva, E. F., Denefle, P., Ehrlich, M. E., Nairn, A. C., Greengard, P., and Girault, J. A. (1995) Biochem. Biophys. Res. Commun. 206 652-658 [DOI] [PubMed] [Google Scholar]
- 72.Boisvert, F. M., van Koningsbruggen, S., Navascues, J., and Lamond, A. I. (2007) Nat. Rev. Mol. Cell. Biol. 8 574-585 [DOI] [PubMed] [Google Scholar]
- 73.Tembe, V., and Henderson, B. R. (2007) Cell. Signal. 19 1113-1120 [DOI] [PubMed] [Google Scholar]
- 74.Lesage, B., Beullens, M., Pedelini, L., Garcia-Gimeno, M. A., Waelkens, E., Sanz, P., and Bollen, M. (2007) Biochemistry 46 8909-8919 [DOI] [PubMed] [Google Scholar]