Abstract
Human epidermal growth factor receptor-2 (HER-2/ErbB2/neu), a receptor tyrosine kinase that is amplified/overexpressed in poor prognosis breast carcinomas, confers resistance to apoptosis by activating cell survival pathways. Here we demonstrate that the cytoplasmic tail of HER-2 is cleaved by caspases at Asp1016/Asp1019 to release a ∼47-kDa product, which is subsequently proteolyzed by caspases at Asp1125 into an unstable 22-kDa fragment that is degraded by the proteasome and a predicted 25-kDa product. Both the 47- and 25-kDa products translocate to mitochondria, release cytochrome c by a Bcl-xL-suppressible mechanism, and induce caspase-dependent apoptosis. The 47- and 25-kDa HER-2 cleavage products share a functional BH3-like domain, which is required for cytochrome c release in cells and isolated mitochondria and for apoptosis induction. Caspase-cleaved HER-2 binds Bcl-xL and acts synergistically with truncated Bid to induce apoptosis, mimicking the actions of the BH3-only protein Bad. Moreover, the HER-2 cleavage products cooperate with Noxa to induce apoptosis in cells expressing both Bcl-xL and Mcl-1, confirming their Bad-like function. Collectively, our results indicate that caspases activate a previously unrecognized proapoptotic function of HER-2 by releasing a Bad-like cell death effector.
Caspases are a highly conserved family of proteases that function as essential effectors of apoptosis by cleaving proteins at Asp residues to induce cell death (1). Caspases are synthesized as inactive zymogens that are activated by two principal pathways: the extrinsic pathway, initiated by cytokines of the tumor necrosis factor (TNF)2-α family, including TNF-α and TNF-related apoptosis-inducing ligand (TRAIL), and the intrinsic pathway, initiated by genotoxic stress. In both pathways, mitochondria play a critical role in amplifying caspase activation. The initiator caspase-2, -8, and -10 induce mitochondrial outer membrane permeabilization (MOMP), which results in the release of cytochrome c and other proapoptotic molecules from the mitochondrial intermembrane space (2-4). Cytosolic cytochrome c induces the Apaf-1-dependent oligomerization and activation of procaspase-9 in the apoptosome (5). Caspase-9 and other initiator caspases activate the executioner caspases, caspase-3, -6, and -7, which catalyze many of the hallmark events of apoptosis (1).
Caspases induce MOMP at least in part by cleaving Bid, a proapoptotic Bcl-2 family member that contains a single Bcl-2 homology domain (BH3-only) and is normally localized in the cytoplasm (2, 4). Caspase cleavage of Bid exposes an amino-terminal Gly residue in the BH3 domain-containing product (truncated Bid (tBid)), which is N-myristoylated, thereby targeting tBID to cardiolipin-rich mitochondrial membranes (6, 7). Unlike tBid, other BH3-only family members (e.g. Bim, Puma, Bad, and Noxa) are activated by transcriptional or non-proteolytic post-transcriptional mechanisms in response to apoptotic stimuli (8). The α-helical BH3 domain in these cell death effectors, which is essential for MOMP and apoptosis induction, inserts into a hydrophobic pocket formed by the BH1, BH2, and BH3 domains of multidomain antiapoptotic Bcl-2 family members (e.g. Bcl-2, Bcl-xL, and Mcl-1), thereby inhibiting their cytoprotective function (9, 10). Recent studies indicate that BH3-only proteins bind and inactivate distinct subsets of antiapoptotic Bcl-2 proteins (11-13). Bim, Bid, and Puma bind to a broad range of these proteins, including Bcl-2, Bcl-xL, and Mcl-1, whereas Bad and Noxa interact with different subsets, Bad with Bcl-2 and Bcl-xL and Noxa with Mcl-1. The multidomain proapoptotic Bcl-2 family members Bax and Bak are essential downstream targets of BH3-only proteins; MEFs deficient in both Bax and Bak are resistant to MOMP and apoptosis induction by tBid and by agents that activate the intrinsic apoptotic pathway (14). In addition to neutralizing antiapoptotic Bcl-2 family members, “activator” BH3-only proteins (Bid, Bim, and possibly Puma) have been reported to directly bind and activate Bax/Bak by inducing its oligomerization, thereby triggering cytochrome c release through a putative mitochondrial pore (11, 13, 15-17). However, other studies have contested this direct activation model (12, 18, 19), suggesting that the mechanisms by which BH3-only proteins activate Bax/Bak have yet to be fully resolved.
Here we report that caspases cleave human epidermal growth factor receptor-2 (HER-2/ErbB2/neu; hereafter referred to as HER-2) and release a BH3-like domain-containing cleavage product that translocates to mitochondria and induces MOMP and apoptosis. HER-2 is a ligandless receptor-tyrosine kinase of the HER/EGFR family that plays a key role in regulating proliferation, migration, adhesion, differentiation, and apoptosis (20). HER-2 transduces signals by autophosphorylating several conserved Tyr residues in its carboxyl-terminal tail that serve as docking sites for signaling molecules. HER-2 is aberrantly activated by gene amplification and/or overexpression in ∼25% of human breast carcinomas, and its expression is associated with poor survival and resistance to chemotherapy (21, 22). HER-2 inhibits apoptosis induced by chemotherapy, TNF-α, and TRAIL, at least in part, by activating the phosphoinositol-3 kinase/Akt survival pathway (23-25).
In the present work, we demonstrate that HER-2 has a novel and previously unrecognized proapoptotic function that is activated by caspases. Although prior studies indicated that HER-2 is cleaved by caspases (26, 27), the detailed mechanisms and functional consequences of HER-2 proteolysis have not been delineated. We now show that the cytoplasmic tail of HER-2 is cleaved by caspases to release two proapoptotic products (47 and 25 kDa), which translocate to mitochondria, release cytochrome c, and induce apoptosis. Importantly, the 47- and 25-kDa HER-2 products share a BH3-like domain, which is required for cytochrome c release in cells and isolated mitochondria and for apoptosis induction. Taken together, our findings reveal a dual role for HER-2 in regulating apoptosis and suggest that HER-2 is a novel BH3-only protein activated by caspases.
EXPERIMENTAL PROCEDURES
Cell Lines—Human MDA-MB-231 and MDA-MB-468 breast carcinoma cells were obtained from ATCC and maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS and penicillin/streptomycin (Invitrogen). Human SKBR3 breast carcinoma cells and Jurkat cells (ATCC) were maintained in RPMI 1640 supplemented with 2 mm l-glutamine, 10% FBS, and penicillin/streptomycin. Human MDA-MB-453 cells (ATCC) were grown in Dulbecco's modified Eagle's medium/F-12 with 10% FBS and penicillin/streptomycin. SV40-immortalized WT and Bax-/-/Bak-/- double knock-out (DKO) MEFs (14, 28) were kindly provided by Dr. Craig Thompson and maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and penicillin/streptomycin. MCF-7 breast cancer cells stably expressing caspase-3 were provided by Dr. Christopher Froelich (29) and grown in modified Eagle's medium supplemented with nonessential amino acids, 10 mm HEPES, 10% fetal bovine serum, and penicillin/streptomycin (Invitrogen).
HER-2 Plasmid Constructs and Mutagenesis—Primer sequences for PCR cloning and mutagenesis are listed in the supplemental materials. All constructs were verified by sequencing. WT full-length human HER-2 cDNA, kindly provided by Dr. Gibbes Johnson (30), was subcloned into the XhoI sites of the retroviral vector pLXSN (Clontech). To create a cDNA encoding caspase truncated HER-2 (amino acids 1-1016), two internal EcoRI sites were destroyed by mutating a single nucleotide at positions 1450 and 3072, without altering the coding sequence, using the QuikChange site-directed mutagenesis kit (Stratagene). This cDNA was PCR-amplified with primers containing EcoRI and XhoI restriction sites and subcloned into pLXSN. cDNAs encoding the 25-kDa (amino acids 1017-1125), 22-kDa (amino acids 1126-1254), and 47-kDa HER-2 products (amino acids 1017-1254) were PCR-amplified from WT HER-2 cDNA using primers containing EcoRI and XhoI restriction sites and subcloned into pcDNA3NFLAG (31) or the bicistronic retroviral vector pBMN-IRES-GFP (25- and 47-kDa cDNAs), a gift from Dr. Hiroaki Kiyokawa. pBABE-EGFP-tBid (32) was kindly provided by Dr. Navdeep Chandel, and pCMV5a-tBID was a gift from Dr. Honglin Li (2). pMIG-HA Bad and pMIG-HA Noxa were generously provided by Dr. David C. S. Huang (12).
Each of four Asp residues at candidate caspase cleavage sites (Asp1016, Asp1019, Asp1087, and Asp1125) in the HER-2 cytoplasmic tail was individually altered to a Glu residue using the QuikChange site-directed mutagenesis kit (Stratagene). Double, triple, and quadruple (4×) mutations were generated by sequential site-directed mutagenesis. This same method was used to create each of the HER-2 BH3 domain mutants (L1120E and D1125E) and the double mutant (2×E).
Small Pool Expression Cloning and Caspase Cleavage of HER-2 in Vitro—A human prostate adenocarcinoma cDNA library (Invitrogen) was screened for cDNAs encoding caspase substrates by small pool expression cloning as described (33-37). cDNA 158-5E, which encodes amino acids 762-1254 of human HER-2 (GenBank™ accession number NM-004448.2), was isolated using this approach. WT or 4× mutant HER-2 158-5E cDNAs were transcribed and translated in vitro in the presence of [35S]methionine using the TNT SP6 coupled transcription/translation system (Promega). 35S-Labeled HER-2 proteins were incubated with buffer or 2.5 or 25 ng of recombinant caspases (Biomol) for 1 h at 37 °C and analyzed as described (34).
Caspase Proteolysis of HER-2 in Vivo—SKBR3 or MDA-MB-453 cells were treated with 2 μg/ml TRAIL and 1 μg/ml cycloheximide or 200 μm etoposide (Calbiochem) for 0-16 h, with or without preincubating cells for 1 h with 50 μm Z-VAD-fmk (ICN Pharmaceuticals) and/or 100 nm epoxomicin (Calbiochem). Cells were lysed in radioimmune precipitation buffer (50 mm Tris, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, and 1 mm EDTA) supplemented with 1 mm phenylmethylsulfonyl fluoride and 1× Protease Inhibitor Mixture I (Sigma). Lysates were examined by immunoblotting using a HER-2 mAb, which recognizes a carboxyl-terminal epitope (amino acids 1242-1254) (BD Biosciences). In parallel experiments, SKBR3 cells were treated as above, and apoptotic nuclei were scored as described under “Apoptosis Assays.”
Creation of MDA-MB-231 Stable Pools by Retroviral Infection—Phoenix cells (ATCC) were transfected with 10 μg of pLXSN vector or pLXSN plasmid encoding WT HER-2, 4× HER-2, or truncated HER-2, or with 10 μg of pBABE vector or pBABE plasmid encoding Bcl-xL, and retroviruses were generated as described (38). MDA-MB-231 breast cancer cells were overlaid with retroviral supernatant for 24 h, and pools were selected by growth in 800 μg/ml G418 for 14 days (pLXSN constructs) or 1 μg/ml puromycin for 10 days (pBABE constructs). Stable expression was confirmed by immunoblotting.
Analysis of Cell Surface Expression of HER-2 Constructs—1 × 106 MDA-MB-231 cells stably expressing HER-2 constructs were incubated with 1 μg of HER-2 Ab2 (extracellular domain epitope; Lab Vision) for 45 min at room temperature. Cells were washed, incubated with fluorescein isothiocyanate-tagged goat anti-mouse Ab (1:50; ICN Pharmaceuticals) for 30 min at room temperature, washed, and fixed in 0.5% paraformaldehyde for 10 min at room temperature. Cells were then analyzed by flow cytometry (Beckman Epics XL).
Apoptosis Assays—MDA-MB-231 pools were treated for 16 h with 0-5 μg/ml recombinant TRAIL (amino acids 95-281), which was produced as described (39). To score apoptosis, cells were fixed with 1.25% glutaraldehyde for 15 min at room temperature, and nuclei were stained with 10 μg/ml bisbenzimide (Hoescht 33258; Sigma) for 10 min at room temperature. The percentage of cells with apoptotic (condensed or fragmented) nuclei was determined by fluorescence microscopy (E400 Nikon Eclipse). At least 200 cells were scored per treatment condition, and all experiments were performed at least three times.
Apoptosis Induction by Ectopically Expressed HER-2 Constructs—To determine whether one or more of the intracellular HER-2 caspase cleavage products induced apoptosis, MDA-MB-231 or MDA-MB-468 cells were transiently co-transfected with 0.2 μg of pEGFP-N1 (Clontech) and 1 μg of pcDNA3NFLAG vector or pcDNA3NFLAG encoding the 47-, 25-, or 22-kDa HER-2 products (or pCMV5a-tBid) using Lipofectamine/PLUS reagent (Invitrogen). Transfections were performed with or without preincubating cells for 1 h with 100 μm Z-VAD-fmk. Twenty-four h later, GFP-positive cells were scored for apoptotic nuclei. To determine the mechanism(s) by which the 47- and 25-kDa products induce apoptosis, MDA-MB-231 cells were transiently co-transfected with 0.2 μg of pEGFP-N1, 0.5 μg of pcDNA3NFLAG encoding the 47- or 25-kDa HER-2, and 0.5 μg of empty vector or plasmid encoding FADD DN, CrmA, p35, or Bcl-2. To determine the Bax/Bak dependence of apoptosis induction, WT or Bax-/-/Bak-/- DKO MEFs were infected with retroviruses co-expressing GFP and vector, 47- or 25-kDa HER-2, or expressing GFP-tagged tBid. Retroviruses were produced by transfecting Phoenix cells with bicistronic pBMN-IRES-GFP retroviral constructs (HER-2 cDNAs) or pBABE-EGFP-tBid (32) as described (38). GFP-positive MEFs were scored for apoptotic nuclei 48 h after infection. To determine whether the 25-kDa HER-2 product acts synergistically with truncated Bid to induce apoptosis, MDA-MB-231 cells stably expressing Bcl-xL were transiently co-transfected with 1 μg of pCMV-tBid and 1 μg of pMIG vector, pMIG-HA-Noxa, pMIG-HA-Bad, or pBMN-25-kDa HER-2 (bicistronic vectors co-expressing GFP) using Lipofectamine/Plus reagent. The percentage of GFP-positive cells with apoptotic nuclei was scored 24 h later. For the Bad and Noxa cooperation experiments, MCF-7 cells stably expressing caspase-3 (29) were transiently co-transfected with 1 μg of pcDNA3NFLAG vector, pcDNA3NFLAG-47-kDa HER-2, pcDNA3NFLAG-25-kDa HER-2, pcDNA3.1-HA-Noxa, and pcDNA3.1-HA-Bad in pairwise combinations (2 μg total) and 0.2 μg of pEGFP-N1 using Lipofectamine/Plus reagent. The percentage of GFP-positive cells with apoptotic nuclei was scored 24 h later.
Colocalization of Ectopically Expressed HER-2 Cleavage Products with Mitochondria—MDA-MB-231 cells on glass coverslips were transiently transfected with 1 μg of pcDNA3NFLAG vector or pcDNA3NFLAG encoding the 47- or 25-kDa HER-2 products (WT or BH3 mutants). Twenty-four h later, cells were incubated with 100 nm Mitotracker Deep Red 633 (Molecular Probes) for 30 min at 37 °C. Cells were then washed, allowed to recover for an additional 30 min, fixed in 4% paraformaldehyde for 10 min at room temperature, and permeabilized with 0.1% Triton X-100 for 5 min at room temperature. After washing, cells were blocked in blocking buffer (10% goat serum and 1% bovine serum albumin in phosphate-buffered saline) for 1 h at room temperature and incubated with 10 μg/ml preabsorbed fluorescein isothiocyanate-conjugated M2 FLAG mAb (Sigma) for 1 h at 37 °C, followed by the addition of 0.5 ng/ml 4′,6-diamidino-2-phenylindole (Sigma) for 15 min at room temperature. Coverslips were mounted using Prolong Anti-fade Gold (Molecular Probes). Images were acquired with a Zeiss LSM510 UV META confocal microscope and analyzed with LSM5 software. Colocalization of fluorescein isothiocyanate-labeled HER-2 proteins with Mitotracker and mitochondrial DNA was scored in at least 200 GFP-positive cells per experiment in three independent experiments.
Mitochondrial Cytochrome c Release by Ectopically Expressed HER-2 Cleavage Products—Cytochrome c release was analyzed as described (40) with modifications. MDA-MB-231 cells stably expressing Bcl-xL or pBABE vector were plated on glass coverslips, preincubated with 100 μm Z-VAD-fmk for 1 h, and then co-transfected with 0.2 μg of pEGFP-N1 and 1 μg of pcDNA3NFLAG encoding the 47- or 25-kDa HER-2 product (or 0.8 μg of pCMV5a-tBid). After transfection, cells were incubated for 24 h in the presence of 100 μm Z-VAD-fmk, washed, and then fixed in 4% paraformaldehyde for 10 min at room temperature and permeabilized with 0.1% Triton X-100 for 5 min at room temperature. After washing, cells were blocked in blocking buffer for 1 h at room temperature and then incubated with a native conformation cytochrome c Ab (1:250; BD Biosciences) for 30 min at 37 °C, washed, and incubated with TRITC donkey anti-mouse Ab (1:50; Jackson ImmunoResearch Laboratories) for 30 min at 37 °C. Images were acquired by confocal microscopy and analyzed as described above. In contrast to the punctate pattern of mitochondrial cytochrome c staining, cells with release of cytochrome c from mitochondria exhibit diffuse or nearly absent cytochrome c staining (40, 41). The percentage of GFP-positive cells (at least 200 cells/experiment) with cytochrome c release was scored in five independent experiments. In addition, cytochrome c release by ectopically expressed HER-2 products was determined by immunoblotting of subcellular fractions as described below, using a cytochrome c Ab (catalogue number 556433; BD Biosciences).
Mitochondrial Localization of the Endogenous 47-kDa HER-2 Cleavage Product—2.2 × 105 SKBR3 cells were treated with 2.5 μg/ml TRAIL and 1 μg/ml cycloheximide for 0-4 h. After washing, cells were pelleted by centrifugation at 120 × g and resuspended in Buffer A (20 mm HEPES, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 4 mm dithiothreitol, and 1× Protease Inhibitor Mixture I (Sigma)) for 1 h at 4 °C. Lysates were centrifuged at 16,000 × g to separate supernatant and heavy membrane fractions. The heavy membrane pellet was incubated with radioimmune precipitation buffer for 30 min at 4 °C and then centrifuged at 16,000 × g for 10 min to remove cellular debris (pellet). The protein concentrations of each fraction were measured with the BCA protein assay kit (Pierce). 20 μg of supernatant (cytosolic) or heavy membrane lysate (mitochondrial) was analyzed by immunoblotting with a carboxyl-terminal HER-2 mAb (BD Biosciences), Cox IV Ab (mitochondrial; Molecular Probes), or α-tubulin Ab (cytosolic; Sigma).
Immunoprecipitations—3 × 105 MDA-MB-231 cells stably overexpressing Bcl-xL were transiently transfected with 2 μg of pcDNA3NFLAG vector or pcDNA3NFLAG-25-kDa HER-2. After overnight incubation, cells were lysed in ice cold radioimmune precipitation buffer for 30 min. Lysates (600 μg) were incubated overnight with rabbit Bcl-xL Ab (1:50; Cell Signaling) prior to incubation with protein A-agarose beads (Pierce) for 1 h at 4 °C. Lysates were then boiled in 2× Laemmli buffer (20% glycerol, 2% SDS, 250 mm Tris, pH 6.8, 10% β-mercaptoethanol, and 0.1% bromphenol blue), separated by SDS-PAGE, and analyzed by immunoblotting using a murine FLAG mAb (M2; Sigma) or Bcl-x mAb (BIOSOURCE).
Isolation of Mitochondria and Cytochrome c Release Assays—Mitochondria were isolated as described (42) with modifications. 2 × 107 Jurkat cells were pelleted at 120 × g and resuspended in ice-cold HIM buffer (200 mm mannitol, 70 mm sucrose, 1 mm EGTA, and 10 mm HEPES, pH 7.5) supplemented with 1 mm phenylmethylsulfonyl fluoride and 1× Protease Inhibitor Mixture I (Sigma). Cells were lysed with 20 strokes of a Dounce homogenizer. Nuclei and unlysed cells were pelleted at 120 × g for 10 min. The supernatant was transferred to a new tube and centrifuged at 7500 × g for 15 min. The mitochondrial pellet was washed in ice-cold HIM buffer and resuspended in MRM buffer (250 mm sucrose, 10 mm HEPES, 1 mm ATP, 5 mm sodium succinate, 0.8 mm ADP, and 2 mm K2HPO4).
High pressure liquid chromatography-purified peptides encoding the BH3 domains and flanking sequences in WT HER-2 (SEDPTVPLPSETDGYVAPLT), 2×E mutant HER-2 (SEDPTVPEPSETEGYVAPLT), and Bid (CIRNIARHLAQVGDSMDRSIPP) were purchased from Abgent. The conserved BH3 domain residues in WT HER-2 and Bid, and the 2 corresponding Glu substitutions in the 2×E mutant, are indicated in boldface type. Peptides were added to mitochondria in MRM buffer and incubated for 3 h at room temperature. Mitochondria were then pelleted at 16,000 × g for 15 min. The supernatant was stored at -80 °C until analyzed for cytochrome c concentration by enzyme-linked immunosorbent assay (R&D Systems).
Statistical Analyses—Statistical significance was determined by ANOVAs with post-tests or two-tailed unpaired t-tests as indicated in the figure legends using Prism 4 (GraphPad Software).
RESULTS
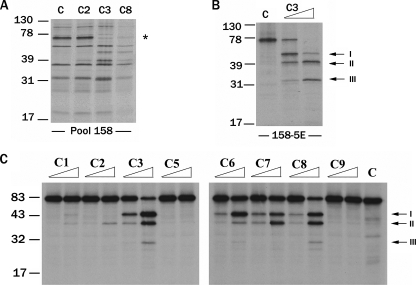
The Cytoplasmic Tail of HER-2 Is Cleaved by Multiple Caspases in Vitro—Using an expression cloning strategy we described previously (33-37), we screened a cDNA library for cDNAs encoding caspase substrates. Small cDNA pools were transcribed and translated in vitro in the presence of [35S]methionine, and the resulting 35S-labeled protein pools were incubated with buffer or recombinant caspases. Pool 158 contained a ∼76-kDa protein band (indicated by an asterisk), which was cleaved by caspase-3 and -8 (Fig. 1A). Subdivision and reanalysis of the corresponding cDNA pool led to the identification of a single cDNA (158-5E) encoding the ∼76-kDa protein, which was cleaved into multiple proteolytic products (labeled I, II, and III that are ∼47, 41, and 32 kDa, respectively) by caspase-3 (Fig. 1B). Sequence analysis revealed that 158-5E (hereafter referred to as 158-5E/WT HER-2) is a partial human HER-2 cDNA encoding amino acids 762-1254, which encompasses the majority of the kinase domain and multiple tyrosine residues known to play a critical role in signaling. 35S-Labeled 158-5E/WT HER-2 was preferentially cleaved by the initiator caspase-8 and executioner caspases-3, -6, and -7 into similar sized products (labeled I, II, and III) (Fig. 1C). These results suggest that the cytoplasmic tail of HER-2 is proteolyzed by multiple caspases at the same sites in vitro.
FIGURE 1.
The cytoplasmic tail of HER-2 is cleaved by multiple caspases in vitro. Small pool expression cloning was used to identify caspase substrates from a prostate adenocarcinoma cDNA library. A, cDNA pools were transcribed and translated in vitro in the presence of [35S]methionine and incubated with control (C) buffer or 25 ng of recombinant caspase-2 (C2), -3 (C3), or -8 (C8). Pool 158 contained a ∼76-kDa protein (indicated by an asterisk), which was cleaved by caspase-3 and -8. B, pool 158 was progressively subdivided and reanalyzed until a single cDNA (158-5E) encoding a ∼76-kDa protein, which was cleaved by caspase-3 into several fragments (indicated by arrows and labeled I-III), was isolated. 158-5E was identified as a partial human HER-2 cDNA encoding amino acids 762-1254. C, 35S-labeled 158-5E protein was incubated with 2.5 or 25 ng of caspase-1, -2, -3, -5, -6, -7, -8, or -9 or buffer (C) for 1 h at 37 °C, and the cleavage products (labeled I-III) were identified.
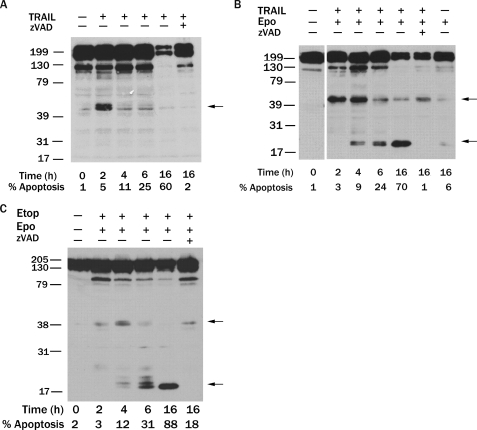
HER-2 Is Proteolyzed by Caspases in HER-2-overexpressing Breast Cancer Cells during the Induction of Apoptosis—To determine whether HER-2 is cleaved by caspases during apoptosis, we treated human SKBR3 breast cancer cells, which overexpress HER-2, with the proapoptotic cytokine TRAIL. Treatment of SKBR3 cells with TRAIL led to the rapid production (within 2 h) of a ∼47-kDa fragment, which was less abundant at later time points (Fig. 2A). Importantly, proteolysis of HER-2 into the 47-kDa product occurred when only 5% of SKBR3 cells were apoptotic, indicating that HER-2 is initially cleaved during the induction of apoptosis and might contribute to its execution. Furthermore, a reduction in the amount of full-length HER-2 was observed in response to TRAIL treatment, and this reduction was suppressed by the broad spectrum caspase inhibitor Z-VAD-fmk, indicating that caspases were responsible for the observed cleavage of HER-2 (Fig. 2A).
FIGURE 2.
HER-2 is proteolyzed by caspases in HER-2-overexpressing breast cancer cells during the induction of apoptosis. A, HER-2-overexpressing SKBR3 human breast cancer cells were treated with 2 μg/ml TRAIL and 1 μg/ml cycloheximide for 0-16 h with or without pretreatment with 50 μm Z-VAD-fmk for 1 h. HER-2 was detected by immunoblotting using a HER-2 mAb that recognizes the carboxyl terminus. B, SKBR3 cells were preincubated with 100 nm epoxomicin and 50 μm Z-VAD-fmk for 1 h as indicated and then treated with 2 μg/ml TRAIL and 1 μg/ml cycloheximide for 0-16 h. C, SKBR3 cells were preincubated with 100 nm epoxomicin and 50 μm Z-VAD-fmk for 1 h as indicated and then treated with 200 μm etoposide for 0-16 h. In A-C, the HER-2 cleavage products are indicated by arrows. Apoptotic nuclei were scored at each time point in parallel experiments, and the percentage of apoptotic cells is indicated.
Because the 47-kDa HER-2 cleavage product was only transiently observed in apoptotic SKBR3 cells, we postulated that it might be proteolyzed by caspases into an unstable product. To test this hypothesis, we preincubated SKBR3 cells with the proteasome inhibitor, epoxomicin (43), and then treated them with TRAIL. Strikingly, epoxomicin led to the stabilization of a previously undetected 22-kDa HER-2 fragment that was accompanied by a reduction in the amount of both the 47-kDa product and full-length HER-2 protein (Fig. 2B), suggesting that the 22-kDa fragment is generated by caspase cleavage of the 47-kDa product and then rapidly degraded by the proteasome. Of note, the unstable 22-kDa HER-2 cleavage product, but not the 47-kDa fragment, was reported recently (26). Although the production of the 47-kDa HER-2 fragment preceded that of the 22-kDa product, the 22-kDa product was generated when <10% of cells were apoptotic. Similar results were observed when SKBR3 cells were treated with the topoisomerase II inhibitors etoposide (Fig. 2C) or doxorubicin or the microtubule agent taxol (data not shown). In addition to the carboxyl-terminal 22-kDa product, caspase proteolysis of the 47-kDa HER-2 fragment produces a predicted 25-kDa amino-terminal product. This latter product was not visualized in cells due to lack of an antibody targeting this epitope, although a similar sized fragment was observed when 35S-labeled HER-2 cytoplasmic tail was cleaved by caspases in vitro (Fig. 1C). Caspase cleavage of HER-2 was also observed in MDA-MB-453 breast cancer cells treated with TRAIL (supplemental Fig. S1). These findings suggest that HER-2 is initially cleaved by caspases during the induction of apoptosis into a 47-kDa product, which is subsequently proteolyzed by caspases into an unstable 22-kDa product that is degraded by the proteasome and a predicted 25-kDa product.
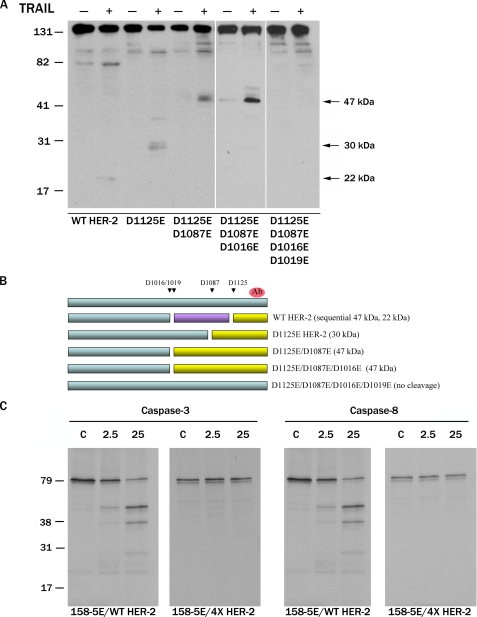
HER-2 Is Cleaved by Caspases at Four Sites in the Cytoplasmic Tail—To delineate the caspase cleavage sites in the cytoplasmic tail of HER-2, we mutated the Asp residues at four putative cleavage sites (DMGD1016, DLVD1019, DVFD1087, and SETD1125) to Glu residues (individually and in combination). Two of these sites (Asp1087 and Asp1125) in HER-2 were recently reported to be cleaved by caspases (26, 27). MDA-MB-231 human breast cancer cells, which express very low levels of HER-2, were transiently transfected with WT or mutant HER-2 cDNAs and treated with TRAIL, and HER-2 cleavage was assessed by immunoblotting. Consistent with our prior observations, WT HER-2 was proteolyzed into a faint 22-kDa product (Fig. 3A). The D1125E HER-2 mutant was cleaved into a ∼30-kDa product. Both the D1125E/D1087E double mutant HER-2 and D1125E/D1087E/D1016E triple mutant HER-2 were cleaved into a 47-kDa product. In contrast, the D1125E/D1087E/D1016E/D1019E quadruple mutant HER-2 (designated 4×) was completely resistant to TRAIL-induced proteolysis. These results are represented schematically in Fig. 3B. Similarly, 35S-labeled 158-5E/4×-HER-2 was resistant to cleavage by recombinant caspase-3 or -8 in vitro (Fig. 3C). These results indicate that HER-2 is cleaved by caspases at four sites (D1016, D1019, D1087, and D1125E) in the cytoplasmic tail in vitro and in vivo.
FIGURE 3.
HER-2 is cleaved by caspases at four sites in the cytoplasmic tail. Four putative caspase cleavage sites (Asp1016, Asp1019, Asp1087, and Asp1125) were mutated to glutamic acid residues, and the mutant HER-2 proteins were examined for sensitivity to caspase proteolysis. A, MDA-MB-231 human breast cancer cells were transiently transfected with WT HER-2 or mutant HER-2 cDNAs as indicated. The next day, cells were preincubated with 100 nm epoxomicin for 1 h and then treated with 0.5 μg/ml TRAIL for 6 h. HER-2 was detected by immunoblotting using a carboxyl-terminal HER-2 mAb (cleavage products are indicated by arrows). B, schematic representation of HER-2 fragments (yellow) resulting from caspase proteolysis at each of the sites identified in A. The caspase cleavage-resistant D1016E/D1019E/D1087E/D1125E HER-2 mutant is designated 4× HER-2. The 25-kDa product (purple) is not detected by the carboxyl-terminal HER-2 Ab (red). C, 35S-labeled 158-5E/WT HER-2 or 158-5E/4× HER-2 proteins were incubated with control (C) buffer or the indicated amounts (ng) of caspase-3 or -8.
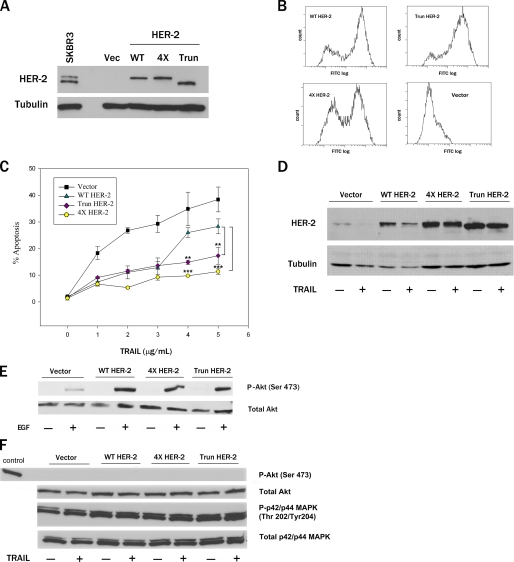
Cleavage-resistant and Truncated HER-2 Confer Greater Protection against Apoptosis than WT HER-2—To determine whether HER-2 proteolysis plays a role in apoptosis, we generated MDA-MB-231 pools stably expressing vector, WT HER-2, a truncated HER-2 encoding the amino-terminal caspase cleavage product (amino acids 1-1016, designated Trun), or caspase cleavage-resistant 4× HER-2. The expression levels of these HER-2 proteins in MDA-MB-231 pools were similar to that of endogenous HER-2 in SKBR3 cells (Fig. 4A). Like WT HER-2, the Trun and 4× HER-2 proteins were expressed on the cell surface (Fig. 4B). Moreover, WT and 4× HER-2 were phosphorylated on Tyr1248 in response to EGF treatment (data not shown). WT HER-2 conferred robust protection against apoptosis at concentrations of TRAIL ≤3 μg/ml but conferred little protection at higher concentrations of TRAIL (Fig. 4C). In contrast, both Trun HER-2 and 4× HER-2 protected MDA-MB-231 pools against TRAIL-induced apoptosis even at the highest concentration tested (5 μg/ml). Importantly, the loss of protective function by WT HER-2 against 5 μg/ml TRAIL was associated with proteolytic cleavage of the WT receptor (Fig. 4D). EGF treatment resulted in comparable Akt activation in MDA-MB-231 pools stably expressing WT, Trun, and 4× HER-2, indicating that the enhanced antiapoptotic activity of Trun and 4× HER-2 was not due to differential activation of Akt (Fig. 4E). Moreover, the levels of active Akt and p42/44 MAPK were similar in TRAIL-treated MDA-MB-231 pools expressing WT, Trun, and 4× HER-2 (Fig. 4F). These observations indicate that caspase proteolysis of HER-2 promotes apoptosis by releasing a proapoptotic carboxyl-terminal product, rather than by disrupting the antiapoptotic function of the full-length receptor.
FIGURE 4.
Cleavage-resistant and truncated HER-2 confer greater protection against apoptosis than WT HER-2. A, immunoblot analysis of MDA-MB-231 pools stably expressing vector, WT, caspase-truncated (amino acids 1-1016, designated Trun), or caspase cleavage-resistant 4× HER-2. B, FACS analysis of cell surface expression of HER-2 proteins in MDA-MB-231 pools. C, MDA-MB-231 pools were treated with 0-5 μg/ml TRAIL for 24 h, and apoptotic nuclei were scored (mean ± S.E., n = 3). **, p < 0.01; ***, p < 0.001 for the indicated comparisons by two-way ANOVA with Bonferroni post-test. D, immunoblot analysis of MDA-MB-231 pools treated with TRAIL (0 or 5 μg/ml) for 24 h. E, MDA-MB-231 pools were serum-starved for 24 h, treated with 100 ng/ml EGF for 10 min, and examined for phospho-Akt (serine 473) and total Akt levels by immunoblotting. F, MDA-MB-231 pools were treated with 5 μg/ml TRAIL for 24 h. Phospho-Akt (serine 473), total Akt levels, phospho-p42/p44 MAPK (Thr202/Tyr204), and total p42/p44 MAPK were determined by immunoblotting. A positive control lysate for phospho-Akt (serine 473; Cell Signaling) was also immunoblotted.
The 47- and 25-kDa HER-2 Products Induce Apoptosis by a Caspase-dependent and Bcl-2-suppressible Mechanism—To determine which of the HER-2 products was proapoptotic, we co-transfected MDA-MB-231 cells with pEGFP-N1 and cDNAs encoding each of the three carboxyl-terminal HER-2 cleavage products: the 47-kDa product (amino acids 1017-1254), and the 25-kDa (amino acids 1017-1125) and 22-kDa (amino acids 1126-1254) fragments generated by caspase proteolysis of the 47-kDa product (Fig. 5A). The 47- and 25-kDa HER-2 cleavage products induced apoptosis in transfected (GFP-positive) cells, whereas the 22-kDa product did not induce apoptosis above levels observed in vector-transfected cells (Fig. 5B). The proapoptotic activity of the 47- and 25-kDa HER-2 products was confirmed in a second breast cancer cell line (supplemental Fig. S2). Moreover, apoptosis induced by the 47- and 25-kDa products was suppressed by Z-VAD-fmk, indicating that the observed cell death was caspase-dependent (Fig. 5B). tBid, a positive control, also induced apoptosis that was inhibited by Z-VAD-fmk, consistent with prior reports (2, 4). The lack of proapoptotic activity of the 22-kDa fragment was not due to its rapid degradation; the addition of an amino-terminal FLAG epitope tag stabilized the 22-kDa product (Fig. 5C).
FIGURE 5.
The 47- and 25-kDa HER-2 products induce apoptosis by a caspase-dependent and Bcl-2-suppressible mechanism. A, schematic representation of the HER-2 products generated by caspase cleavage. B, MDA-MB-231 cells were co-transfected with cDNAs encoding FLAG-tagged HER-2 cleavage products (25, 22, or 47 kDa) or FLAG vector and pEGFP-N1 with or without a 1-h pretreatment with 100 μm Z-VAD-fmk. GFP-positive cells were scored for apoptotic nuclei 24 h later (mean ± S.E., n = 3). ***, p < 0.001 versus vector by two-way ANOVA with Bonferroni post-test. C, immunoblot confirms expression of an amino-terminal FLAG-tagged 22-kDa HER-2 cDNA in transfected MDA-MB-231 cells. D, MDA-MB-231 cells were co-transfected with cDNAs encoding the 25-kDa HER-2 (left) or the 47-kDa HER-2 cleavage product (right), pEGFP-N1, and vector, dominant negative (DN) FADD, CrmA, Bcl-2, or p35 cDNA. Apoptosis was scored as in B. **, p < 0.01 versus vector by one-way ANOVA with Tukey's multiple comparison post-test.
To delineate the mechanism(s) by which the 47- and 25-kDa HER-2 products induce apoptosis, we co-transfected MDA-MB-231 cells with each of these cDNAs, one of several antiapoptotic cDNAs, and pEGFP-N1. Neither a FADD dominant negative mutant (DN) nor CrmA, both of which inhibit activation of the initiator caspase-8 (44, 45), suppressed apoptosis induced by the 25- or 47-kDa products (Fig. 5D). In contrast, the baculoviral p35 protein, a broad spectrum caspase inhibitor, and Bcl-2, an antiapoptotic protein that antagonizes cytochrome c release from mitochondria (46, 47), suppressed apoptosis induced by both HER-2 products. These results confirm the caspase dependence of apoptosis induced by these HER-2 products and suggest that they may act on mitochondria to trigger cytochrome c release.
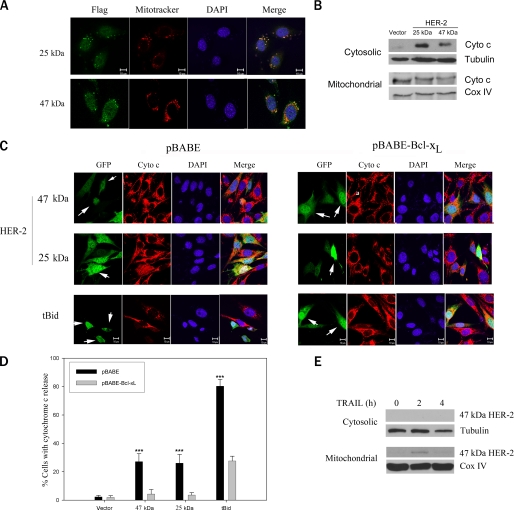
The 47- and 25-kDa HER-2 Products Localize to Mitochondria and Induce Cytochrome c Release—To determine whether the HER-2 products might act directly on mitochondria, we transfected MDA-MB-231 cells with cDNAs encoding the FLAG-tagged 47- or 25-kDa products. Confocal microscopy revealed that the FLAG-tagged 47- and 25-kDa products (green) colocalized with mitochondria visualized by Mitotracker staining (red); colocalization (yellow) is shown in the merged image (Fig. 6A). MDA-MB-231 cells transiently transfected with the 25- or 47-kDa HER-2 product (but not empty vector) expressed robust cytochrome c levels in cytosolic fractions, as determined by immunoblotting, consistent with mitochondrial release of cytochrome c (Fig. 6B). We next asked whether these HER-2 products induced cytochrome c release in MDA-MB-231 cells stably expressing pBABE vector or pBABE-Bcl-xL. Cells were transiently co-transfected with cDNAs encoding the FLAG-tagged 47- or 25-kDa products and pEGFP-N1 in the presence of Z-VAD-fmk, and the subcellular distribution of cytochrome c (red) was determined in GFP-positive cells (green) by confocal microscopy. Both the 47- and 25-kDa HER-2 products induced cytochrome c release in pBABE-expressing cells, albeit less robustly than tBid (Fig. 6, C (left) and D). Transfected cells with cytochrome c release (indicated by arrows in the GFP panels) exhibited diffuse or nearly absent cytochrome c staining, consistent with previous reports (40, 41). In contrast, Bcl-xL antagonized cytochrome c release by the 47- and 25-kDa HER-2 products and tBid (Fig. 6, C (right) and D). Transfected cells with intact mitochondrial cytochrome c (indicated by arrows in the GFP panels) showed punctate cytoplasmic staining. To determine the subcellular localization of the endogenous 47-kDa caspase cleavage product, we treated SKBR3 cells with TRAIL and isolated cytosolic and mitochondria-enriched heavy membrane fractions. Immunoblot analysis revealed that the 47-kDa product was present exclusively in the mitochondrial fraction (Fig. 6E). As noted previously, we are unable to detect the 25-kDa product with available antibodies and therefore cannot determine whether the endogenous 25-kDa product is also present in the mitochondrial fraction. Our results indicate that caspase cleavage of HER-2 releases two proapoptotic products that translocate to the mitochondria and trigger cytochrome c release.
FIGURE 6.
The 47- and 25-kDa HER-2 products localize to mitochondria and induce cytochrome c release. A, confocal images of MDA-MB-231 cells transfected with cDNAs encoding FLAG-tagged 25- or 47-kDa HER-2 cleavage products. Transfected MDA-MB-231 cells were immunostained with FLAG mAb (green). Mitochondria were labeled with Mitotracker Deep Red 633 (red), and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Colocalization (yellow) is shown in the merged image. Bar, 10 μm. B, immunoblot analysis of cytochrome c levels in cytosolic and mitochondrial fractions of MDA-MB-231 cells transfected with vector or HER-2 products. C, confocal analysis of cytochrome c release. MDA-MB-231 cells stably expressing pBABE vector (left) or pBABE-Bcl-xL (right) were co-transfected with the FLAG-tagged 25- or 47-kDa HER-2 product (or tBid) and pEGFP-N1 in the presence of Z-VAD-fmk. GFP-positive cells (green), cytochrome c immunostaining (red), and 4′,6-diamidino-2-phenylindole staining (DAPI, blue) are shown. In the MDA-MB-231/pBABE cells (left), cells with cytochrome c release (indicated by an arrow in the GFP panels) exhibit diffuse or nearly absent cytochrome c staining. In the MDA-MB-231/pBABE-Bcl-xL (right), cells with intact mitochondrial cytochrome c (indicated by an arrow in the GFP panels) show punctate staining. D, quantitation of the cytochrome c release data shown in C (mean ± S.E., n = 5). ***, p < 0.001 versus vector by two-way ANOVA with Bonferroni post-test. E, immunoblots of cytosolic and mitochondrial fractions from HER-2-overexpressing SKBR3 cells treated with 2.5 μg/ml TRAIL and 1 μg/ml cycloheximide for the indicated number of hours.
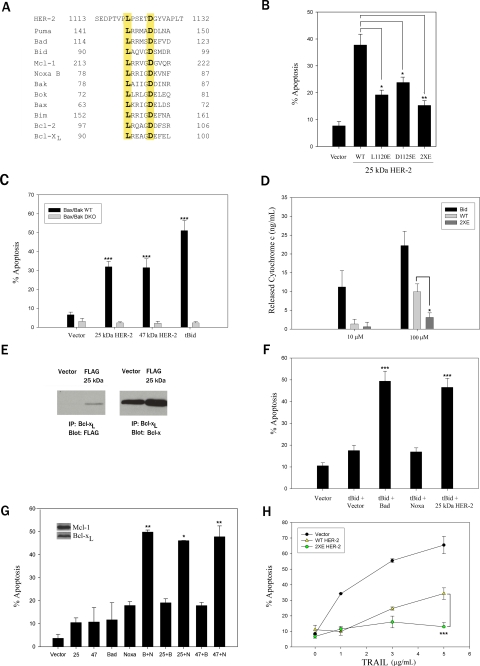
Caspase-cleaved HER-2 Contains a BH3-like Domain and Mimics Bad—Because the 47- and 25-kDa HER-2 products mimic actions of proapoptotic BH3-only proteins, we postulated that they might contain a BH3 domain. Alignment of the protein sequence shared by the 47- and 25-kDa HER-2 products with the BH3 domains of Bcl-2 family members revealed conservation of leucine (L) and aspartic acid (D) residues (highlighted in yellow in Fig. 7A) that play a critical role in apoptosis induction (11, 16, 48). These Leu (Leu1120) and Asp (Asp1125) residues in the HER-2 BH3-like domain are absolutely conserved across many species (data not shown). Substitution of either Leu1120 or Asp11125 with a Glu residue or substitution of both residues, as in the double mutant L1120E/D1125E (designated 2×E), reduced the proapoptotic activity of the 25-kDa HER-2 product (Fig. 7B). These results underscore the importance of the BH3-like domain in the HER-2 products for apoptosis induction. In contrast, mutation of the HER-2 BH3 domain did not impair mitochondrial localization (supplemental Fig. S3). Both the 25- and 47-kDa products (and tBid) induced apoptosis in retrovirally infected WT MEFs but not Bax-/-/Bak-/- DKO MEFs (Fig. 7C), indicating that apoptosis induction by these products is Bax/Bak-dependent, a hallmark of cell death induced by BH3-only proteins (14).
FIGURE 7.
Caspase-cleaved HER-2 contains a BH3-like domain and mimics the actions of Bad. A, sequence alignment of the 47- and 25-kDa HER-2 cleavage products and the BH3 domains of Bcl-2 family members. The absolutely conserved Leu and Asp residues in the BH3 domain that are required for proapoptotic activity are highlighted in yellow. B, the conserved Leu1120 and Asp1125 in human HER-2 were mutated to glutamic acid, either alone (L1120E or D1125E) or in combination (L1120E/D1125E, designated 2×E). MDA-MB-231 cells were co-transfected with cDNAs encoding the 25-kDa HER-2 product (WT or BH3 mutants) and pEGFP-N1. GFP-positive cells were scored for apoptotic nuclei 24 h later. C, apoptosis induced by the HER-2 products is Bax/Bak-dependent. WT or Bax/Bak DKO MEFs were infected with a bicistronic retrovirus encoding GFP and vector, GFP and HER-2 product (25 or 47 kDa), or GFP-tagged tBid for 48 h, and the percentage of GFP-positive cells with apoptotic nuclei was scored. D, 20-mer peptides (amino acids 1113-1132 of human HER-2) containing the WT HER-2 BH3 domain, a mutant 2×E HER-2 BH3 domain, or a Bid BH3 domain were incubated with isolated Jurkat cell mitochondria, and the released cytochrome c in the supernatant was analyzed by enzyme-linked immunosorbent assay. Readings were normalized to DMSO treatment. E, the 25-kDa HER-2 cleavage product binds to Bcl-xL. MDA-MB-231 pools stably expressing Bcl-xL or empty vector were immunoprecipitated (IP) with a rabbit Bcl-xL antibody and immunoblotted with a mouse FLAG (left) or mouse Bcl-x antibody (right). F, the 25-kDa HER-2 acts synergistically with tBid to induce apoptosis, mimicking the actions of Bad. MDA-MB-231 cells stably expressing Bcl-xL were transiently co-transfected with tBid and a bicistronic plasmid co-expressing GFP and vector, BAD, Noxa, or the 25-kDa HER-2 product. GFP-positive cells were scored for apoptotic nuclei 24 h later. G, the 25- and 47-kDa HER-2 products cooperate with Noxa to induce apoptosis. MCF-7 cells stably expressing caspase-3 were co-transfected with cDNAs encoding the 25- or 47-kDa HER-2 product, Bad (B), Noxa (N), or empty vector (alone and in combination) and pEGFP-N1. GFP-positive cells were scored for apoptotic nuclei 24 h later. H, MDA-MB-231 cells were transiently co-transfected with pEGFP-N1 and empty vector or cDNAs encoding full-length WT or mutant 2×E HER-2. Twenty-four h later, cells were treated with TRAIL for 24 h, and GFP-positive cells were scored for apoptotic nuclei. In B-D and F-H, data are the mean ± S.E. (n = 3). B and D,*, p < 0.05; **, p < 0.01 for the indicated comparisons by two-tailed unpaired t test. C, ***, p < 0.001 versus vector by two-way ANOVA with Bonferroni post-test. F, ***, p < 0.001 versus tBid + vector by one-way ANOVA with Bonferroni post-test. G,*, p < 0.05; **, p < 0.01 versus Noxa by one-way ANOVA with Bonferroni post-test. H, ***, p < 0.001 versus WT HER-2 by two-way ANOVA with Bonferroni post-test.
To determine whether the HER-2 BH3-like domain was sufficient to induce MOMP in a cell-free system, we added peptides (amino acids 1113-1132 of human HER-2) containing the WT HER-2 BH3 sequence, a mutant 2×E HER-2 BH3 sequence, or a Bid BH3 sequence to isolated mitochondria from Jurkat cells and measured cytochrome c release by an enzyme-linked immunosorbent assay. The WT HER-2 BH3 peptide, but not the 2×E peptide, induced cytochrome c release, albeit less robustly than the Bid peptide (Fig. 7D). Because the 25-kDa product and tBid bind to Bcl-xL (Fig. 7E) (11), we postulated that the 25-kDa product would inhibit Bcl-xL and act synergistically with tBid to induce apoptosis in MDA-MB-231 cells stably expressing Bcl-xL. These Bcl-xL-overexpressing cells were resistant to tBid-induced apoptosis (Fig. 7F). Co-expression of the 25-kDa product or Bad, but not Noxa, sensitized these cells to tBid-induced apoptosis (Fig. 7F), consistent with the binding of the 25-kDa product and Bad with Bcl-xL and the inability of Noxa to bind to Bcl-xL (Fig. 7E) (data not shown).
We next examined whether the 25- and 47-kDa HER-2 products cooperated with Bad or Noxa to induce apoptosis in MCF-7 breast cancer cells, which express endogenous Bcl-xL (inhibited by Bad) and Mcl-1 (inhibited by Noxa). Because parental MCF-7 cells lack caspase-3 and are inherently resistant to apoptosis, MCF-7 cells stably expressing caspase-3 (29) were used for these studies. As predicted by virtue of their expression of both Bcl-xL and Mcl-1, these cells were resistant to apoptosis induction by ectopic expression of Bad or Noxa alone (or either of the HER-2 products) (Fig. 7G). In contrast, ectopic expression of both Bad and Noxa robustly induced apoptosis. Similarly, the 25- and 47-kDa HER-2 products cooperated with Noxa, but not Bad, to induce apoptosis (Fig. 7G), thereby providing additional evidence that the HER-2 products are Bad-like cell death effectors.
To further delineate the functional relevance of the HER-2 BH3-like domain, we examined the sensitivity to apoptosis of MDA-MB-231 cells transfected with full-length WT or BH3 mutant 2×E HER-2. Although both the full-length WT and 2×E HER-2 proteins were cleaved into the expected 47-kDa product in response to TRAIL treatment (data not shown), the mutant 2×E HER-2 conferred greater protection against apoptosis than WT HER-2 due to the lack of a proapoptotic BH3-like domain in the mutant protein (Fig. 7H). These results indicate that caspases activate a previously unrecognized proapoptotic function of HER-2 by releasing a BH3-like cell death effector.
DISCUSSION
Ever since the recognition of its pathogenic role in a subset of poor prognosis breast carcinomas (21), HER-2 has been one of the most intensely scrutinized molecules in cancer biology. Indeed, HER-2 has been directly linked to many of the hallmarks of cancer, including dysregulated proliferation, migration, invasion, and suppression of apoptosis (20). Here we describe a new and seemingly paradoxical function for HER-2 as a cell death effector activated by caspases. Specifically, we have demonstrated that the cytoplasmic tail of HER-2 is cleaved at four sites (Asp1016, Asp 1019, Asp1087, and Asp1125) by caspases in vitro and in multiple breast cancer cell lines treated with multiple apoptotic stimuli. These results suggest that HER-2 proteolysis is neither cell typenor stimulus-specific. Caspases initially cleave HER-2 at Asp1016/Asp1019 to release a carboxyl-terminal 47-kDa fragment that is subsequently proteolyzed by caspases at Asp1125 into an unstable 22-kDa product, which is rapidly degraded by the proteasome, and a predicted 25-kDa product. In addition, mutation of the Asp1125 caspase cleavage site uncovered a fourth cleavage site (Asp1087). Mutation of all four caspase cleavage sites (as in the 4× mutant) is necessary to generate a caspase cleavage-resistant HER-2 protein. Although caspase proteolysis of HER-2 at Asp1087 and Asp1125 has been reported (26, 27), our findings indicate that the apoptotic proteolysis of HER-2 is more complex than previously recognized and has profound functional consequences for cell death regulation.
We have also demonstrated that caspase proteolysis of HER-2 promotes apoptosis by a novel mechanism, namely by releasing a BH3-like cell death effector. Several lines of evidence support this mechanism. First, stable expression of the 4× caspase cleavage-resistant HER-2 protein or the amino-terminal caspase cleavage product (Trun HER-2) protects breast cancer cells against apoptosis more robustly than WT HER-2, strongly suggesting that caspase cleavage of HER-2 promotes apoptosis by releasing a proapoptotic product rather than by suppressing the antiapoptotic function of WT HER-2. Second, ectopic expression of the 47- and 25-kDa HER-2 cleavage products (but not the 22-kDa fragment) is sufficient to induce apoptosis by a caspase-dependent, Bax/Bak-dependent, and Bcl-2-suppressible mechanism. Of note, both the 47- and 25-kDa products induce comparable amounts of apoptosis, indicating that caspase cleavage of the 47-kDa product is not required to activate its proapoptotic function. Third, the 47-kDa product is present in the mitochondrial fraction in cells undergoing apoptosis. Fourth, ectopic expression of the 47- and 25-kDa products leads to their mitochondrial localization and Bcl-xL-suppressible cytochrome c release. Fifth, both the 47- and 25-kDa products share a BH3-like domain that is required for apoptosis induction and cytochrome c release from isolated mitochondria; mutation of one or both highly conserved Leu or Asp residues in this BH3-like domain inhibits these activities. Sixth, the 25-kDa HER-2 product interacts with Bcl-xL. Finally, a full-length HER2 protein containing the 2×E BH3 domain mutant is cleaved by caspases but confers greater protection against apoptosis than WT HER-2 due to the lack of a proapoptotic BH3-like domain in the mutant protein, thereby underscoring the functional relevance of the HER-2 BH3-like domain.
Although our results strongly suggest that HER-2 contains a BH3-like domain, an unusual feature is that caspase cleavage truncates the BH3-like domain at Asp1125 in the 25-kDa product (which contains the truncated domain 1120LPSETD1125 in its carboxyl terminus) but does not abrogate its proapoptotic function. Similarly, BokS, an alternatively spliced form of the BH3-only protein Bok contains a truncated BH3 domain (lacking the corresponding Asp residue and carboxyl-terminal BH3 domain residues) fused to a BH1 domain, but retains its proapoptotic function (49). Clearly, crystal structure analyses are needed to assess whether and how the HER-2 BH3-like domain (full-length and truncated) inserts into the hydrophobic pocket of antiapoptotic Bcl-2 family members. Importantly, there is a growing family of novel BH3-only proteins, which like the HER-2 products, have limited homology to the canonical BH3 domain, are weakly proapoptotic and bind to antiapoptotic Bcl-2 family members (50, 51). Furthermore, we provide functional evidence for the BH3-like nature of the HER-2 cleavage products. Specifically, we demonstrate in two independent systems that the HER-2 products are functionally interchangeable with the well characterized BH3-only protein Bad with regard to their ability to cooperate with Noxa and/or tBid to induce apoptosis. Of note, the observed proapoptotic actions of the HER-2 BH3-like domain are distinct from those of a previously reported HER-2 kinase domain fragment that induces caspase-independent cell death (52). Intriguingly, HER-4/ErbB4 has recently been shown to contain a BH3-like intracellular domain, which is released by γ-secretase and triggers Bax/Bak-dependent MOMP (53). These findings suggest that proteolytic activation of intracellular BH3-like domains in transmembrane receptors may be a more generalized mechanism of caspase amplification.
In many respects, the activation of the BH3-like domain in HER-2 by caspases is highly reminiscent of the activation of the BH3-only protein Bid. In both cases, caspase cleavage is required for the mitochondrial translocation of a truncated BH3-only product that is sufficient to induce MOMP and apoptosis. Notably, the BH3 domains in both proteins and in the BH3-only protein Puma are dispensable for mitochondrial localization (supplemental Fig. S3) (15, 54). In the case of Bid, caspase proteolysis exposes an amino-terminal Gly residue in tBid, which undergoes post-translational N-myristoylation, an event that triggers the translocation of tBid from the cytoplasm to mitochondrial membranes (6). Two other caspase-truncated proteins that are targeted to mitochondria (actin and gelsolin) have been shown to be N-myristoylated on amino-terminal Gly residues, although mitochondrial targeting of gelsolin was not N-myristoylation-dependent (55, 56). In contrast, neither the 47- nor 25-kDa HER-2 products contain an amino-terminal Gly residue, suggesting that mechanisms other than N-myristoylation are likely to be responsible for targeting the HER-2 products to mitochondria. Compared with tBid, the HER-2 products are less potent at inducing cytochrome c release and apoptosis. Moreover, we demonstrated that the 25-kDa HER-2 product acts synergistically with tBid to induce apoptosis in breast cancer cells overexpressing Bcl-xL, mimicking the actions of Bad (which binds Bcl-xL and Bcl-2) but not Noxa (which binds Mcl-1) (11-13). These findings are consistent with previous reports indicating synergy between tBid and Bad (11, 13, 16) and suggest that the HER-2 product is more Bad-like than Bid-like.
Additional evidence for the Bad-like nature of the HER-2 products comes from our observation that these products, like Bad, cooperate with Noxa to induce apoptosis in breast cancer cells that express endogenous Bcl-xL and Mcl-l. Our results are also consistent with both models of BH3 ligand-induced activation of Bax/Bak that have been proposed (57). Caspase-cleaved HER-2 promotes apoptosis by neutralizing Bcl-xL, acting either as a Bad-like “sensitizer” BH3-only protein, which displaces activator BH3-only proteins (e.g. tBid) from the activator BH3-Bcl-xL complex (“direct activation” model), or as a Bad-like inhibitor of Bcl-xL that displaces Bax/Bak (“indirect activation” model) from Bcl-xL (11, 18). Moreover, the ability of the HER-2 BH3 peptide to induce cytochrome c release in isolated mitochondria derived from T-cell leukemia Jurkat cells is similar to that reported for a Bad peptide using mitochondria derived from leukemia cells (17). This group has suggested that cancer cells may be exquisitely vulnerable to BH3 ligands because they may be “primed for death” due to high occupancy of antiapoptotic Bcl-2 proteins with BH3-only proteins. Viewed from this context, caspase cleavage of HER-2 may also prime cancer cells for death by generating an intracellular Bad-like molecule. Intriguingly, both Bad and the HER-2 BH3-like products are unable to inactivate Mcl-1, which is commonly overexpressed in HER-2-positive breast tumors (58), suggesting that breast tumors may evade this proapoptotic mechanism by overexpressing Mcl-1. Collectively, our results suggest that HER-2 is a novel BH3-like protein, which is activated by caspases, thereby releasing a Bad-like cell death effector.
HER-2, then, plays a dual role in regulating apoptosis. Full-length HER-2 protects cells from apoptosis by activating survival pathways, such as phosphoinositol 3-kinase/Akt (23-25). Our results indicate that HER-2 is a dormant proapoptotic molecule that harbors a functional BH3-like domain in its cytoplasmic tail, which is released by caspase cleavage, thereby unmasking a previously unrecognized proapoptotic function of HER-2. Several other antiapoptotic proteins have been shown to be converted into proapoptotic molecules by caspases, including Bcl-2, Bcl-xL, and Mcl-1 (59-61). Caspase proteolysis of Bcl-2 and Bcl-xL removes an amino-terminal BH4 domain and generates a carboxyl-terminal product that induces apoptosis by a BH3-dependent mechanism. Together with the data presented here, these findings suggest that the cell death machinery is elegantly parsimonious, with some molecules performing both anti- and proapoptotic functions. In addition, our results have important clinical implications, because they indicate that caspase cleavage of HER-2 is an early and functionally important event for apoptosis induction in HER-2-expressing cancer cells.
Supplementary Material
Acknowledgments
We are indebted to Craig Thompson, David C. S. Huang, Gibbes Johnson, Navdeep Chandel, Hiroaki Kiyokawa, Honglin Li, Christopher Froelich, and Richard Kitsis for providing reagents and/or reading the manuscript. Technical assistance was provided by Nimrod Deiss-Yehiely.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA097198 (to V. L. C.). This work was also supported by Department of Defense Breast Cancer Research Program Grants DAMD25-02-1-0526 (to V. L. C.) and DMAD25-00-1-0386 (to A. M. S.), a grant from the Breast Cancer Research Foundation (to V. L. C.), and a Katten Muchin Rosenman Travel Scholarship (to A. M. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3.
Footnotes
The abbreviations used are: TNF, tumor necrosis factor; TRAIL, TNF-related apoptosis-inducing ligand; MOMP, mitochondrial outer membrane permeabilization; DKO, double knock-out; MEFs, murine embryonic fibroblasts; Trun, truncated; WT, wild type; Z, benzyloxycarbonyl; fmk, fluoromethyl ketone; Ab, antibody; mAb, monoclonal antibody; GFP, green fluorescent protein; TRITC, tetramethylrhodamine isothiocyanate; ANOVA, analysis of variance; EGF, epidermal growth factor; tBid, truncated Bid.
References
- 1.Cryns, V., and Yuan, J. (1998) Genes Dev. 12 1551-1570 [DOI] [PubMed] [Google Scholar]
- 2.Li, H., Zhu, H., Xu, C. J., and Yuan, J. (1998) Cell 94 491-501 [DOI] [PubMed] [Google Scholar]
- 3.Lassus, P., Opitz-Araya, X., and Lazebnik, Y. (2002) Science 297 1352-1354 [DOI] [PubMed] [Google Scholar]
- 4.Luo, X., Budihardjo, I., Zou, H., Slaughter, C., and Wang, X. (1998) Cell 94 481-490 [DOI] [PubMed] [Google Scholar]
- 5.Srinivasula, S. M., Ahmad, M., Fernandes-Alnemri, T., and Alnemri, E. S. (1998) Mol. Cell 1 949-957 [DOI] [PubMed] [Google Scholar]
- 6.Zha, J., Weiler, S., Oh, K. J., Wei, M. C., and Korsmeyer, S. J. (2000) Science 290 1761-1765 [DOI] [PubMed] [Google Scholar]
- 7.Lutter, M., Fang, M., Luo, X., Nishijima, M., Xie, X., and Wang, X. (2000) Nat. Cell Biol. 2 754-761 [DOI] [PubMed] [Google Scholar]
- 8.Puthalakath, H., and Strasser, A. (2002) Cell Death Differ. 9 505-512 [DOI] [PubMed] [Google Scholar]
- 9.Wang, K., Yin, X. M., Chao, D. T., Milliman, C. L., and Korsmeyer, S. J. (1996) Genes Dev. 10 2859-2869 [DOI] [PubMed] [Google Scholar]
- 10.Sattler, M., Liang, H., Nettesheim, D., Meadows, R. P., Harlan, J. E., Eberstadt, M., Yoon, H. S., Shuker, S. B., Chang, B. S., Minn, A. J., Thompson, C. B., and Fesik, S. W. (1997) Science 275 983-986 [DOI] [PubMed] [Google Scholar]
- 11.Letai, A., Bassik, M. C., Walensky, L. D., Sorcinelli, M. D., Weiler, S., and Korsmeyer, S. J. (2002) Cancer Cell 2 183-192 [DOI] [PubMed] [Google Scholar]
- 12.Chen, L., Willis, S. N., Wei, A., Smith, B. J., Fletcher, J. I., Hinds, M. G., Colman, P. M., Day, C. L., Adams, J. M., and Huang, D. C. (2005) Mol. Cell 17 393-403 [DOI] [PubMed] [Google Scholar]
- 13.Kim, H., Rafiuddin-Shah, M., Tu, H. C., Jeffers, J. R., Zambetti, G. P., Hsieh, J. J., and Cheng, E. H. (2006) Nat. Cell Biol. 8 1348-1358 [DOI] [PubMed] [Google Scholar]
- 14.Wei, M. C., Zong, W. X., Cheng, E. H., Lindsten, T., Panoutsakopoulou, V., Ross, A. J., Roth, K. A., MacGregor, G. R., Thompson, C. B., and Korsmeyer, S. J. (2001) Science 292 727-730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei, M. C., Lindsten, T., Mootha, V. K., Weiler, S., Gross, A., Ashiya, M., Thompson, C. B., and Korsmeyer, S. J. (2000) Genes Dev. 14 2060-2071 [PMC free article] [PubMed] [Google Scholar]
- 16.Kuwana, T., Bouchier-Hayes, L., Chipuk, J. E., Bonzon, C., Sullivan, B. A., Green, D. R., and Newmeyer, D. D. (2005) Mol. Cell 17 525-535 [DOI] [PubMed] [Google Scholar]
- 17.Certo, M., Del Gaizo Moore, V., Nishino, M., Wei, G., Korsmeyer, S., Armstrong, S. A., and Letai, A. (2006) Cancer Cell 9 351-365 [DOI] [PubMed] [Google Scholar]
- 18.Willis, S. N., Chen, L., Dewson, G., Wei, A., Naik, E., Fletcher, J. I., Adams, J. M., and Huang, D. C. (2005) Genes Dev. 19 1294-1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willis, S. N., Fletcher, J. I., Kaufmann, T., van Delft, M. F., Chen, L., Czabotar, P. E., Ierino, H., Lee, E. F., Fairlie, W. D., Bouillet, P., Strasser, A., Kluck, R. M., Adams, J. M., and Huang, D. C. (2007) Science 315 856-859 [DOI] [PubMed] [Google Scholar]
- 20.Hynes, N. E., and Lane, H. A. (2005) Nat. Rev. Cancer 5 341-354 [DOI] [PubMed] [Google Scholar]
- 21.Slamon, D. J., Clark, G. M., Wong, S. G., Levin, W. J., Ullrich, A., and McGuire, W. L. (1987) Science 235 177-182 [DOI] [PubMed] [Google Scholar]
- 22.Gusterson, B. A., Gelber, R. D., Goldhirsch, A., Price, K. N., Save-Soderborgh, J., Anbazhagan, R., Styles, J., Rudenstam, C. M., Golouh, R., Reed, R., Martinez-Tello, F., Tiltman, A., Torhorst, J., Grigolato, P., Bettelheim, R., Neville, A. M., Burki, K., Castiglione, M., Collins, J., Lindtner, J., and Senn, H.-J. (1992) J. Clin. Oncol. 10 1049-1056 [DOI] [PubMed] [Google Scholar]
- 23.Zhou, B. P., Hu, M. C., Miller, S. A., Yu, Z., Xia, W., Lin, S. Y., and Hung, M. C. (2000) J. Biol. Chem. 275 8027-8031 [DOI] [PubMed] [Google Scholar]
- 24.Zhou, B. P., Liao, Y., Xia, W., Zou, Y., Spohn, B., and Hung, M. C. (2001) Nat. Cell Biol. 3 973-982 [DOI] [PubMed] [Google Scholar]
- 25.Cuello, M., Ettenberg, S. A., Clark, A. S., Keane, M. M., Posner, R. H., Nau, M. M., Dennis, P. A., and Lipkowitz, S. (2001) Cancer Res. 61 4892-4900 [PubMed] [Google Scholar]
- 26.Tikhomirov, O., and Carpenter, G. (2001) J. Biol. Chem. 276 33675-33680 [DOI] [PubMed] [Google Scholar]
- 27.Benoit, V., Chariot, A., Delacroix, L., Deregowski, V., Jacobs, N., Merville, M. P., and Bours, V. (2004) Cancer Res. 64 2684-2691 [DOI] [PubMed] [Google Scholar]
- 28.Lindsten, T., Ross, A. J., King, A., Zong, W. X., Rathmell, J. C., Shiels, H. A., Ulrich, E., Waymire, K. G., Mahar, P., Frauwirth, K., Chen, Y., Wei, M., Eng, V. M., Adelman, D. M., Simon, M. C., Ma, A., Golden, J. A., Evan, G., Korsmeyer, S. J., MacGregor, G. R., and Thompson, C. B. (2000) Mol. Cell 6 1389-1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang, X. H., Sladek, T. L., Liu, X., Butler, B. R., Froelich, C. J., and Thor, A. D. (2001) Cancer Res. 61 348-354 [PubMed] [Google Scholar]
- 30.Wong, L., Deb, T. B., Thompson, S. A., Wells, A., and Johnson, G. R. (1999) J. Biol. Chem. 274 8900-8909 [DOI] [PubMed] [Google Scholar]
- 31.Kamradt, M. C., Chen, F., and Cryns, V. L. (2001) J. Biol. Chem. 276 16059-16063 [DOI] [PubMed] [Google Scholar]
- 32.Majewski, N., Nogueira, V., Robey, R. B., and Hay, N. (2004) Mol. Cell Biol. 24 730-740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lustig, K. D., Stukenberg, P. T., McGarry, T. J., King, R. W., Cryns, V. L., Mead, P. E., Zon, L. I., Yuan, J., and Kirschner, M. W. (1997) Methods Enzymol. 283 83-99 [DOI] [PubMed] [Google Scholar]
- 34.Cryns, V. L., Byun, Y., Rana, A., Mellor, H., Lustig, K. D., Ghanem, L., Parker, P. J., Kirschner, M. W., and Yuan, J. (1997) J. Biol. Chem. 272 29449-29453 [DOI] [PubMed] [Google Scholar]
- 35.Byun, Y., Chen, F., Chang, R., Trivedi, M., Green, K. J., and Cryns, V. L. (2001) Cell Death Differ. 8 443-450 [DOI] [PubMed] [Google Scholar]
- 36.Chen, F., Kamradt, M., Mulcahy, M., Byun, Y., Xu, H., McKay, M. J., and Cryns, V. L. (2002) J. Biol. Chem. 277 16775-16781 [DOI] [PubMed] [Google Scholar]
- 37.Chen, F., Arseven, O. K., and Cryns, V. L. (2004) J. Biol. Chem. 279 27542-27548 [DOI] [PubMed] [Google Scholar]
- 38.Moyano, J. V., Evans, J. R., Chen, F., Lu, M., Werner, M. E., Yehiely, F., Diaz, L. K., Turbin, D., Karaca, G., Wiley, E., Nielsen, T. O., Perou, C. M., and Cryns, V. L. (2006) J. Clin. Invest. 116 261-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamradt, M. C., Lu, M., Werner, M. E., Kwan, T., Chen, F., Strohecker, A., Oshita, S., Wilkinson, J. C., Yu, C., Oliver, P. G., Duckett, C. S., Buchsbaum, D. J., Lobuglio, A. F., Jordan, V. C., and Cryns, V. L. (2005) J. Biol. Chem. 280 11059-11066 [DOI] [PubMed] [Google Scholar]
- 40.Kennedy, S. G., Kandel, E. S., Cross, T. K., and Hay, N. (1999) Mol. Cell Biol. 19 5800-5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madesh, M., Antonsson, B., Srinivasula, S. M., Alnemri, E. S., and Hajnoczky, G. (2002) J. Biol. Chem. 277 5651-5659 [DOI] [PubMed] [Google Scholar]
- 42.Gross, A., Yin, X. M., Wang, K., Wei, M. C., Jockel, J., Milliman, C., Erdjument-Bromage, H., Tempst, P., and Korsmeyer, S. J. (1999) J. Biol. Chem. 274 1156-1163 [DOI] [PubMed] [Google Scholar]
- 43.Meng, L., Mohan, R., Kwok, B. H., Elofsson, M., Sin, N., and Crews, C. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 10403-10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muzio, M., Chinnaiyan, A. M., Kischkel, F. C., O'Rourke, K., Shevchenko, A., Ni, J., Scaffidi, C., Bretz, J. D., Zhang, M., Gentz, R., Mann, M., Krammer, P. H., Peter, M. E., and Dixit, V. M. (1996) Cell 85 817-827 [DOI] [PubMed] [Google Scholar]
- 45.Chinnaiyan, A. M., Tepper, C. G., Seldin, M. F., O'Rourke, K., Kischkel, F. C., Hellbardt, S., Krammer, P. H., Peter, M. E., and Dixit, V. M. (1996) J. Biol. Chem. 271 4961-4965 [DOI] [PubMed] [Google Scholar]
- 46.Bump, N. J., Hackett, M., Hugunin, M., Seshagiri, S., Brady, K., Chen, P., Ferenz, C., Franklin, S., Ghayur, T., Li, P., Licari, J., Mankovich, L., Shi, L., Greenberg, A. H., Miller, L. K., and Wong, W. W. (1995) Science 269 1885-1888 [DOI] [PubMed] [Google Scholar]
- 47.Kluck, R. M., Bossy-Wetzel, E., Green, D. R., and Newmeyer, D. D. (1997) Science 275 1132-1136 [DOI] [PubMed] [Google Scholar]
- 48.Wang, K., Gross, A., Waksman, G., and Korsmeyer, S. J. (1998) Mol. Cell Biol. 18 6083-6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu, S. Y., and Hsueh, A. J. (1998) J. Biol. Chem. 273 30139-30146 [DOI] [PubMed] [Google Scholar]
- 50.Liu, H., Toman, R. E., Goparaju, S. K., Maceyka, M., Nava, V. E., Sankala, H., Payne, S. G., Bektas, M., Ishii, I., Chun, J., Milstien, S., and Spiegel, S. (2003) J. Biol. Chem. 278 40330-40336 [DOI] [PubMed] [Google Scholar]
- 51.Rodolfo, C., Mormone, E., Matarrese, P., Ciccosanti, F., Farrace, M. G., Garofano, E., Piredda, L., Fimia, G. M., Malorni, W., and Piacentini, M. (2004) J. Biol. Chem. 279 54783-54792 [DOI] [PubMed] [Google Scholar]
- 52.Tikhomirov, O., Dikov, M., and Carpenter, G. (2005) Oncogene 24 3906-3913 [DOI] [PubMed] [Google Scholar]
- 53.Naresh, A., Long, W., Vidal, G. A., Wimley, W. C., Marrero, L., Sartor, C. I., Tovey, S., Cooke, T. G., Bartlett, J. M., and Jones, F. E. (2006) Cancer Res. 66 6412-6420 [DOI] [PubMed] [Google Scholar]
- 54.Nakano, K., and Vousden, K. H. (2001) Mol. Cell 7 683-694 [DOI] [PubMed] [Google Scholar]
- 55.Utsumi, T., Sakurai, N., Nakano, K., and Ishisaka, R. (2003) FEBS Lett. 539 37-44 [DOI] [PubMed] [Google Scholar]
- 56.Sakurai, N., and Utsumi, T. (2006) J. Biol. Chem. 281 14288-14295 [DOI] [PubMed] [Google Scholar]
- 57.Fletcher, J. I., and Huang, D. C. (2006) Cell Death Differ. 13 1268-1271 [DOI] [PubMed] [Google Scholar]
- 58.Henson, E. S., Hu, X., and Gibson, S. B. (2006) Clin. Cancer Res. 12 845-853 [DOI] [PubMed] [Google Scholar]
- 59.Cheng, E. H., Kirsch, D. G., Clem, R. J., Ravi, R., Kastan, M. B., Bedi, A., Ueno, K., and Hardwick, J. M. (1997) Science 278 1966-1968 [DOI] [PubMed] [Google Scholar]
- 60.Clem, R. J., Cheng, E. H., Karp, C. L., Kirsch, D. G., Ueno, K., Takahashi, A., Kastan, M. B., Griffin, D. E., Earnshaw, W. C., Veliuona, M. A., and Hardwick, J. M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 554-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weng, C., Li, Y., Xu, D., Shi, Y., and Tang, H. (2005) J. Biol. Chem. 280 10491-10500 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.