Abstract
Toll-IL-1 receptor (TIR) domain-containing adaptor molecule-1 (TICAM-1, also named TIR domain-containing adaptor-inducing interferon (IFN)-β or TRIF)) is a signaling adaptor of Toll-like receptor (TLR) 3/4 that activates the transcription factors, interferon regulatory factor-3 (IRF-3) and NF-κB leading to inducing IFN-β production. The mechanisms by which TICAM-1 is activated by TLR3/4 to serve as a signaling platform are unknown. In this study, we show that homo-oligomerization of TICAM-1 is critical for TICAM-1-mediated activation of NF-κB and IRF-3. Both TIR and C-terminal domain of TICAM-1 mediated TICAM-1 oligomerization. Pro434 located in the TIR domain and the C-terminal region, with the exception of the RIP homotypic-interacting motif, were determinants of TICAM-1 oligomerization. Mutation of TIR domain (P434H) or deletion of C-terminal domain greatly reduced TICAM-1-mediated NF-κB and IFN-β promoter activation. TICAM-1 oligomerization at either the TIR domain or the C-terminal region resulted in recruitment of tumor necrosis factor receptor-associated factor 3, a downstream signaling molecule essential for TICAM-1-mediated IRF-3 activation, but not recruitment of the IRF-3 kinase complex, NF-κB-activating kinase-associated protein 1 and TANK-binding kinase 1. In addition, RIP homotypic-interacting motif mutant, which possesses two oligomerization motifs but not the RIP1 binding motif, also failed to recruit NF-κB-activating kinase-associated protein 1 and TANK-binding kinase 1. Thus, full activation and formation of TICAM-1 signalosomes requires oligomerization induced at two different sites and RIP1 binding.
The innate immune system senses bacterial and viral infections using Toll-like receptors (TLRs)5 and cytoplasmic pattern-recognition receptors. Endosomal TLRs and cytoplasmic DEAD/H box RNA helicases, retinoic acid-inducible gene-I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), play key roles in anti-viral immunity by inducing transcription of type I interferon (IFN) and IFN-regulatory genes (1, 2).
Among these receptors, TLR3 has distinct properties that allow recognition of extracellular virus-derived double-stranded RNA (dsRNA) and induction of type I IFN/cytokine production and dendritic cell maturation that results in activation of natural killer cells and cytotoxic T lymphocytes (3-6). TLR3 signaling is mediated via an adaptor molecule, Toll-IL-1 receptor (TIR) domain-containing adaptor molecule-1 (TICAM-1, also named TIR domain-containing adaptor inducing IFN-β (TRIF)), which activates the transcription factors interferon regulatory factor-3 (IRF-3), NF-κB, and AP-1 (7, 8). TICAM-1 consists of a proline-rich N-terminal region, a TIR domain, and a C-terminal region. The TIR domain of TICAM-1 is essential for binding to the TIR domain of TLR3 as well as to the TLR4 adaptor molecule, TICAM-2 (also called TRIF-related adaptor molecule) (9, 10). The N-terminal region is crucial for TICAM-1-mediated IRF-3 activation via recruitment of IRF-3-activating kinases, TANK-binding kinase 1 (TBK1) and inhibitor of nuclear factor κB kinase ε (IKKε, also called IKKι) (7, 11-13). The C-terminal region is involved in NF-κB activation and induction of apoptosis via binding of the receptor interacting protein (RIP) 1 to the RIP homotypic-interacting motif (RHIM) domain (14, 15). The NF-κB-activating kinase-associated protein 1 (NAP1) and tumor necrosis factor receptor-associated factor 3 (TRAF3) are found downstream of TICAM-1 and are involved in the TICAM-1-mediated activation of IRF-3 (16-18).
NAP1 and TRAF3 are also involved in RIG-I/MDA5-mediated production of IFN-β through interactions with the adaptor molecule, IFN-β promoter stimulator 1 (also called MAVS, Cardif, or VISA) (19-24). IFN-β promoter stimulator 1 localizes to the mitochondrial membrane where it is activated by RIG-I/MDA5 through caspase recruitment domain (CARD)-CARD interactions (20). Although TLR- and RIG-I/MDA5-mediated signaling is transmitted via interaction of unique domain structures, TIR and CARD, respectively, the detailed mechanisms of signal transduction regulated by these domains as well as the spatiotemporal regulation of signaling have not been studied.
TICAM-1 localizes diffusely in the cytosol of resting cells. Once TLR3 is activated by dsRNA, TICAM-1 transiently co-localizes with TLR3, then dissociates from the receptor and forms speckle-like structures with RIP1 and NAP1 (25). In addition, overexpressed TICAM-1 strongly activates the IFN-β promoter in a TLR3-independent manner. The mechanisms by which TICAM-1 is activated by receptor ligation or spontaneously following overexpression are unknown. In this study, we constructed various TICAM-1 mutants and used a yeast two-hybrid system and immunoprecipitation studies to identify the critical regions involved in TICAM-1 oligomerization. We show that full activation and formation of TICAM-1 signalosomes require oligomerization induced at two different sites and RIP1 binding. Furthermore, we demonstrate that, during TLR3/4-TICAM-1-signaling, the conserved proline residue within the BB loop of the upstream TIR domain determines the interaction with the downstream TIR domain, which in turn leads to signal transduction.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents—HEK293 cells and HEK293FT cells were maintained in Dulbecco's Modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (Invitrogen) and antibiotics. HeLa cells were maintained in MEM (Nissui, Tokyo, Japan) supplemented with 10% heat-inactivated fetal calf serum. Anti-FLAG M2 mAb, anti-HA pAb, 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) and z-VAD-fmk were purchased from Sigma-Aldrich. Alexa Fluor®-conjugated secondary antibodies were from Invitrogen. Anti-Myc mAb was from Neomarkers (Lab Vision Corp., Fremont, CA).
Plasmids—Complementary DNAs for human TICAM-1, RIP1, and TRAF3 were cloned in our laboratory by reverse transcription-PCR and were ligated into the cloning site of the expression vectors, pEF-BOS and p3xFLAG-CMV-14 (C-terminal 3×FLAG tag). The HA tag was inserted into the C-terminal of each pEF-BOS expression vector for the TICAM-1 mutant. The pCDNA3.1/NAP1-Myc and pCDNA3.1/TBK1-FLAG expression vectors were kindly provided by Dr. M. Nakanishi (Nagoya City University, Nagoya, Japan). Point mutations in the TIR domain (P434H, where Pro434 was replaced with His) and RHIM domain (687VQLG690 were replaced with four Ala) were generated by site-directed mutagenesis. The truncated TICAM-1 mutant (N+TIR, 1-566 aa) was generated by PCR using specific primers.
Yeast Two-hybrid Assay—The yeast two-hybrid assay was performed as described previously (7). The yeast AH109 strain (Clontech, Palo Alto, CA) was transformed using bait (pGBKT7) and prey (pGADT7) plasmids. The transformants were streaked onto plates and incubated for 3-5 days. BD and AD in the figures represent the bait and prey plasmid, respectively. The various BD- and AD-TICAM-1 mutants were constructed by inserting each cDNA fragment into the pGBKT7 (bait) or pGADT7 (prey) plasmids (Clontech). BD-TLR3-TIR and AD-TLR3-TIR were constructed by inserting the cDNA fragment encoding the TIR domain of TLR3 into the pGBKT7 or pGADT7 plasmids. BD-TICAM-2 and AD-TICAM-2 were constructed by inserting the full-length TICAM-2 cDNA into the pGBKT7 or pGADT7 plasmids. SD-WLH is a yeast synthetic dextrose medium that lacks Trp, Leu, and His amino acids. SD-WLHA lacks adenine in addition to Trp, Leu, and His.
Confocal Microscopy—HeLa cells (1.0 × 105 cells/well) were plated onto micro cover glasses (Matsunami, Tokyo, Japan) in a 24-well plate. The following day, cells were transfected with the indicated plasmids using FuGENE HD (Roche Diagnostics). The total amount of DNA (0.5 μg/well) was kept constant by adding empty vector. In the cells transfected with wild-type TICAM-1 and the P434H mutant, z-VAD-fmk (20 μm) was added to the cells before transfection to inhibit apoptosis. Twenty-four hours after transfection, cells were fixed using 10% formaldehyde in PBS and permeabilized with PBS containing 0.2% Triton X-100 for 15 min. Fixed cells were blocked in PBS containing 1% bovine serum albumin and labeled with the indicated primary Abs (5 μg/ml) for 60 min at room temperature. Alexa-conjugated secondary Abs (1:400) were used to visualize staining of the primary Abs. Nuclei were stained with DAPI (2 μg/ml) in PBS for 10 min before mounting onto glass slides using PBS containing 2.3% 1,4-diazabicyclo[2.2.2]octane and 50% glycerol. Cells were visualized at a magnification of ×63 with an LSM510 META microscope (Zeiss, Jena, Germany).
Reporter Gene Assay—HEK293 cells (4 × 104 cells/well) cultured in 96-well plates were transfected with the expression vectors for wild-type TICAM-1, TICAM-1 mutants, or empty vector together with the reporter plasmid (30 ng/well) and an internal control vector, phRL-TK (Promega, Madison, WI) (1 ng/well) using the calcium phosphate method as described previously (26). The p-125 luc reporter contained the human IFN-β promoter region (-125 to +19) was provided by Dr. T. Taniguchi (University of Tokyo, Tokyo, Japan). The reporter plasmid containing the ELAM-1 promoter was previously constructed in our laboratory (27). The total amount of DNA (200 ng/well) was kept constant by adding empty vector. After 24 h, cells were lysed in lysis buffer (Promega), and the Firefly and Renilla luciferase activities were determined using a dual-luciferase reporter assay kit (Promega). The Firefly luciferase activity was normalized by Renilla luciferase activity and is expressed as the -fold stimulation relative to the activity in vector-transfected cells.
Immunoprecipitation and Immunoblotting—HEK293FT cells (5 × 105 cells/well) cultured in 6-well plates were transfected with the indicated plasmids using the calcium phosphate method. For the wild-type TICAM-1 and the TICAM-1 P434H mutant, z-VAD-fmk (20 μm) was added to the cells before transfection to inhibit apoptosis. The total amount of DNA (4.0 μg/well) was kept constant by adding empty vector. After 24 h, cells were lysed in lysis buffer (20 mm Tris-HCl (pH 7.5) containing 150 mm NaCl, 1% Nonidet P-40, 10 mm EDTA, 25 mm iodoacetamide, 2 mm phenylmethylsulfonyl fluoride, and a protease inhibitor mixture (Roche Applied Science)). Lysates were clarified by centrifugation, pre-cleared with Protein G-Sepharose (GE Healthcare, Buckinghamshire, UK), and incubated with 0.5-2.5 μg of Abs. The immunocomplexes were recovered by incubation with Protein G-Sepharose, washed three times with lysis buffer, and resuspended in denaturing buffer. Samples were analyzed by SDS-PAGE (7.5-10%) under reducing conditions followed by immunoblotting with anti-tag Abs.
RESULTS
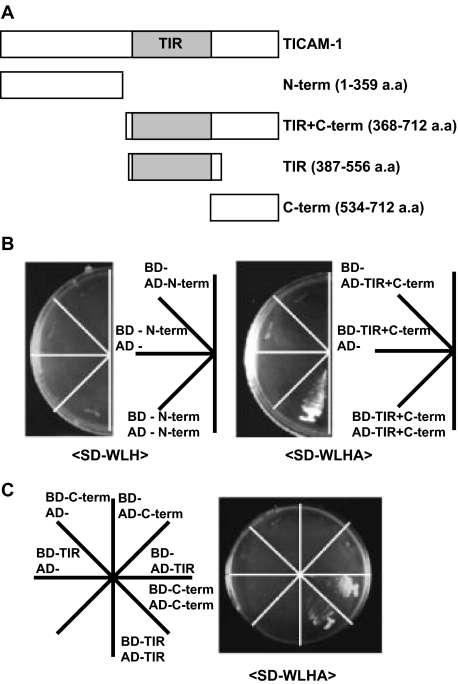
TICAM-1 Homodimerizes via the TIR Domain and C-terminal Region in Yeast—Upon dsRNA stimulation, TICAM-1 transiently associates with TLR3 before forming a speckle-like structure with downstream signaling molecules (25). The topological dynamics of TICAM-1 secondary to activation by dsRNA-TLR3 remains unknown, although currently accepted view is that the interaction between TLR3-TIR and TICAM-1-TIR is important for clustering the adaptor molecules around the receptor. Furthermore, overexpressed TICAM-1 forms speckle-like signalosomes in a TLR3-independent manner (25). To examine TICAM-1 oligomerization, we generated several truncated TICAM-1 constructs and assayed their homodimerization activities using the yeast two-hybrid system (Fig. 1A). The construct containing the TIR domain and C-terminal region of TICAM-1 induced homodimerization, whereas the construct containing the N-terminal region alone did not (Fig. 1B). Of the constructs that had been truncated further, only the TIR domain or C-terminal region was homodimerized in the yeast system (Fig. 1C) suggesting that TICAM-1 oligomerization is mediated through the TIR domain and C-terminal region.
FIGURE 1.
The homodimerization motifs of TICAM-1 are located in both the TIR domain and C-terminal region. A, construction of TICAM-1 mutants for the yeast two-hybrid assay. Each truncated TICAM-1 construct, N-term (1-359 aa), TIR+C-term (368-712 aa), TIR (387-556 aa), or C-term (534-712 aa), was inserted into the pGBKT7 (bait) and the pGADT7 (prey) vectors. The shaded box represents the TIR domain of TICAM-1 (394-533 aa). B and C, homodimerization of the TICAM-1 mutants in yeast. The TICAM-1 mutant containing the N-terminal region (N-term) failed to homodimerize (B, left panel), whereas the TICAM-1 mutant containing the TIR domain and C-terminal region (TIR+C-term) was homodimerized in yeast (B, right panel). The TICAM-1 mutants containing only the TIR domain or C-terminal region were homodimerized in the WLHA plate (C). BD and AD represent the bait and prey plasmids, respectively. SD-WLH reflects a weak interaction; SD-WLHA reflects a strong interaction.
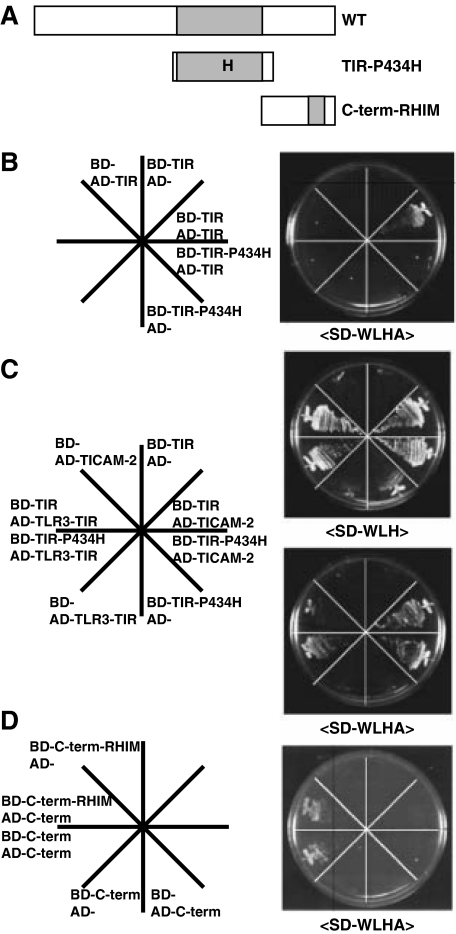
TICAM-1 Dimerization Requires the Pro434 Residue in the TIR Domain and the C-terminal Region with the Exception of the RHIM Domain—We have previously reported that the TIR domain TIR-P434H mutant acts as a dominant-negative form during TLR3-mediated signaling (7). In the yeast two-hybrid system, TIR-P434H failed to interact with the TIR domain of TICAM-1 but bound tightly to the tail of TLR3 or TICAM-2 (Figs. 2, A-C). These findings suggest that Pro434 is critical for the TICAM-1 signaling that mediates TICAM-1 dimerization via the TIR domain. To further identify residues required for mediating TICAM-1 dimerization through the C-terminal region, we generated a TICAM-1 construct consisting of the C-terminal region with a mutated-RHIM domain (Fig. 2A). The RHIM domain is essential for TICAM-1-mediated NF-κB activation and induction of apoptosis via binding of RIP1 (14, 15). Mutation of the RHIM motif did not affect the dimerization of the C-terminal region (Fig. 2D) suggesting that the RHIM motif is dispensable for TICAM-1 dimerization via the C-terminal region.
FIGURE 2.
Proline 434 in the TIR domain is critical for homodimerization of TICAM-1 but not for association with TLR3 or TICAM-2. A, construction of TICAM-1 mutants for the yeast two-hybrid assay. The constructs of the TICAM-1 mutant, TIR-P434H (387-556 aa, Pro434 in the TIR domain is replaced by His) and C-term-RHIM (534-712 aa, 587VQLG690 within the RHIM domain are replaced with four Ala) were inserted into the pGBKT7 (bait) and the pGADT7 (prey) vectors. B and C, interactions between the TICAM-1mutant (TIR-P434H) and wild-type TICAM-1-TIR (TIR), TLR3-TIR, or TICAM-2 in the yeast two-hybrid system. No association was detected between the mutated TIR (TIR-P434H) and wild-type TICAM-1-TIR (B), whereas strong association was observed between the TIR-P434H mutant and the TIR of TLR3 (755-904 aa) or TICAM-2 (C). D, interaction between the C-terminal of TICAM-1 with its RHIM mutant. Association between the RHIM mutant and wild-type C-terminal of TICAM-1 was observed. BD and AD represent the bait and prey plasmids, respectively.
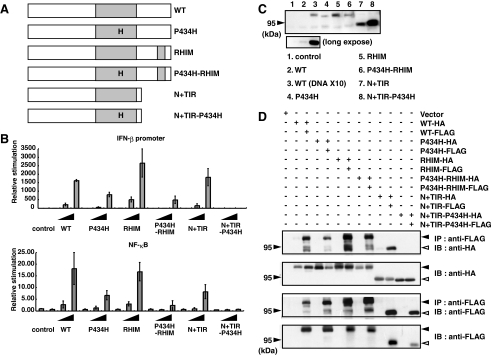
TICAM-1-mediated IRF-3 and NF-κB Activation Is Reduced by Mutations in the TIR or RHIM Domain—Based on the results of the yeast-two hybrid assay, we constructed several TICAM-1 mutants and examined their activities in mammalian cells (Fig. 3A). Only the TICAM-1 N+(TIR-P434H) mutant failed to activate both the IFN-β promoter and NF-κB, even after transfection of high concentrations of plasmid (Fig. 3B). The other mutants retained their IFN-β promoter and NF-κB activating abilities. However, transfection of the same concentration of plasmid resulted in increased protein expression of all TICAM-1 mutants compared with wild-type TICAM-1 protein expression (Fig. 3C). Subsequent normalization of the protein expression levels revealed that the signaling activities of these mutants were very weak compared with wild-type TICAM-1.
FIGURE 3.
Pro434 in the TIR domain and the C-terminal RHIM domain are required for full activation of TICAM-1. A, constructs of various TICAM-1 mutants were used. The mutant P434H construct contains a mutated TIR domain in which Pro434 is substituted with His. The mutant RHIM construct contains a mutated RHIM domain (587VQLG690 are replaced with four Ala). The mutant P434H-RHIM construct has two mutated sites, Pro434 and RHIM. The mutant N+TIR (1-566 aa) contains the N-terminal region and the TIR domain and the mutant N+(TIR-P434H) contains the N-terminal region with the mutated TIR-P434H domain. B, Pro434 in the TIR domain and the RHIM domain play an important role in TICAM-1-mediated NF-κB and IFN-β promoter activation. HEK293 cells in 96-well plates were transfected with the indicated expression plasmids (0.2, 2.0, and 20.0 ng) together with the IFN-β promoter reporter (upper panel, 100 ng) or NF-κB reporter plasmids (lower panel, 100 ng), and phRL-TK (1.0 ng). Twenty-four hours after transfection, the luciferase reporter activities were measured. The average activities from three independent assays are shown as relative stimulation. C, HEK293 cells in 24-well plates were transfected with expression plasmids for HA-tagged wild-type TICAM-1 (20 and 200 ng) or each TICAM-1 mutant (20 ng) in the presence of an apoptosis inhibitor. Protein expression levels were determined by immunoblotting using anti-HA pAb. The protein levels of the TICAM-1 mutants are significantly higher than that of wild-type TICAM-1 using the same concentration of DNA plasmid. Lower panel, long exposed membrane. D, the homodimerizing ability of the TICAM-1 mutants was assessed using a co-immunoprecipitation assay. HEK293 cells were transfected with the indicated plasmids for HA- or FLAG-tagged TICAM-1 mutants, and cell lysates were immunoprecipitated with anti-FLAG mAb. Immunoprecipitants were resolved on SDS-PAGE under reducing conditions followed by immunoblotting with anti-HA pAb or anti-FLAG mAb. The protein expression in the total cell lysates was analyzed by immunoblotting with anti-HA pAb or anti-FLAG mAb (IB). Only the N+(TIR-P434H) mutant did not homodimerized (upper panel). Closed arrowheads indicate full-length TICAM-1, and open arrowheads indicate the truncated form of TICAM-1 (N+TIR).
To examine the relationship between the reduced NF-κB and IFN-β promoter activation of each mutant and their homodimerizing activity, immunoprecipitation studies using HA- and FLAG-tagged constructs were performed. The homodimerizing activity was completely diminished in the dysfunctional N+(TIR-P434H) mutant (Fig. 3D). As observed in the yeast system, either the TIR domain or the C-terminal region enabled the TICAM-1 mutants to homodimerize in HEK293 cells. However, lack of either domain reduced the signaling activities of these mutants. These results suggest that full activation of TICAM-1 requires homodimerization at both the TIR domain and the C-terminal region.
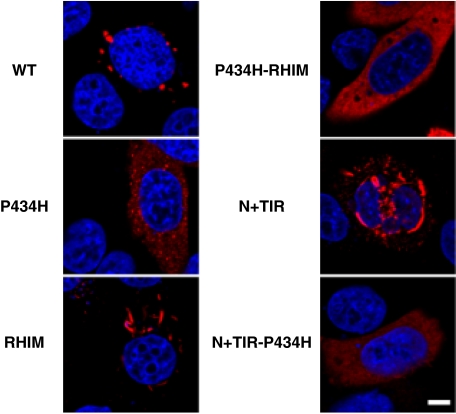
Mutations of TICAM-1 Affect Its Subcellular Localization—To clarify the functional differences between wild-type TICAM-1 and the TICAM-1 mutants, we examined the subcellular localization of each FLAG-tagged mutant by confocal microscopy. As reported previously, we found that overexpression of wild-type TICAM-1 in HeLa cells leads to spontaneous activation of TICAM-1 and formation of speckle-like signalosomes (25) (Fig. 4, upper left panel). In contrast, the TICAM-1 P434H mutant localized diffusely in the cytosol with slight speckle formation (Fig. 4, middle left panel). A fiber-like staining pattern was observed in cells overexpressing the RHIM and N+TIR mutants, which contain the intact TIR domain but not the RHIM domain (lower left and middle right panels). The double P434H-RHIM mutant also localized diffusely with slight fiber-like formation (upper right panel). Finally, overexpression of the dysfunctional N+(TIR-P434H) mutant resulted in a completely diffuse staining pattern (lower right panel). These results suggest that Pro434 in the TIR domain is a critical for accumulation of TICAM-1 and formation of the fiber-like structures, whereas the RHIM domain is involved in the speckle formation of TICAM-1.
FIGURE 4.
Subcellular localization of TICAM-1 mutants. HeLa cells were transfected with the indicated plasmids for FLAG-tagged wild-type TICAM-1 and TICAM-1 mutants and stained with anti-FLAG mAb followed by Alexa568-labeled goat anti-mouse Ab (red). Mutation of Pro434 in the TIR domain or mutation of the C-terminal RHIM domain affects the subcellular localization of TICAM-1. Nuclei were stained with DAPI (blue). Bar, 10 μm.
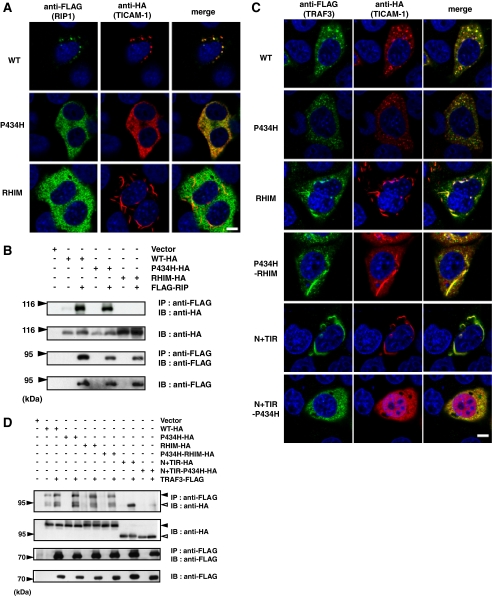
Association of RIP1 and TRAF3 with TICAM-1—To examine the relationship between the reduced activity of the TICAM-1 mutants and changes in their subcellular localization, we analyzed the co-localization of the downstream signaling molecules, RIP1 and TRAF3, with the TICAM-1 mutants by confocal microscopy and immunoprecipitation studies. RIP1 directly associates with TICAM-1 via the RHIM domain leading to activation of NF-κB (14, 15). When HeLa cells were co-transfected with FLAG-tagged RIP1 together with each of the HA-tagged TICAM-1 mutants, we found that RIP1 co-localized with wild-type TICAM-1 and the P434H mutant but not with the RHIM mutant (Fig. 5A). The association of RIP1 with these TICAM-1 mutants was confirmed by immunoprecipitation (Fig. 5B). As expected, the TICAM-1 mutants containing the mutated RHIM domain (P434H-RHIM) or a deleted C-terminal region (N+TIR) did not interact with RIP1 (data not shown).
FIGURE 5.
Association of RIP1 and TRAF3 with the TICAM-1 mutants. A, confocal images of HeLa cells co-expressing FLAG-tagged RIP1 and HA-tagged TICAM-1 mutants. Cells were fixed and stained with anti-FLAG mAb and anti-HA pAb, followed by Alexa488-labeled goat anti-rabbit Ab and Alexa568-labeled goat anti-mouse Ab. RIP1 co-localizes with wild-type TICAM-1 and the P434H mutant. Green, RIP1; red, wild-type and mutated TICAM-1; blue, DAPI-stained nuclei. Bar, 10 μm. B, the RHIM domain of TICAM-1 is essential for association with RIP1. 293FT cells were transfected with the indicated HA-tagged TICAM-1 mutants and FLAG-tagged RIP1. After 24 h, cells were lysed and RIP1 was immunoprecipitated with anti-FLAG mAb. The immunoprecipitants were resolved on SDS-PAGE followed by immunoblotting with anti-HA pAb or anti-FLAG mAb. Total cell lysates were subjected to immunoblotting with anti-HA pAb or anti-FLAG mAb to detect protein expression (IB). C, confocal images of HeLa cells expressing FLAG-tagged TRAF3 and HA-tagged TICAM-1 mutants. HeLa cells were transfected with the indicated HA-tagged TICAM-1 mutants and FLAG-tagged TRAF3, and then stained with anti-FLAG mAb and anti-HA pAb, followed by Alexa488-labeled goat anti-rabbit Ab and Alexa568-labeled goat anti-mouse Ab. All TICAM-1 mutants except for N+TIR-P434H associate with TRAF3. Green, TRAF3; red, wild-type and mutated TICAM-1; blue, DAPI-stained nuclei. Bar, 10 μm. D, co-immunoprecipitation of TRAF3 with the TICAM-1 mutants. 293FT cells were transfected with the indicated HA-tagged TICAM-1 mutants and FLAG-tagged TRAF3. After 24 h, cells were lysed and TRAF3 was immunoprecipitated using an anti-FLAG mAb. Samples were resolved on SDS-PAGE followed by immunoblotting with anti-HA Ab or anti-FLAG mAb. Total cell lysates were subjected to immunoblotting with anti-HA pAb or anti-FLAG mAb to detect protein expression (IB).
Next we analyzed the association of the TICAM-1 mutants with TRAF3, a downstream signaling molecule that is essential for TICAM-1-mediated IRF-3 activation (17, 18). In contrast to RIP1, all of the TICAM-1 mutants with the exception of the dysfunctional N+(TIR-P434H) mutant, co-localized strongly with TRAF3 irrespective of their various localization patterns (Fig. 5C). Similar association profiles were observed in the immunoprecipitation studies (Fig. 5D), indicating that the mutation of the TIR and RHIM domains barely affected the recruitment of TRAF3. Our data suggest that, once TICAM-1 is oligomerized either through the TIR domain or the C-terminal region, TRAF3 is recruited to the N-terminal region of TICAM-1. Thus, association of TICAM-1 with RIP1 and TRAF3 is necessary but not sufficient for TICAM-1-mediated signaling.
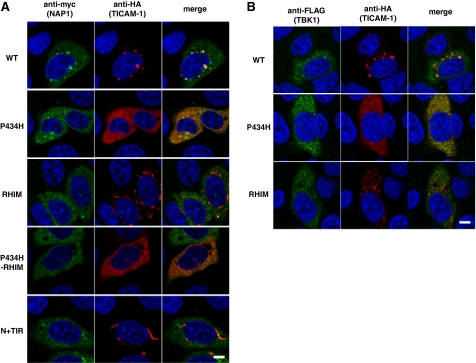
NAP1 and TBK1 Are Recruited to the TICAM-1 Speckle-like Signalosomes—NAP1 has been identified as an essential molecule for TLR3- and RIG-1/MDA-5-mediated IRF-3 activation (16, 23). Because NAP1 directly binds to TBK1, an IRF-3-activating kinase (28), it is possible that NAP1 functions downstream of TRAF3. We examined the localization profile of NAP1 and TBK1 in cells co-expressing the TICAM-1 mutants. Importantly, NAP1 co-localized partially with only wild-type TICAM-1 (Fig. 6A). None of the TICAM-1 mutants used in this study co-localized with NAP1 even if the mutants were able to recruit TRAF3 (Fig. 6A). In addition, TBK1 also co-localized only with wild-type TICAM-1 (Fig. 6B). These results suggest that oligomerization induced by the TIR domain and the C-terminal region in conjunction with RIP1 binding is required for speckle formation and recruitment of the IRF-3 kinase complexes that lead to effective activation of IRF-3 and NF-κB.
FIGURE 6.
Pro434 and the RHIM domain are indispensable for recruitment of NAP1 and TBK1 to TICAM-1. Confocal images show HeLa cells co-expressing HA-tagged TICAM-1 mutants and Myc-tagged NAP1 (A) or FLAG-tagged TBK1 (B). A, HeLa cells, transfected with the indicated HA-tagged TICAM-1 mutants and Myc-tagged NAP1, were stained with anti-Myc mAb and anti-HA pAb, followed by Alexa488-labeled goat anti-rabbit Ab and Alexa568-labeled goat anti-mouse Ab. B, HeLa cells, transfected with the indicated HA-tagged TICAM-1 mutants and FLAG-tagged TBK1, were stained with anti-FLAG mAb and anti-HA pAb, followed by Alexa488-labeled goat anti-rabbit Ab and Alexa568-labeled goat anti-mouse Ab. Green, NAP1 (A) and TBK1 (B); red, wild-type and mutated TICAM-1; blue, DAPI-stained nuclei. Bar, 10 μm.
DISCUSSION
TLR3-TICAM-1 signaling results in transcriptional activation of various genes, including the type I IFN and IFN-regulatory genes. In this study, we examined the molecular mechanism of TICAM-1-mediated signaling and demonstrated that TICAM-1 acts as a platform for recruitment of signaling molecules after homo-oligomerization at the TIR domain and C-terminal region. We show that Pro434 in the BB loop of the TIR domain is critical for TICAM-1 homo-oligomerization. Furthermore, the C-terminal region plays an important role in TICAM-1 signaling by inducing homo-dimerization and RIP1 recruitment to the RHIM domain. These results bring new insight into the molecular dynamics of TICAM-1 signaling and regulation of TLR3/4-mediated type I IFN production.
TLR signaling is mediated through TIR-containing adaptor molecules. The conserved proline residue in the BB loop of the TLR-TIR domain is crucial for binding the adaptor molecule (29). The TLR4 mutant P714H fails to bind TICAM-2 resulting in signaling off of TLR4-mediated signaling (9), whereas the dysfunctional TLR3 mutant A795H looses its TICAM-1-recruiting ability (7). Importantly, the TICAM-2-TIR mutant C117H bound the TLR4-TIR but failed to assemble homo (TICAM-2-TICAM-2)- and hetero (TICAM-2-TICAM-1)-dimers (9). Here, we show that Pro434 in the TIR domain of TICAM-1 is crucial for mediating interactions with TICAM-1-TIR but not with TLR3-TIR or TICAM-2-TIR. Thus, during TLR3/4-TICAM-1 signaling the BB loop of the upstream TIR determines the association with the downstream TIR domain.
TIR-containing adaptor molecules exhibit different molecular dynamics in response to stimulation by different TLRs. MyD88 is recruited to the plasma membrane or endosome and forms a signaling complex with receptors (30-32). Mal/TIRAP and TICAM-2/TRIF-related adaptor molecule, which associate with the plasma membrane via a phosphatidylinositol 4,5-bisphosphate-binding domain or myristoylation, respectively, act as a sorting adaptor to recruit MyD88 or TICAM-1 (33-35). During TLR4-mediated signaling, TICAM-1 may dissociate from the upstream adaptor TICAM-2 to form speckle-like signalosomes that induce type I IFN as observed during TLR3 signaling (25). Here, we show that TICAM-1 has two homo-dimerization motifs in the TIR domain and the C-terminal region. MyD88 also has the protein-protein interaction domain, the TIR and N-terminal death domain (36). Low molecular weight compounds that disrupt the TIR-TIR interaction of MyD88 have been synthesized to inhibit MyD88 signaling (37, 38). Homo-dimerization motifs of TICAM-1 would be good targets for designing chemical compounds that specifically block TICAM-1 signaling.
Several reports suggest that the C-terminal region of TICAM-1 is important for NF-κB activation and apoptosis in association with RIP1 binding to the RHIM domain but dispensable for IRF-3 activation (14, 15). In our study, decreased NF-κB and IRF-3 activation was observed in cells overexpressing the TICAM-1 mutants with either a mutated RHIM motif or a deleted C-terminal region compared with cells overexpressing wild-type TICAM-1. These observations are consistent with a previous study using RIP1-deficient cells (39). In the presence of an apoptosis inhibitor, transfection of the same concentration of plasmid resulted in increased protein expression levels of these TICAM-1 mutants compared with wild-type TICAM-1. Why protein expression of the TICAM-1 mutants is higher than that of wild-type TICAM-1 remains question. However, it is possible that wild-type TICAM-1 is rapidly degraded via some protein modifications. RIP1 binding appears to be required for not only NF-κB activation but also IRF-3 activation. Indeed, our immunofluorescence data demonstrate that RIP1 has a critical role in the recruitment of NAP1 and TBK1 to the TICAM-1 speckle-like signalosomes. Because RIP1 associated with the TICAM-1 P434H mutant, it is possible that C-terminal region-mediated homodimerization may trigger RIP1 recruitment.
The TICAM-1 mutant (N+(TIR-P434H)) that lacks both homodimerization motifs failed to homodimerize resulting in abolishment its activity. However, mutants that lacked one homodimerization motif (P434H, P434H-RHIM, and N+TIR) were able to induce homodimerization. Remarkably, these mutants as well as the RHIM mutant recruited TRAF3 but not NAP1 or TBK1. TRAF3 has recently been identified as an essential molecule for TICAM-1-medated IRF-3 activation but not for NF-κB activation (17, 18). Direct interactions between TRAF3 and TICAM-1 were not observed in the yeast two-hybrid system (data not shown). Because TICAM-1 has a TRAF6 binding site at the N-terminal and TRAF2 also directly interacts with the N-terminal region of TICAM-1 (11, 40),6 TRAF3 appears to form a molecular complex with TICAM-1 via TRAF2/6. In contrast to TRAF3, NAP1 and TBK1 co-localized partially with only wild-type TICAM-1. NAP1 has been identified as a TBK1-binding protein (28) that is involved in the recruitment of TBK1 to the N-terminal region of TICAM-1 (16). Because the TICAM-1 mutants, P434H and RHIM, did not co-localize with NAP1 and TBK1, oligomerization at two different sites in conjunction with RIP1 binding should be required for recruitment of the IRF-3 kinase complex. According to a proteolytic analysis, TICAM-1 possessed the protease-resistant structural domain at the N terminus. This domain was followed by the regions interacted with TRAF6, TRAF2, and TBK1. The TICAM-1 mutant that deleted the structural domain retained full IFN-inducing activity compared with wild-type TICAM-1.7 One possible explanation why oligomerized TICAM-1 rather than monomer TICAM-1 can recruit downstream molecules is that in the monomer TICAM-1 the region associating with downstream signaling molecules is covered with the N-terminal structural domain. Once TICAM-1 is oligomerized at the TIR and C-terminal domains, the N-terminal region is exposed to interact with downstream molecules. The mechanism of how TICAM-1 recruits the IRF-3 kinase complex requires further investigation. Recent report suggests that TRAF3 ubiquitination facilitates the recruitment of TBK1 (41). We have also observed TICAM-1 phosphorylation and ubiquitination.6 Structural analysis of the TICAM-1 molecule and characterization of protein modifications of signaling components are important for understanding the molecular topology for initiation of TICAM-1-mediated signaling.
Acknowledgments
We are grateful to M. Shingai, T. Ebihara, A. Ishii, and A. Matsuo for critical discussion. Thanks are also due to Dr. M. Nakanishi (Nagoya City University, Nagoya) and Dr. T. Taniguchi (Tokyo University, Tokyo) for providing plasmids.
This work was supported by Core Research for Evolutional Science and Technology, Japan Science and Technology Corporation, and by Grants-in-Aid from the Ministry of Education, Science, and Culture (Specified Project for Advanced Research) and the hepatitis C virus project of the National Institutes of Health of Japan, and by the Uehara Memorial Foundation, Mitsubishi Foundation, Takeda Foundation, and NorthTec Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TLR, Toll-like receptor; CARD, caspase recruitment domain: DAPI, 4′,6-diamidine-2′-phenylindole; dsRNA, double-stranded RNA; HA, hemagglutinin; IFN-β, interferon-β; IRF, IFN regulatory factor; MDA5, melanoma differentiation-associated gene 5; NAP1, NF-κB-activating kinase-associated protein 1; RHIM, RIP homotypic interacting motif; RIG-I, retinoic acid-inducible gene I; RIP1, receptor-interacting protein 1; TBK1, TANK-binding kinase 1; TICAM-1, TIR-containing adaptor molecule-1; TIR, Toll-IL-1 receptor; TRAF, tumor necrosis factor receptor-associated factor; TRIF, TIR domain-containing adaptor-inducing IFN-β; Ab, antibody; mAb, monoclonal antibody; pAb, polyclonal antibody; z, benzyloxycarbonyl; fmk, fluoromethyl ketone; aa, amino acid(s); PBS, phosphate-buffered saline.
M. Sasai, H. Oshiumi, M. Matsumoto, and T. Seya, manuscript in preparation.
M. Matsumoto, M. Sasai, and T. Seya, unpublished data.
References
- 1.Akira, S., Uematsu, S., and Takeuchi, O. (2006) Cell 124 783-801 [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S., and Fujita, T. (2004) Nat. Immunol. 5 730-737 [DOI] [PubMed] [Google Scholar]
- 3.Alexopoulou, L., Holt, A. C., Medzhitov, R., and Flavell, R. A. (2001) Nature 413 732-738 [DOI] [PubMed] [Google Scholar]
- 4.Matsumoto, M., Kikkawa, S., Kohase, M., Miyake, K., and Seya, T. (2002) Biochem. Biophys. Res. Commun. 293 1364-1369 [DOI] [PubMed] [Google Scholar]
- 5.Schulz, O., Diebold, S. S., Chen, M., Naslund, T. I., Nolte, M. A., Alexopoulou, L., Azuma, Y. T., Flavell, R. A., Liljestrom, P., and Reis e Sousa, C. (2005) Nature 433 887-892 [DOI] [PubMed] [Google Scholar]
- 6.Akazawa, T., Ebihara, T., Okuno, M., Okuda, Y., Shingai, M., Tsujimura, K., Takahashi, T., Ikawa, M., Okabe, M., Inoue, N., Okamoto-Tanaka, M., Ishizaki, H., Miyoshi, J., Matsumoto, M., and Seya, T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 252-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oshiumi, H., Matsumoto, M., Funami, K., Akazawa, T., and Seya, T. (2003) Nat. Immunol. 4 161-167 [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto, M., Sato, S., Hemmi, H., Hoshino, K., Kaisho, T., Sanjo, H., Takeuchi, O., Sugiyama, M., Okabe, M., Takeda, K., and Akira, S. (2003) Science 301 640-643 [DOI] [PubMed] [Google Scholar]
- 9.Oshiumi, H., Sasai, M., Shida, K., Fujita, T., Matsumoto, M., and Seya, T. (2003) J. Biol. Chem. 278 49751-49762 [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald, K. A., Rowe, D. C., Barnes, B. J., Caffrey, D. R., Visintin, A., Latz, E., Monks, B., Pitha, P. M., and Golenbock, D. T. (2003) J. Exp. Med. 198 1043-1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato, S., Sugiyama, M., Yamamoto, M., Watanabe, Y., Kawai, T., Takeda, K., and Akira, S. (2003) J. Immunol. 171 4304-4310 [DOI] [PubMed] [Google Scholar]
- 12.Sharma, S., tenOever, B. R., Grandvaux, N., Zhou, G. P., Lin, R., and Hiscott, J. (2003) Science 300 1148-1151 [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald, K. A., McWhirter, S. M., Faia, K. L., Rowe, D. C., Latz, E., Golenbock, D. T., Coyle, A. J., Liao, S. M., and Maniatis, T. (2003) Nat. Immunol. 4 491-496 [DOI] [PubMed] [Google Scholar]
- 14.Meylan, E., Burns, K., Hofmann, K., Blancheteau, V., Martinon, F., Kelliher, M., and Tschopp, J. (2004) Nat. Immunol. 5 503-507 [DOI] [PubMed] [Google Scholar]
- 15.Kaiser, W. J., and Offermann, M. K. (2005) J. Immunol. 174 4942-4952 [DOI] [PubMed] [Google Scholar]
- 16.Sasai, M., Oshiumi, H., Matsumoto, M., Inoue, N., Fujita, F., Nakanishi, M., and Seya, T. (2005) J. Immunol. 174 27-30 [DOI] [PubMed] [Google Scholar]
- 17.Hacker, H., Redecke, V., Blagoev, B., Kratchmarova, I., Hsu, L. C., Wang, G. G., Kamps, M. P., Raz, E., Wagner, H., Hacker, G., Mann, M., and Karin, M. (2006) Nature 439 204-207 [DOI] [PubMed] [Google Scholar]
- 18.Oganesyan, G., Saha, S. K., Guo, B., He, J. Q., Shahangian, A., Zarnegar, B., Perry, A., and Cheng, G. (2006) Nature 439 208-211 [DOI] [PubMed] [Google Scholar]
- 19.Kawai, T., Takahashi, K., Sato, S., Coban, C., Kumar, H., Kato, H., Ishii, K. J., Takeuchi, O., and Akira, S. (2005) Nat. Immunol. 6 981-988 [DOI] [PubMed] [Google Scholar]
- 20.Seth, R. B., Sun, L., Ea, C. K., and Chen, Z. J. (2005) Cell 122 669-682 [DOI] [PubMed] [Google Scholar]
- 21.Meylan, E., Curran, J., Hofmann, K., Moradpour, D., Binder, M., Bartenschlager, R., and Tschopp, J. (2005) Nature 437 1167-1172 [DOI] [PubMed] [Google Scholar]
- 22.Xu, L. G., Wang, Y. Y., Han, K. J., Li, L. Y., Zhai, Z., and Shu, H. B. (2005) Mol. Cell 19 727-740 [DOI] [PubMed] [Google Scholar]
- 23.Sasai, M., Shingai, M., Funami, K., Yoneyama, M., Fujita, T., Matsumoto, M., and Seya, T. (2006) J. Immunol. 177 8676-8683 [DOI] [PubMed] [Google Scholar]
- 24.Saha, S. K., Pietras, E. M., He, J. Q., Kang, J. R., Liu, S. Y., Oganesyan, G., Shahangian, A., Zarnegar, B., Shiba, T. L., Wang, Y., and Cheng, G. (2006) EMBO J. 25 3257-3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Funami, K., Sasai, M., Ohba, Y., Oshiumi, H., Seya, T., and Matsumoto, M. (2007) J. Immunol. 179 6867-6872 [DOI] [PubMed] [Google Scholar]
- 26.Chen, C., and Okayama, H. (1987) Mol. Cell. Biol. 7 2745-2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsujita, T., Tsukada, H., Nakao, M., Oshiumi, H., Matsumoto, M., and Seya, T. (2004) J. Biol. Chem. 279 48588-48597 [DOI] [PubMed] [Google Scholar]
- 28.Fujita, F., Taniguchi, Y., Kato, T., Narita, Y., Furuya, A., Ogawa, T., Sakurai, H., Joh, T., Itoh, M., Delhase, M., Karin, M., and Nakanishi, M. (2003) Mol. Cell. Biol. 23 7780-7793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu, Y., Tao, X., Shen, B., Horng, T., Medzhitov, R., Manley, J. L., and Tong, L. (2000) Nature 408 111-115 [DOI] [PubMed] [Google Scholar]
- 30.Latz, E., Visintin, A., Lien, E., Fitzgerald, K. A., Monks, B. G., Kurt-Jones, E. A., Golenbock, D. T., and Espevik, T. (2002) J. Biol. Chem. 277 47834-47843 [DOI] [PubMed] [Google Scholar]
- 31.Latz, E., Schoenemeyer, A., Visintin, A., Fitzgerald, K. A., Monks, B. G., Knetter, C. F., Lien, E., Nilsen, N. J., Espevik, T., and Golenbock, D. T. (2004) Nat. Immunol. 5 190-198 [DOI] [PubMed] [Google Scholar]
- 32.Honda, K., Yanai, H., Mizutani, T., Negishi, H., Shimada, N., Suzuki, N., Ohba, Y., Takaoka, A., Yeh, W. C., and Taniguchi, T. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15416-15421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kagan, J. C., and Medzhitov, R. (2006) Cell 125 943-955 [DOI] [PubMed] [Google Scholar]
- 34.Rowe, D. C., McGettrick, A. F., Latz, E., Monks, B. G., Gay, N. J., Yamamoto, M., Akira, S., O'Neill, L. A., Fitzgerald, K. A., and Golenbock, D. T. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 6299-6304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fitzgerald, K. A., and Chen, Z. J. (2006) Cell 125 834-836 [DOI] [PubMed] [Google Scholar]
- 36.Burns, K., Martinon, F., Esslinger, C., Pahl, H., Schneider, P., Bodmer, J. L., Di Marco, F., French, L., and Tschopp, J. (1998) J. Biol. Chem. 273 12203-12209 [DOI] [PubMed] [Google Scholar]
- 37.Davis, C. N., Mann, E., Behrens, M. M., Gaidarova, S., Rebek, M., Rebek, J., Jr., and Bartfai, T. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 2953-2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loiarro, M., Capolunghi, F., Fanto, N., Gallo, G., Campo, S., Arseni, B., Carsetti, R., Carminati, P., De Santis, R., Ruggiero, V., and Sette, C. (2007) J. Leukoc. Biol. 82 801-810 [DOI] [PubMed] [Google Scholar]
- 39.Cusson-Hermance, N., Khurana, S., Lee, T. H., Fitzgerald, K. A., and Kelliher, M. A. (2005) J. Biol. Chem. 280 36560-36566 [DOI] [PubMed] [Google Scholar]
- 40.Jiang, Z., Mak, T. W., Sen, G., and Li, X. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 3533-3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kayagaki, N., Phung, Q., Chan, S., Chaudhari, R., Quan, C., O'Rourke, K. M., Eby, M., Pietras, E., Cheng, G., Bazan, J. F., Zhang, Z., Arnott, D., and Dixit, V. M. (2007) Science 318 1628-1632 [DOI] [PubMed] [Google Scholar]