Exploration of artificial, nanometer-scale features to mimic natural matrices has revealed a powerful influence of nano-topography over cellular behavior. In this study, aligned nanofibers composed of thermally responsive hydroxybutyl chitosan were electrospun to create a robust scaffold for the production of aligned cell sheets. With a median fiber diameter of 436 nm, fiber alignment created repetitively spaced nanometer-scale topographical features on the scaffold surface. When cultured on these surfaces, human mesenchymal stem cells (hMSC) showed alignment and elongation in both cell body and nucleus. In addition to morphological changes, topographical features induced expression of genes indicative of myogenic induction of hMSC cultured in proliferative, non-differentiating medium. Thermal reversibility of the fibers allows for the dissolution of the polymer from the cell/scaffold construct without disruption of cytoskeletal structure and cell–cell interactions. For cell-based regenerative medicine applications, thermally reversible nanofibers can thus produce polymer-free cell layers engineered with exposure to nano-topographical cues.
Although the use of polymeric scaffolds in the building of engineered tissues has become a necessary foundation, the production of polymer-free tissue constructs has gained much attention in recent literature, mainly in the form of cell sheet engineering. Okano et al., have published extensively on the use of a temperature-responsive polymer, poly(N-isopropyl-acrylamide) (PIPAAm), to create surfaces that can release cell sheets for use in tissue repair.[1–3] By eliminating the scaffold component prior to implantation, these cell sheets circumvent much of the problematic aspects of polymer presence in engineered tissues in vivo, such as lack of biocompatibility, non-ideal degradation profiles, hindrance to mass transport and cell–cell contact, and mismatched mechanical properties. However, this system is limited to surface grafting of PIPAAm and lacks the flexibility to include topographical cues during the culture period, prior to cell sheet formation. Many tissues are comprised of layered structures with cells highly organized within each layer, such as the myocardium, vascular wall, and corneal epithelium. Therefore, the introduction of topographical cues to create an aligned cell sheet may have a tremendous impact on engineering of tissue constructs for regenerative medicine applications.
Natural matrices have been characterized to contain topographical features at the nanometer scale.[4,5] Artificial surfaces with nanometer-scale topographical cues have been shown to be potent effectors of cellular behavior.[6–9] Electrospinning has proved a versatile technique in creating nanometer-scale fibers with a wide range of artificial and natural polymers.[10–16] These electrospun fibers can be aligned by manipulating the electrical field or by collecting the fibers on a rotating target, creating a culture surface endowed with topographical cues at the nanometer-scale.
With electrospinning, we were able to fabricate aligned nanofibrous scaffolds composed of a thermally responsive hydroxybutyl chitosan (HBC). HBC is a modified form of chitosan, and previous work in our lab has shown it to be a biocompatible scaffold suitable for tissue engineering applications.[17] In addition to the topographical cues, the temperature responsiveness allows for dissolution of the fibers once they are cooled to temperatures below its lower critical solution temperature (LCST). We used these scaffolds to provide, for hMSC, a culture environment capable of presenting nanometer-scale topographical cues with potential to modulate cell behavior. Cellular alignment is seen as a critical factor in providing functional competence to many tissue types. Cell and nuclear shapes have been reported to be intimately associated with intracellular signaling cascades for control of gene expression.[18–20] Control of stem cell differentiation has been a challenge in regenerative medicine. With our unique scaffold, we investigated the possibility that modulating cell shape could contribute to directing differentiative fate. After the stem cells are exposed to such an environment and phenotypically and genotypically altered, it may be possible to recover them as a cell sheet for implantation and tissue augmentation. In this study we characterize the hMSC behavior cultured on the HBC nanofibrous scaffold.
Imaging the fibers with scanning electron microscopy (SEM) revealed a scaffold of relatively uniform fiber diameters exhibiting a textured surface in contrast to that of HBC films (Fig. 1). Collagen type I could be blended into the HBC solution and spun into fibers in order to present a more biologically active surface as compared to the HBC fiber. These nanofibers exhibit diameters ranging from less than 200 nm to 900 nm. HBC fibers and HBC/collagen blended fibers show a median diameter of 463 nm and 606 nm, respectively. The addition of collagen increases the average diameter of the fibers by several hundreds of nanometers and reduces the surface tension of the scaffold by nearly 50 % (data not shown). Fibers with diameters greater than 900 nm appear to be the result of fusion of two or more fibers. A larger proportion of fibers within the collagen blended HBC fiber scaffolds show signs of fusion as compared to HBC alone. This may be attributed to the addition of aqueous collagen solution leading to slower evaporation of solvent during fiber deposition.
Figure 1.
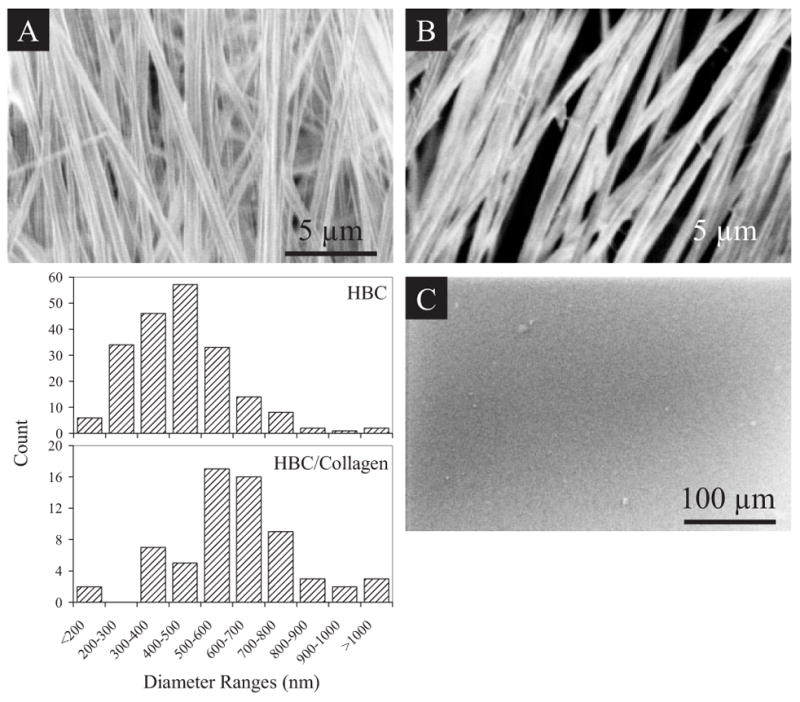
Morphologic and surface analysis of HBC and HBC/collagen blend nanofibrous matrices. SEM images of A) HBC nanofibers, B) HBC/collagen blend nanofibers, and C) HBC film are shown. Both scaffolds can be handled without tearing. Histogram shows differences in the distribution of fiber sizes between HBC and HBC/collagen fibers, with collagen blended fibers exhibiting an increase in diameter as compared to the HBC fibers. Fibers with diameters greater than 900 nm appear to be resultant of fiber fusion during deposition.
The HBC fibers can be dissolved immediately upon contact with a medium that is cooled to 4 °C to leave behind a polymer-free cell sheet. Figure 2A–D shows the appearance of the cell sheet before and after cooling away of the HBC (Fig. 2A and B) and HBC/collagen blended (Fig. 2C and D) fibers. The hMSC in the polymer-free cell sheet retain their elongated cell morphology and cytoskeletal alignment (Fig. 2E–H). After 2 weeks of culture on the nanofibers, hMSC exhibit dramatic elongation of the cell body as well as nuclei. No preferential direction of cell alignment is seen with cells cultured on HBC film (Fig. 2I) or glass (Fig. 2J). Induction of cell elongation occurs on both the HBC and HBC/collagen blended fiber surfaces, indicating that the nanofiber topography itself is the primary factor in inducing cell elongation, irrespective of the presence of collagen. The major difference seen between these two types of scaffolds is in the initial numbers of cells attaching to these surfaces, with more cells attaching to the HBC/collagen blended surface. In addition, the cells cultured on HBC fibers appear ribbon-like and loosely associated with neighboring cells; whereas, the cells on HBC/collagen blended fibers appear more densely packed (data not shown).
Figure 2.
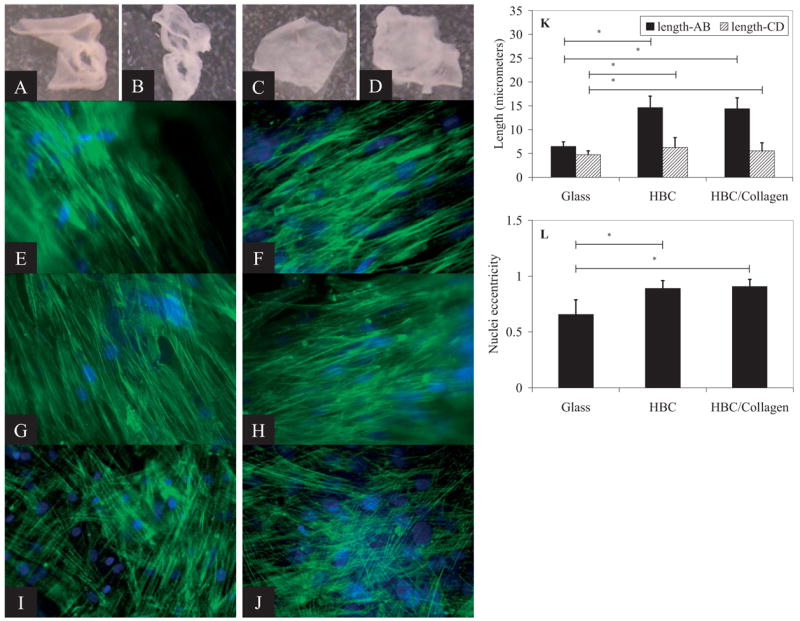
Cell sheet production and cellular morphology on HBC and HBC/collagen fiber surfaces after 2 week culture. hMSC on HBC fibers before cooling (A) and after cooling (B). hMSC on HBC/collagen fibers before cooling (C) and after cooling (D). Corresponding F-actin (green) and nucleic acid (blue) stained images for hMSC on HBC fibers before (E) and after cooling (G), and HBC/collagen fibers before (F) and after cooling (H). Images of hMSC on HBC (I) and glass (J) surfaces show random arrangement of cell cytoskeleton. Lengths of long and short axis of nuclei (K). Asterisks represent p-values ≪ 0.0001. Eccentricity values for hMSC on glass and HBC fibers (L). Scale bars: 100 μm.
Nuclear shape has been shown to have a significant impact on gene regulation, therefore, differences in nuclear shape were investigated between hMSC cultured on glass coverslips versus HBC and HBC/collagen fiber surfaces. The shape of the normal eukaryotic nucleus, in a 2D projection under epi-fluorescence, can best be approximated as an ellipse. Given an elliptical shape, one is able to characterize its eccentricity using the following equation.
where CD is defined as the length of the short axis and AB is the length of the long axis of the ellipse. Values of eccentricity range from 0 to 1, with 0 being a perfect circle and 1 an extremely stretched ellipse (most closely approximated by a line). A confluent monolayer of hMSC cultured on glass show signs of cell body elongation as well as alignment with neighboring cells, in a non-preferential direction, within any given localized region. However, the nuclei do not show any sign of alignment and exhibit a heterogeneous population with average eccentricity values of 0.66 ± 0.13, long axis lengths of (6.47 ± 0.97) μm, and short axis length of (4.72 ± 0.84) μm (Fig. 2). In contrast, almost all of the cells cultured on the aligned nanofiber surfaces exhibit cell body alignment as well as significant nuclear elongation and alignment in the direction of fiber alignment. Nuclei display eccentricity values approaching 1, with 0.89 ± 0.07 and 0.91 ± 0.06 for cells cultured on HBC and HBC/collagen surfaces, respectively. The nuclei of cells cultured on HBC and HBC/collagen surfaces are nearly double in length of the long axis, as compared to cells cultured on glass, with average values of (14.64 ± 2.39) μm and (14.39 ± 2.32) μm, respectively. The short axis lengths are (6.25 ± 2.07) μm and (5.57 ± 1.68) μm for cells on HBC and HBC/collagen surfaces, respectively. One can infer that the nuclear widths, mostly along the perpendicular direction to the aligned fibers, span across approximately 8 to 16 individual nanofibers. The width of these nuclei nearly match the cell width along the short axes of these elongated cells, thereby suggesting restricted expansion of nuclear volume in the direction perpendicular to fiber alignment.
The expression profile of genes representative of three differentiated lineages of hMSC were evaluated: osteogenic, chondrogenic, and myogenic (Fig. 3). Gene expression for these lineages were explored in order to determine if the cell and nuclear elongation, alone, can affect hMSC induction into a differentiated phenotype even when cultured in non-inductive, proliferative medium. Runx2 and Osteocalcin expression were taken as markers of osteogenesis. Collagen type II, TGF-β3, Sox-9, and Aggrecan were used to confirm chondrogenesis. Pax3 and Pax7 are present in myogenic progenitor cells.[21] MyoD is necessary for commitment to the myogenic lineage and remain transcriptionally inactive until a differentiative signal is sensed.[22,23] Other myogenic markers, Myogenin, Desmin, and Collagen IV, were also assayed.
Figure 3.
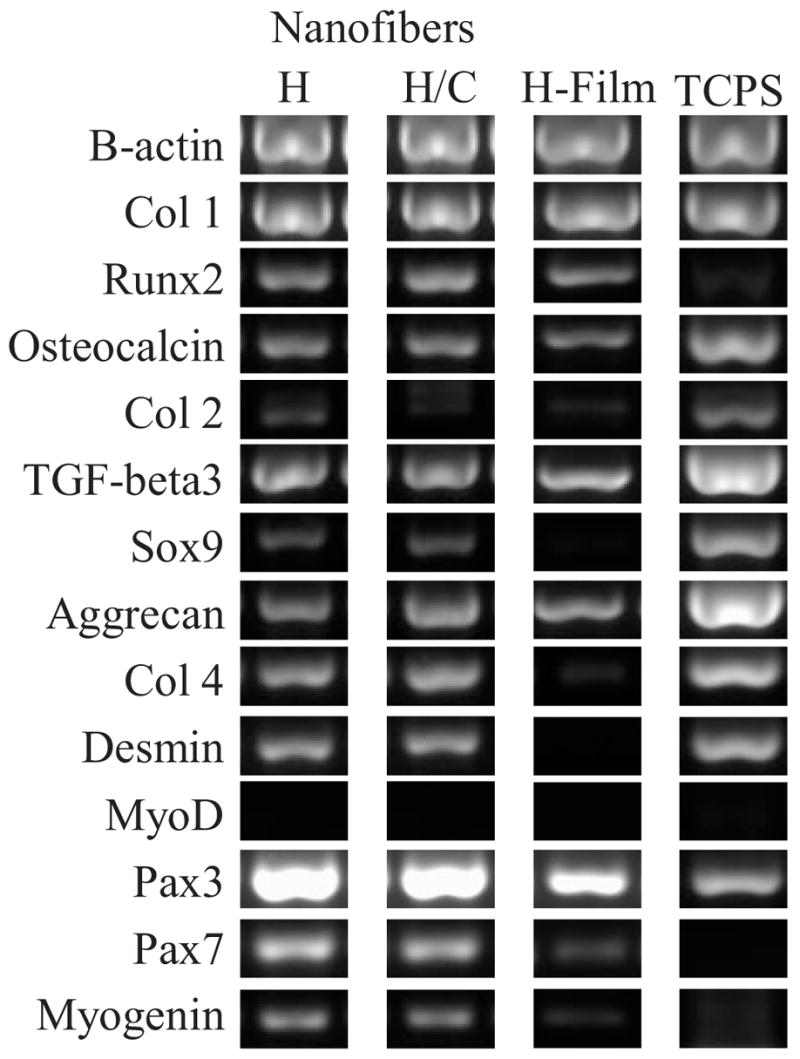
Gene expression analysis of hMSC after 2 week culture period. H = HBC nanofibers. H/C = HBC/collagen blend nanofibers. H-film = HBC films. hMSC cultured on HBC films and TCPS are shown here as control surfaces.
Expression of genes from all three differentiated lineages was detected from hMSC cultured on both HBC and HBC/collagen blended fibers. However, when compared to genes expressed by hMSC cultured on HBC films and TCPS, a definitive upregulation of myogenic genes is apparent (Collagen IV, Desmin, Pax-3, Pax-7, Myogenin). Although MyoD expression was not detected, elevated levels of Myogenin, a gene involved in muscle differentiation and downstream of MyoD expression, suggests myogenic commitment.[24] The combined expression of these genes can be taken to imply that the hMSC cultured on the fiber surfaces have been induced into the myogenic lineage as a result of cell and nuclear elongation. Given the same composition of the two surfaces, film versus fiber, but different only in topography, the results suggest that the aligned fiber topography and induction of elongated nuclear shape are the major factors in myogenic induction. However, the presence of genes of multiple differentiated lineages indicate it may be possible that the induced myogenic commitment may be transient and introduction of further myogenic signals may be necessary to further commit these cells to muscle differentiation. This gene expression data represents a single timepoint from a population of hMSCs that may not be uniformly responsive to its environment and it is possible that only a select group of cells are expressing genes representative of a myogenic commitment while others of different genetic phenotypes.
Mesenchymal stem cells are responsive to a variety of exogenous factors. Soluble growth factors, mechanical stimulation, and matrix stiffness have all been implicated as having potential for inducing differentiation of MSCs.[25–29] We present data here to implicate another factor, cell and nuclear elongation inducible by an aligned nanofibrous surface, that may be stimulative for MSC differentiation. Both the film and fibrous HBC surfaces present a chemically and mechanically (elastic moduli ranging from 10–15 kPa) similar culture surfaces with the major difference being its topological features. Although no definitive genotypic changes for a single differentiative pathway was detected, there were significant differences between cells cultured on these two different surfaces, mainly with genes indicative of myogenic induction. Further exploration of long term cultures and systems that combine other exogenous factors for stem cell differentiation, in addition to topographical cues, may be more fruitful in inducing definitive myogenesis in MSCs. Studies exploring potential changes in nuclear matrix proteins and focal adhesion complexes due to cytoskeletal and nuclear shape changes induced by a nanofibrous topology may also provide cues to potential mediators in MSC differentiation.
The intranuclear environment is rich in structurally interconnected nuclear matrix elements responsible for modulation of gene expression and silencing.[30–32] Therefore, the stretched morphology of nuclei in these aligned cells may be altering the internal matrix structure sufficiently enough to stimulate the expression of otherwise silent genes. Published works on the connectivity of cytoskeletal and nuclear structures[33–35] compel further investigation of changes to cellular function induced by matrix features at the nanoscale. Focal adhesion complexes, in combination with the cell cytoskeleton, largely contribute to the dynamic maintenance of cell morphology.[36–39] Although these are macromolecular sized intracellular structures, individual components of these assemblies are at the nanoscale.[40,41] The nanofibers may be creating a topography that restricts integrin diffusion across the cell membrane and the final size or shape of these focal adhesion assemblies at the level of these nanoscale molecules. Since individual fiber diameters are at a subcellular scale and cells span across several fibers, the limitation in lateral expansion of cell bodies may be due to the repetition of the nanoscale pattern rather than a physical barrier by individual fibers. Further analysis will be needed to correlate between fiber features and restrictions, if any, on the complexation of key players in the cytoskeletal signaling cascade.
These aligned nanofibrous scaffolds composed of a thermally reversible polymer, hydroxybutyl chitosan, are multifunctional scaffolds for the production of engineered cell systems. The aligned nanofibers provide topographical cues to induce cell alignment with potential to have direct affects on influencing gene expression. These fibers can provide a culture surface of adjustable mechanical properties that can better mimic those of natural tissue. Bioactive components, such as natural matrix components and growth factors, can be incorporated into the fibers during fabrication to present a surface capable of providing biochemical cues to enhance cellular function. The convenient removal of the fibers can create a cell sheet conditioned with the desired levels of cell maturation and tissue response to the biologically and topographically active surface of the scaffold. Combined, this multifunctional scaffold system can offer an elegantly controlled microenvironment for the production of tissue-engineered cell sheets for use in regenerative medicine.
Experimental
HBC was purchased from Dainichiseika Color & Chemicals Mgf. Co. Ltd. Purification was performed as previously published [17]. A stock solution of 10 wt % HBC17 in 1,1,1,3,3,3-hexafluoro-2-propanol (HFP; Sigma-Aldrich) was diluted to 7 wt % of HBC17 in HFP for HBC fibers. The stock solution was diluted to 7 wt % of HBC17 in neutralized bovine collagen I solution (BD Biosciences), prepared as outlined in the manufacturer’s protocol. For electrospinning, the polymer solution was placed in a syringe with a 19G blunt needle mounted onto a syringe pump (New Era Pump Systems) and dispensed at a flow rate of 3.5 mL h−1. The needle tip was charged to 11–12 kV using a HV power supply (Gamma High Voltage Research). Fibers were collected on a foil strip mounted onto the shaft of a dc motor (Grainger) set to spin at 4000 rpm. The distance between the needle tip and foil target was maintained at 4 cm. Fiber scaffolds were spun to an average thickness of 200 μm. Spun fibers were stored under desiccation at room temperature until use. To prepare fibers for cell culture, dried fibers were washed twice in PBS (at 37 °C). Excess PBS was removed and fibers were left to dry under UV overnight and used the next day for cell culture. Fibers were coated with 4 nm of and samples were imaged with a LEO field emission SEM with the electron beam set to 1 kV.
For fabrication of HBC films, HBC17 was diluted to a 2 % wt% solution in serum-free culture medium and 200 μL of HBC solution spread onto pre-sterilized 22 0 22 mm2 coverglass. These coverglasses were placed into a humidified incubator, held at 37 °C for 30 min to allow for gelation. After 30 min, the films were left to air-dried, under UV. Once dried, the films were washed twice in PBS warmed to 37 °C and left to air dry, under UV, overnight and used the next day for cell culture.
Human MSCs purchased from Cambrex, Inc. were cultured as directed in the manufacturer’s directions. For culture on HBC fibers, cells were suspended in culture medium at a concentration of 2 0 106 cells mL−1. 100 μL of cell solution was placed onto the dried fiber. For culture on HBC films, cells were suspended in full culture medium at a concentration of 2 0 106 cells mL−1. A 150 μL drop of cell solution was placed in the center of each film and the medium was allowed to absorb into the HBC film.
Cells were cultured on the HBC surface until cell confluence was reached and sufficient matrix produced, which typically took 2 weeks. Cell-seeded scaffolds were loosened from the bottom of the culture dish and transferred to wells with culture medium cooled to 4 °C to dissolve away the HBC. In order to observe cell morphology prior to HBC cooling, cells were fixed with 4 % paraformaldehyde and HBC was removed with immersion in cooled PBS. FITC-phalloidin and DAPI (Invitrogen) was used to visualize the cytoskeleton and nuclei, respectively. The entire cell/fiber scaffold sample was placed into lysis buffer cooled to 4 °C followed by total RNA extraction using RNeasy Kit (Qiagen) according to manufacturer’s protocol. RT-PCR reaction master mix was prepared using the OneStep RT-PCR kit (Qiagen) according to manufacturer’s protocol. All reactions were run on the Biorad iCycler thermal cycler (Biorad) for 35 cycles. Primer sequences used are listed in Supporting Information Table 1.
The lengths of long and short axes of nuclei and nuclear area were determined only on nuclei with clear boundaries. 331, 116, and 75 cells were analyzed for hMSC on glass, HBC nanofibers, and HBC/Collagen blend nanofibers, respectively.
Footnotes
Support for this work is provided by NIH (EB003447). We would like to thank Dr. Sing Yian Chew and I-Chien Liao for their assistance with the electrospinning protocol. Supporting Information is available online from Wiley InterScience or from the author.
Contributor Information
Jiyoung M. Dang, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21205 (USA)
Kam W. Leong, Department of Biomedical, Engineering, Duke University, 136 Hudson Hall, Box 90281, Durham, NC 27708 (USA) E-mail: kam.leong@duke.edu
References
- 1.Miyahara Y, Nagaya N, Kataoka M, Yanagawa B, Tanaka K, Hao H, Ishino K, Ishida H, Shimizu T, Kangawa K, Sano S, Okano T, Kitamura S, Mori H. Nat Med. 2006;12:459. doi: 10.1038/nm1391. [DOI] [PubMed] [Google Scholar]
- 2.Nishida K, Yamato M, Hayashida Y, Watanabe K, Yamamoto K, Adachi E, Nagai S, Kikuchi A, Maeda N, Watanabe H, Okano T, Tano Y. N Engl J Med. 2004;351:1187. doi: 10.1056/NEJMoa040455. [DOI] [PubMed] [Google Scholar]
- 3.Yang J, Yamato M, Kohno C, Nishimoto A, Sekine H, Fukai F, Okano T. Biomaterials. 2005;26:6415. doi: 10.1016/j.biomaterials.2005.04.061. [DOI] [PubMed] [Google Scholar]
- 4.Abrams GA, Schaus SS, Goodman SL, Nealey PF, Murphy CJ. Cornea. 2000;19:57. doi: 10.1097/00003226-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 5.Brody S, Anilkumar T, Liliensiek S, Last JA, Murphy CJ, Pandit A. Tissue Eng. 2006;12:413. doi: 10.1089/ten.2006.12.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen VJ, Smith LA, Ma PX. Biomaterials. 2006;27:3973. doi: 10.1016/j.biomaterials.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 7.Karuri NW, Liliensiek S, Teixeira AI, Abrams G, Campbell S, Nealey PF, Murphy CJ. J Cell Sci. 2004;117:3153. doi: 10.1242/jcs.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norman JJ, Desai TA. Ann Biomed Eng. 2006;34:89. doi: 10.1007/s10439-005-9005-4. [DOI] [PubMed] [Google Scholar]
- 9.Yim EK, Reano RM, Pang SW, Yee AF, Chen CS, Leong KW. Biomaterials. 2005;26:5405. doi: 10.1016/j.biomaterials.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badami AS, Kreke MR, Thompson MS, Riffle JS, Goldstein AS. Biomaterials. 2006;27:596. doi: 10.1016/j.biomaterials.2005.05.084. [DOI] [PubMed] [Google Scholar]
- 11.Bhattarai N, Edmondson D, Veiseh O, Matsen FA, Zhang M. Biomaterials. 2005;26:6176. doi: 10.1016/j.biomaterials.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 12.Chew SY, Wen J, Yim EK, Leong KW. Biomacromolecules. 2005;6:2017. doi: 10.1021/bm0501149. [DOI] [PubMed] [Google Scholar]
- 13.Greiner A, Wendorff JH, Yarin AL, Zussman E. Appl Microbiol Biotechnol. 2006;71:387. doi: 10.1007/s00253-006-0356-z. [DOI] [PubMed] [Google Scholar]
- 14.Li M, Mondrinos MJ, Gandhi MR, Ko FK, Weiss AS, Lelkes PI. Biomaterials. 2005;26:5999. doi: 10.1016/j.biomaterials.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 15.Pham QP, Sharma U, Mikos AG. Tissue Eng. 2006;12:1197. doi: 10.1089/ten.2006.12.1197. [DOI] [PubMed] [Google Scholar]
- 16.Zong X, Bien H, Chung CY, Yin L, Fang D, Hsiao BS, Chu B, Entcheva E. Biomaterials. 2005;26:5330. doi: 10.1016/j.biomaterials.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 17.Dang JM, Sun DD, Shin-Ya Y, Sieber AN, Kostuik JP, Leong KW. Biomaterials. 2006;27:406. doi: 10.1016/j.biomaterials.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 18.Beqaj S, Jakkaraju S, Mattingly RR, Pan D, Schuger L. J Cell Biol. 2002;156:893. doi: 10.1083/jcb.200107049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CD, Curtis AS. Exp Cell Res. 2003;284:274. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 20.Relan NK, Yang Y, Beqaj S, Miner JH, Schuger L. J Cell Biol. 1999;147:1341. doi: 10.1083/jcb.147.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M. J Cell Biol. 2006;172:91. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berkes CA, Tapscott SJ. Semin Cell Dev Biol. 2005;16:585. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Sartorelli V, Caretti G. Curr Opin Genet Dev. 2005;15:528. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi X, Garry DJ. Genes Dev. 2006;20:1692. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- 25.Alsberg E, von Recum HA, Mahoney MJ. Expert Opin Biol Ther. 2006;6:847. doi: 10.1517/14712598.6.9.847. [DOI] [PubMed] [Google Scholar]
- 26.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 27.Le Blanc K, Pittenger M. Cytotherapy. 2005;7:36. doi: 10.1080/14653240510018118. [DOI] [PubMed] [Google Scholar]
- 28.Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 29.Vogel V, Sheetz M. Nat Rev Mol Cell Biol. 2006;7:265. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 30.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. Nat Rev Mol Cell Biol. 2005;6:21. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 31.Handwerger KE, Gall JG. Trends Cell Biol. 2006;16:19. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Zaidi SK, Young DW, Choi JY, Pratap J, Javed A, Montecino M, Stein JL, van Wijnen AJ, Lian JB, Stein GS. EMBO Rep. 2005;6:128. doi: 10.1038/sj.embor.7400337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. J Cell Biol. 2006;172:41. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maniotis AJ, Chen CS, Ingber DE. Proc Natl Acad Sci USA. 1997;94:849. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young KG, Pinheiro B, Kothary R. Exp Cell Res. 2006;312:121. doi: 10.1016/j.yexcr.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Braren R, Hu H, Kim YH, Beggs HE, Reichardt LF, Wang R. J Cell Biol. 2006;172:151. doi: 10.1083/jcb.200506184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaazempur Mofrad MR, Abdul-Rahim NA, Karcher H, Mack PJ, Yap B, Kamm RD. Acta Biomater. 2005;1:281. doi: 10.1016/j.actbio.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Li S, Guan JL, Chien S. Annu Rev Biomed Eng. 2005;7:105. doi: 10.1146/annurev.bioeng.7.060804.100340. [DOI] [PubMed] [Google Scholar]
- 39.Romer LH, Birukov KG, Garcia JG. Circ Res. 2006;98:606. doi: 10.1161/01.RES.0000207408.31270.db. [DOI] [PubMed] [Google Scholar]
- 40.Goffin JM, Pittet P, Csucs G, Lussi JW, Meister JJ, Hinz B. J Cell Biol. 2006;172:259. doi: 10.1083/jcb.200506179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Biochim Biophys Acta. 2004;1692:103. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]


