Abstract
Thirty adults were tested for humoral and cellular immune responses following immunization with the trivalent inactivated influenza vaccine. Modest but significant inverse correlations between the baseline and the fold changes in the number of IFNγ producing cells and the levels of neutralizing antibodies were observed. Specific increases in proliferative responses in the CD8 CD45RA+ population were noted after vaccination. Minimal correlations between neutralizing antibody titers and the number of IFNγ producing cells in terms of prevaccination levels or fold increases were observed. These results show specific increases in a CD8 T cell subset and discordant T and B responses induced by the inactivated influenza vaccine.
Keywords: B cells, T cells, influenza
1. Introduction
Human influenza is a highly contagious acute respiratory illness that is responsible for significant morbidity and excess mortality in the elderly and the very young worldwide. Though effective antiviral medications targeting the neuraminidase (NA) glycoprotein are available, prevention of influenza morbidity and mortality is primarily through the immunization of target groups at high risk for mortality or hospitalization [1]. Annual worldwide epidemics of influenza A and the recent emergence of zoonotic infections with highly pathogenic H5N1 and H9N2 avian influenza strains have heightened efforts to understand the role of both humoral and cell mediated immunity in the control of influenza virus infection [2].
Current vaccine approaches depend on the induction of antibodies to the viral surface proteins hemagglutinin and neuraminidase that neutralize the infectivity of the virus and interfere with the release of newly replicated virus from the host cell [2, 3].When the vaccine virus closely matches the challenge infecting virus, the vaccines are effective. The virus however undergoes frequent mutations at antibody combining sites and the vaccine is less effective. This is a much bigger problem when a new subtype of influenza virus emerges eg. the H5N1 avian derived viruses and there are no cross-reactive antibody sites on the vaccine virus hemagglutinin (HA) and the new subtype of virus. Ideally, influenza vaccines would also be expected to induce influenza specific CD8 T cell mediated responses that may contribute to protective immunity. Studies in murine models of influenza demonstrated that CD8 T cells were effective in reducing viral titers and aided in recovery [4–9]. These models have also demonstrated that delayed influenza virus clearance occurs in CD8 T cell deficient mice [10]and that memory T lymphocytes can act independently of a humoral immune response in order to confer resistance to influenza infection [11, 12]. Influenza virus specific cytotoxic T lymphocytes (CTL) have also been shown to limit influenza A virus replication and protect against lethal viral challenge [7, 8, 13]. The role of CD4 T cells in influenza infection is less defined. CD4 virus-specific T cells may help compensate for the absence of CD8 CTL because the virus can be cleared in CD8 -deficient mice; however, mice lacking both CD4 and CD8 T cells do not clear virus or survive [14, 15]. Betz et al. demonstrated that responses in the secondary lymphoid organs of CD4 T cell deficient mice infected with influenza were defective while responses in bronchoalveolar lavage were similar in CD4 T cell deficient mice and wild type mice. This suggested that CD4 T cells may not be required for the primary response to influenza but may be important in the generation of memory CD8 T cells[16].
In contrast, there have been few studies on cellular immunity to influenza in humans. Knowledge about human CD8 T cell immune responses has been more well developed than human CD4 T cell responses[17, 18]. Studies on the cytotoxic T lymphocyte repertoire to influenza A viruses indicate that influenza memory T cell responses are directed to a number of epitopes on a variety of proteins including the nucleoprotein, nonstructural protein 1 (NS1), and the matrix protein 1(M1) [19, 20]. Most of these highly conserved cross reactive epitopes have been found to be conserved in H5N1 viruses from recent outbreaks [17]. Therefore, cell- mediated immunity appears to be important in both restricting influenza A virus replication and reducing disease severity, and appears to offer a more potentially cross- reactive vaccine approach for the prevention of pandemic or epidemic influenza.
In this study, we evaluated the human memory T cell and the serum antibody responses of healthy subjects following immunization with the licensed 2005–2006 trivalent inactivated influenza vaccine in order to better understand the role that both T and B cells may play in the protection afforded by the vaccine. Using CarboxyFluoroscein Succinimidyl Ester (CFSE) , Enzyme-linked immunosorbent spot (ELISPOT) and neutralization assays to examine responses to the individual influenza A H1 and H3 viral strains ,we were particularly interested in the following questions: 1) the effect of prevaccination levels of T cell immunity to both influenza A virus strains on the cellular immune responses generated by subsequent vaccination 2) the phenotype(s) of the T cells responding to vaccine and 3) the relationships between the antibody and T cell components of the host immune response to influenza vaccine.
2. Materials and Methods
2.1. Viruses
Influenza A viruses H1N1 A/New Caledonia/20/99 (Lot # 3XANA060818B) and H3N2 A/Wisconsin/67/05 (Lot # 3XAWN060818B) were purchased from Charles River Laboratories (North Franklin, Connecticut) for use in CFSE, ELISPOT, and proliferation assays. These viruses were propagated in the allantoic cavity in Specific Pathogen Free (SPF) eggs with the final hemagglutinin (HA) titers for the A/New Caledonia strain and the A/Wisconsin strains 1:512 per 0.05 ml and 1: 16 per 0.05 ml, respectively. The following egg adapted virus strains- Influenza A/New Caledonia/20/99 IVR-166 and A/Wisconsin/67/2005X–161B,which contained 5.5 × 107 and 2.0 × 108 PFU/ml respectively, were used in microneutralization assays and were kindly provided by Dr. Michel De Wilde and Dr. Robert Ryall from Sanofi Pasteur. Titers were performed by Plaque assay in Madin-Darby Canine Kidney (MDCK) epithelial cells
2.2. Human sera and peripheral blood mononuclear cells (PBMC)
Blood specimens were obtained from 30 normal healthy donors (5 males and 25 females) under an Institutional Review Board (IRB) approved protocol. Inclusion criteria for volunteers include 1) Healthy adults > 18 years of age, 2) Volunteers are designated to be vaccinated according to the recommendations of the US Department of Health and Human Services Advisory Committee on Immunization Practice 3) Willing to return to donate samples of blood at designated time points 4) Informed Consent obtained. Exclusion criteria for volunteers include 1) History of HIV (human immunodeficiency virus), IVDA (intravenous drug abuse), Hepatitis B or C, or other current viral or bacterial infection 2) Vaccination in the last 60 days 3) Recent (past 5 days) corticosteroid use 4) Immunosuppressive Therapy 5) Treatment with Immunoglobulin for any reason in the past 60 days. The age of these subjects ranged from 26– 58 years with an average age of 44 years. This study population received the licensed 2005–2006 trivalent inactivated vaccine comprised of H1N1 A/ New Caledonia/20/99 and H3N2 A/ California/07/2004 strains manufactured by Sanofi Pasteur. Blood samples were obtained before vaccination, at approximately 2 weeks to 1 month (13–39 days) post vaccination and at approximately 2 months (63–77 days) post vaccination. PBMC were purified by Ficoll-Hypaque density gradient centrifugation as previously described [21]. Serum was obtained from each donor at each of the timepoints for use in microneutralization assays.
2.3. ELISPOT assay
Cryopreserved PBMC were tested in ELISPOT assays as described previously [22]. The H3N2 A/Wisconsin/67/05 and the H1N1 A/New Caledonia/20/99 strain were used to stimulate PBMC at final dilutions of 1:8 and 1:32, respectively. The optimal concentrations of the two influenza strains were determined in preliminary experiments using PBMC from an individual with substantial T cell and antibody responses to these influenza strains. We used 96 well filtration plates (Millipore, Bedford, MA) coated with murine anti human- interferon γ (IFNγ) Ab (Pharmingen, San Diego, CA). Cryopreserved PBMC were thawed, washed, and added to the plates at 2.5 ×105/well in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin, glutamine, and HEPES. Cells were incubated at 37° C for 15 hours with virus. Medium was used as a negative control and Phytohemagluttinin (PHA), (final concentration in assay = 20µg/ml) (Sigma # L-9132) and a Cytomegalovirus (CMV), Epstein Barr virus (EBV), and flu (CEF) peptide pool was used as a positive control and was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: CEF Control Peptide Pool from DAIDS, NIAID [23]. This reagent contains HLA class I restricted T cell epitopes of CMV, EBV, and influenza A virus and was used at a final concentration of 2µg/ml per peptide. The plates were washed and then incubated with biotinylated murine anti- human IFNγ Ab (Pharmingen). Spots were developed using fresh substrate buffer (0.3mg/ml of 3- amino-9ethylcarbazole and 0.015% H2O2 in 0.1M sodium acetate, pH=5). The frequency of peptide-specific IFNγ producing cells was calculated as (average number of spots in the virus wells – average number of spots in medium wells number of cells/ well) and converted to spot forming units (SFU) per106 PBMC. Average background spots in medium wells were 2.3 spots (range 0– 15).
2.4. CFSE labeling and staining of cells
PBMC were labeled with 1.5uM CFSE in PBS 7.2−0.1% bovine serum albumin (BSA) (Invitrogen # C1157) in the dark. Cells were incubated at 37°C for 15 minutes in a 96 well plate before being washed in RPMI-1640 with Penicillin- Streptomycin and L-glutamine with 10% FBS (Hyclone). CFSE labeled PBMC (2 × 106/ml) were incubated with the influenza H1N1 A/New Caledonia/20/99 (Lot 3XANA060840B) at 1:200 final dilution, or were allowed to rest in medium (RPMI) for 6 days at 37°C. On Day 1, PHA (Sigma # L-9132 Lot 113K8926) at a final concentration of 1:1000 was added to appropriate wells. The concentrations of virus used and the 6 day stimulation period were chosen based on preliminary experiments using the PBMC of the same individual used in the previous ELISPOT assays. On Day 6, these expanded and medium rested cells were prepared for flow cytometry. Cells were spun at 1100 rpm for 5 minutes, then the supernatants decanted and the cells washed with cold Fluorescence-Activated Cell Sorting (FACS) buffer [PBS 7.2 w/1%BSA 1% sodium azide]. Cells were then stained with the following surface antibodies anti human CD3 Ab – Alexa Flour 700 (Clone UCHT1 BD #557943), anti human CD4 Ab – Pacific Blue (Clone OKT4 e-Biosciences #57-0048-73), anti human CD8 Ab -APC (Clone OKT8 e-Biosciences #17- 0086-73), and anti human CD45 RA Ab-PE (Clone HI100 BD# 555489) for 30 minutes at 4°C. Cells were washed and resuspended in Cytofix diluted 1:4 in FACS wash buffer and sent for FACS analysis. Data were acquired on a FACS Aria machine (BD Biosciences) and analyzed using Version 6.1 of FlowJo software (Tree Star). The number of events collected for each sample varied between 150,000 and 250,000. Gating was set on medium-stimulated samples and then applied to virus stimulated samples.
2.5. Influenza microneutralization assay
Microneutralization antibody titers were determined using minor modifications of a previously reported method [24, 25]. Ten 2 fold dilutions of serum, starting at 1:10 and ending at 1:5120, were made in culture medium [modified Eagle’s medium ( MEM) −1% BSA with Penicillin Streptomycin] and 50 µl of each dilution was mixed with 50 µl containing 50% Tissue Culture Infective Dose of the H1N1 A/New Caledonia/20/99 or H3N2 A/Wisconsin/67/2005 viruses. After incubation of plates at 37°C for 1 hour, an equal volume of MDCK cells in MEM-1% BSA with Penicillin Streptomycin and trypsin (final concentration 0.2 µg/ml) was added to the serum- virus antigen mixture. Replicate wells of four were used for each dilution of antibody. After 4 days at 37° C, supernatant fluids were removed and the cells stained with crystal violet for 15 minutes. Wells were read for cytopathic effect using a microscope or an ELISPOT reader. The serum neutralizing titer of a given sample was determined as the reciprocal of the last serum dilution with < 50% cytopathic effect.
2.6. Data analysis and statistics
All statistical analyses were performed using the ANALYSIS TOOLPAK software in MICROSOFT EXCEL 2003. Mean values were compared between the prevaccination and each of the post vaccination timepoints using Analysis Of Variance between groups (ANOVA) single factor analysis. Comparison between prevaccination and the peak post-vaccination responses were done using a paired 2 sample two tailed Student’s T test. Relationships between prevaccination and peak change (fold change) responses and relationships between H3N2 A/Wisconsin/67/2005 or H1N1 A/New Caledonia/20/99 virus responses were determined using linear regression analysis. Statistically significant comparisons are denoted with p values <.05. Log10 was used in all calculations involving neutralizing titers.
3. Results
3.1. IFNγ producing cell responses to influenza vaccination
We used ELISPOT assays to quantitate the number of IFNγ- producing cells in PBMC specific for the influenza A H1N1 subtype strain A/New Caledonia/20/99 and the H3N2 subtype strain A/Wisconsin/67/05. The H3N2 A/Wisconsin/67/05 virus is a very closely related antigenic variant of the 2005–06 vaccine strain H3N2A/California/07/04. There was an overall moderate but not a statistically significant increase in the number of IFN-γ-producing cells post vaccination after stimulation with either of the wild type strains H3N2 A/Wisconsin/67/05 (prevaccination 599.8±421.4 to ~ 2 weeks post vaccination 660.1±416.0 to ~ 2 months post vaccination 693.7±454.3) or H1N1 A/New Caledonia A/20/99 (prevacccination 818.9±496.3 to ~ 2 weeks post vaccination 881.1±487.2 to ~ 2 months post vaccination 913.4±504.4) using ANOVA analysis. We noted variability in influenza specific interferon gamma (IFNγ) responses with some donors showing the highest responses at their first post vaccination time point while others developing their highest responses at the second post vaccination time point. Given this variability in the kinetics of individual responses, we compared the numbers of IFN-γ-producing cells between the prevaccination and the post vaccination timepoint that showed the highest responses. Statistically significant increases were seen between prevaccination and peak post vaccination responses for both viruses (H1N1A/New Caledonia p=0.019; H3N2 A/Wisconsin p= 0.011 by paired student t test) (Data not shown).
We also noted another characteristic of the kinetics of influenza specific responses. Individuals can be grouped into those with low baseline numbers of IFNγ producing cells followed by high responses post vaccination and those with high baseline numbers of IFNγ producing cells followed by low responses post vaccination. Using regression analysis, a statistically significant inverse correlation was detected between the baseline and fold changes in SFU/106 IFNγ cells to H3N2 A/Wisconsin/67/05 or to H1N1 A/New Caledonia/20/99 at both the first time point and the second time point as well as the peak timepoint post vaccination (H1N1 A/New Caledonia r2 = 0.39 p= 0.0002; H3N2 A/Wisconsin r2 = 0.24 p= 0.005) (Figure 2).
Figure 2.
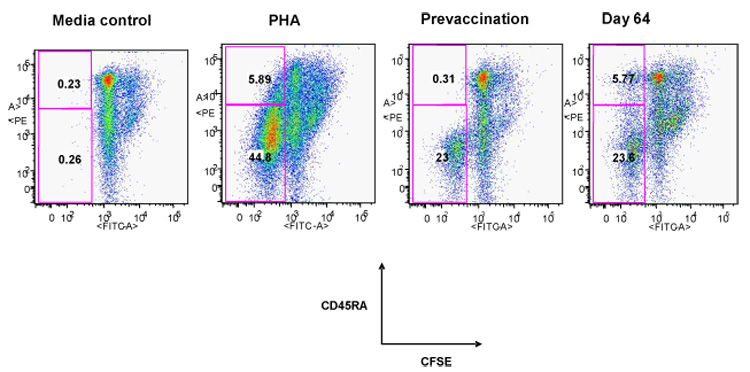
Increase in CD8 CD45RA+ cells capable of virus specific proliferation after influenza vaccination. Typical flow cytometry plot using carboxyfluorescein succinimidyl ester (CFSE) based assay for an individual donor at prevaccination and at Day 64 post vaccination. Media and Phytohemagluttinin controls are shown. CFSE labeled PBMC were stimulated with A/New Caledonia virus/20/99 for 6 days and then surface stained for CD3,CD8 and CD45RA.Cells were analyzed on Flow Jo software by gating on live lymph cells, then CD3 vs side scatter, then CD8 vs. side scatter and then CD45RA vs CFSE. Bolded numbers in the quadrants refers to the percentage of CD8/CFSE+/CD45RA+ or CD8/CFSE+/CD45RA− cells.
3.2. Increases in proliferating CD4 and CD8 T cells to A/New Caledonia/20/99 virus after influenza vaccination
The prevaccination PBMC from these 30 donors showed variable but substantial levels of baseline proliferation to each of the viruses (H3N2 A/Wisconsin SI range 8– 153 and H1N1 A/New Caledonia SI range 6–145) in standard tritiated thymidine (3H )incorporation assays, reflecting previous exposure to influenza virus through natural infection and/or immunization ( data not shown). Increases in mean proliferative responses in PBMC to the H3N2 A/Wisconsin/67/05 and the H1N1 A/New Caledonia/20/99 strains from the prevaccination to the postvaccination timepoint were seen which were not statistically significant ( Data not shown). We used CFSE staining to determine if different subsets of T cells showed an increase in proliferation after immunization. After CFSE labeling and 6 days of in vitro stimulation with live influenza H1N1 A/New Caledonia/20/99, virus stimulated and medium treated cells were stained for detection of CD4, CD8 and CD45 RA surface expression to characterize the different proliferating populations. Representative CFSE plots are shown in Figure 2. Overall, the mean percentages of total CD4 and CD8 T cells as well as the corresponding CD45RA− subsets that proliferated after vaccination were lower while the mean percentage of the CD4 CD45RA+ and the CD8 CD45RA+ cells that proliferated after vaccination increased. Neither result was statistically significant (Table 1). However, taking into account variability in individual responses, we compared the prevaccination and the post-vaccination timepoint with the highest response and found a significant increase in the proliferating CD8 CD45RA+ T cell population post vaccination (0.25% to 1.1% p=0.03) (Table 1).
Table 1.
Mean % CFSE proliferation in different CD4 and CD8 subsets prevaccination and post-vaccination
| Analysis | Prevaccination | First timepoint ~ 2 weeks post vaccination | Second timepoint ~ 2 month post vaccination | Peak post-vaccination timepoint | |
|---|---|---|---|---|---|
| (%)a | (%)a | (%)a | (%)a | ||
| Entire | |||||
| cohort | CD8 | 5.15 | 4.9 | 4.3 | 6.4 |
| CD8RA+ | 0.25 | 0.82 | 0.6 | 1.1 ( p=.030) * | |
| CD8RA− | 5.5 | 4.9 | 4.8 | 6.1 | |
| CD4 | 5.3 | 5.6 | 4.7 | 7 | |
| CD4RA+ | 0.3 | 0.6 | 0.48 | 0.73 | |
| CD4RA− | 6.3 | 6.5 | 5.8 | 7.4 |
Each value represents the mean percentage of cells in the population that proliferated in response to the live influenza A/New Caledonia/20/99 virus subtracting the medium background.
Student T test value- statistically significant if p<.05 CFSE= CarboxyFluoroscein Succinimidyl Ester
3.3. Serum titers of neutralizing antibodies pre and post immunization
Serum hemagglutination inhibition titers of 40 or greater are associated with significant protection against influenza [26, 27] Prior to immunization, 100% of subjects had antibody titers ≥ 1:40 against the H1N1 A/New Caledonia/20/99 and the H3N2 A/Wisconsin/67/05 influenza strains. Only seven individuals developed a ≥ 4 fold increase in antibody titer either against H3N2 A/Wisconsin/67/05 or the H1N1 A/New Caledonia/20/99 viruses at either the first or the second post vaccination timepoint. Log10 neutralizing antibody titers increased after vaccination for all viruses (p<0.05) although the fold increases were moderate (about 2 fold). Not surprisingly, there was a statistically significant inverse correlation between neutralizing antibody titers pre vaccination and the fold increase post vaccination with the strongest correlation seen with the H1N1 A/New Caledonia/20/99 strain (Data not shown).
3.4. Correlation between neutralizing antibody levels and IFNγ producing cells
We examined several correlations between neutralizing antibody levels and the number of influenza specific IFNγ producing cells. Prevaccination, neutralizing antibody titers and the number of IFNγ producing cells to the H1N1 A/New Caledonia/20/99 strain correlated but those to H3N2 A/Wisconsin/67/05 did not (Figure 3a). There were no statistically significant correlations between the fold increases in Log10 neutralizing antibody titers and the numbers of IFNγ producing cells against either H1N1 A/New Caledonia/20/99 or H3N2 A/Wisconsin/67/05 viruses (Figure 3b). We showed above the inverse relationship between the number of prevaccination IFNγ producing cells and the fold increase in IFNγ producing cells post vaccination. A higher level of neutralizing antibodies could explain the lack of response seen in individuals with high baseline IFNγ responses. However, we found no correlation between prevaccination neutralizing antibody levels and the highest fold increase in IFNγ producing cells (Figure 3c). Similarly, we found only minimal correlation between prevaccination number of IFNγ producing cells and the highest fold increase in neutralizing antibody levels for either the H1N1 A/New Caledonia/20/99 ( r2 = 0.13 p= 0.048) or H3N2 A/Wisconsin/67/05 strains r2 = 0.058 p= 0.20) (Figure 3d).
Figure 3.
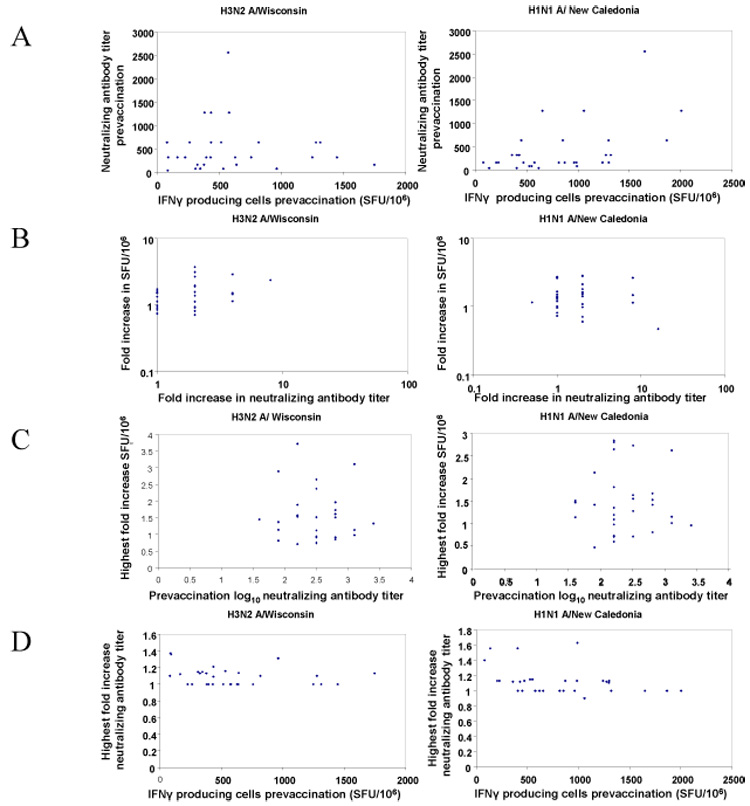
Relationships between frequencies of IFNγ producing cells and the neutralizing antibody titers. All analyses were performed using linear regression analysis.
A- Correlations between the prevaccination numbers of IFNγ producing cells and prevaccination neutralizing antibody titers for H3N2 A/Wisconsin/67/05 prevaccination ( left, r2 = 0.003 p= 0.74 )and H1N1 A/New Caledonia virus/20/99 (right, r2 = 0.25 p=0.0049). SFU = spot forming units.
B- Correlations between the peak post-vaccination fold increases in the number of IFNγ producing cells and peak post-vaccination fold increases in neutralizing antibody titers for H3N2 A/Wisconsin/67/05 (left, r2 = 0.102 p= 0.083) and H1N1 A/New Caledonia virus/20/99 ( right, r2 = 0.001 p= 0.85). SFU = spot forming units.
C– Correlations between prevaccination neutralizing antibody titers and peak fold increase in IFNγ producing cells for H3N2 A/Wisconsin/67/05 (left, r2 = 0.005 p= 0.70) and H1N1 A/New Caledonia virus/20/99 (right, r2 = 0.00005 p= 0.97). SFU = spot forming units.
D- Correlations between prevaccination IFNγ producing cells and peak fold increase in neutralizing antibody titers for H3N2 A/Wisconsin/67/05 (left, r2 = 0.058 p= 0.196)and H1N1 A/New Caledonia virus/20/99 (right, r2 = 0 .132 p= 0.048). SFU = spot forming units
4. Discussion
Our study examined the pattern of cellular responses in influenza vaccinated individuals using three different T cell assays, ELISPOT to quantitate the numbers of specific IFNγ producing cells and 3H thymidine and CFSE assays to quantitate the number of proliferating T cells, in relation to B cell responses examined using microneutralization assays. We studied the responses to two influenza A subtype viruses-H1N1 A/New Caledonia/20/99 and H3N2 A/Wisconsin/67/05 which contain the HA and NA proteins in the 2006–2007 inactivated trivalent influenza vaccine. We found that the kinetics of human influenza immune responses was variable and that preexisting immunity to this virus affected the ability of vaccination to induce significant increases in responses. A most interesting finding in this study was the discordance between cellular and humoral responses in influenza vaccinated individuals.
Given that the majority of participants worked in health care settings and received frequent influenza vaccinations, proliferation in prevaccination PBMC to live influenza viruses in thymidine assays was not unexpected. A more detailed analysis using CFSE flow cytometric assays detected small percentages (~5%) of proliferating CD4 CD45RA− and CD8 CD45RA− T cells consistent with previous exposure through natural infection and/or vaccination (Table 1). When examining peak responses elicited over the post vaccination period, significant increases were seen only in the CD8 CD45 RA+ subset. Traditionally associated with a naive phenotype, CD45RA+ cells have been shown in CMV and EBV systems to include cells capable of effector functions [28–31]. Inactivated vaccines are not thought to primarily induce CD8 T cell responses given that the antigen processing pathway required by CD8 T cells is bypassed. The expansion of the CD8 CD45RA+ population may represent a reexpression of CD45RA+ on previously CD45RA− cells in an assay expanded for 6 days rather a true expansion of naive CD45RA+ cells. However, the inactivated influenza vaccine may have activated memory CD4 T cells and through their cytokine producing functions and ability to co- stimulate CD8 T cells, these CD4 cells were able to stimulate the proliferation of CD8 CD45RA+ T cells. Our data expand on the results from a recent, open, randomized, comparative clinical trial comparing an adjuvanted and non-adjuvanted subunit influenza vaccine, in which CFSE assays detected a significant increase in the frequency of the proliferating CD3 CD4 T cell population ( CD3/CD8 T cells were not analyzed) [32]. Similarly, in a study of eight individuals who received the influenza vaccine, Long et al. found no increases in the frequencies of NK or CD8 T cells as a proportion of lymphocyte subset but did find, using intracellular cytokine staining, that there were increases in IFNγ producing CD8 T cells detected post vaccination after stimulation with virus [33] Our results would seem to similarly suggest that the overall population of CD8 T cells is stable post vaccination but that there is a subset of cells (CD45RA+), that is altered after vaccination. Ideally, studies to verify these increases in CD8 T cell frequency using more quantitative assays such as tetramer staining with CD8 T cell epitopes or through the use of intracellular cytokine assays in our larger cohort would be helpful but availability of PBMC samples was limited.
Similar to the study of He et al [34], we found statistically significant inverse correlations between the numbers of prevaccination IFNγ cells in ELISPOT assays to the individual H1 and H3 viruses and the magnitude of responses post-vaccination. In high responders post vaccination, IFNγ producing T cells may be responding to the HA and the NA proteins contained in the inactivated vaccine. Recent reports have found a considerable amount of nucleoprotein and M1 protein in the inactivated influenza vaccine; these may be additional targets for CD8 T cell responses [35–37]. The presence of high baseline neutralizing antibodies may contribute to the lack of responses in individuals with high baseline numbers of IFNγ producing cells. However, we found no significant correlations between levels of prevaccination neutralizing antibodies and fold increases in the number of IFNγ producing cells. This suggests that the lack of an increase in the number of influenza – specific IFNγ-producing cells in the PBMC of these individuals with the highest prevaccination frequencies may be the presence of CD8 and CD4 influenza specific T cells which might rapidly eliminate antigen presenting cells containing vaccine antigens, limiting subsequent activation of T cells upon reexposure.
Vaccines that enhance T cell immunity in addition to robust antibody responses are likely to be the most protective. Understanding the relationships between influenza specific T cells and antibody responses elicited by vaccination will enable a more rational design of future vaccines to influenza virus. A previous study by McElhaney et al. in older adults found positive correlations between antibody levels and Th1/Th2 cytokine ratios in older adults who received a single dose of vaccine; a negative correlation was seen between these parameters in those who received a booster dose [38]. A relationship between CD4 T cell and antibody responses would have been expected as this subset of T cells is involved in helping antibody producing B cells as well as helping CD8 T cells to proliferate. In our study, we found no correlations between neutralizing antibody titers and the number of IFNγ producing cells in terms of either prevaccination levels or fold increases. This discordance may be related to the high prevaccination immune responses present in these individuals. Though influenza virus causes an acute self limited infection, exposure to this virus occurs repeatedly, either through vaccination or through natural infection. Because of this, it was not unexpected that increases in T cell or B cell immune responses detected through ELISPOT assays or microneutralization assays were modest, findings which could account for some lack of correlation seen in our analysis. The time range of the blood samples taken may account for these results with the second sample ( 13–39 days) which falls within the likely peak of the CD8 T cell response having a more significant effect on the results than the last timepoint sampled. Studies have shown that age impacts T cell responses to influenza vaccination, with elderly individuals (>65 yrs old) showing a decline in type 1 T cell responses which may be a factor in these results; however, the average age of our subjects was 44 years old with no individuals over the age of 60 [39, 40]. The role of innate immunity components such as cytokines, dendritic cells, and toll like receptors in initiating influenza specific responses may also contribute to the discordance between these two arms of the immune system.
Current vaccine approaches depend on the induction of antibodies to the viral surface proteins hemagglutinin and neuraminidase that neutralize the infectivity of the virus and interfere with the release of newly replicated virus from the host cell. Given the fact that these surface proteins undergo frequent mutations, vaccines are reformulated each year to reflect the predominant circulating viruses of the previous influenza season. Current influenza vaccines are unable to elicit cross subtype reactive antibodies to new subtypes of virus such as the avian H5N1 virus which is of concern. Trivalent inactivated influenza vaccines should theoretically induce T cell responses at least to the HA and NA proteins present which contribute to overall immunity to influenza virus. However, our data and others suggest that given the high level of preexisting immunologic memory to influenza virus present in the population, current influenza vaccines are only able to modestly increase either T or B cell immunity to the virus [34]. Induction of cellular immune responses to the more conserved internal proteins as opposed to the more variable surface proteins of the influenza virus contained in current formulations of the inactivated vaccine would be a more attractive candidate for long lasting immunity and potentially cross reactive responses to influenza virus. More detailed investigations on the T cell responses to these internal conserved proteins of the influenza virus and their functional qualities will be helpful in this regard.
Figure 1.
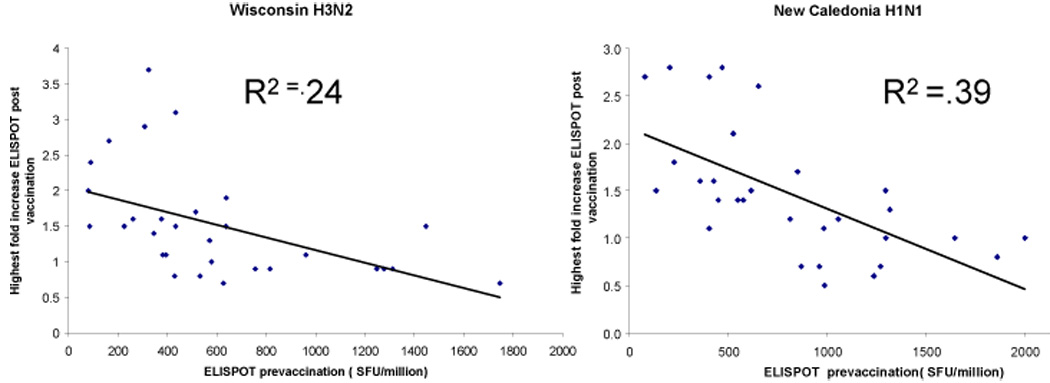
Relationship between the number of IFNγ producing cells at baseline and peak fold increase after immunization using ELISPOT assay. Linear regression analysis was performed using all data available for the three time points for the H3N2 A/Wisconsin/67/05 strain (left panel) and the H1N1 A/New Caledonia virus/20/99 (right panel). SFU= spot forming units
Acknowledgements
We thank Dr. Jeffrey S. Kennedy for helping in providing the human PBMC samples that were used in this study. We thank Christine Turcotte for assistance with HLA typing and Ping Liu for preliminary experiments using plaque assays as a readout for the neutralization assay.
Footnotes
Financial support: This work was supported by Funding from the National Institute of Allergy and Infectious Diseases/National Institutes of Health Grant U19 AI-057319
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used in this paper- CEF, ELISPOT, SFU, MDCK, CTL, PBMC, BSA, FACS
References
- 1.Harper SA, Fukuda K, Uyeki TM, Cox NJ, Bridges CB. Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2004 May 28;53(RR6):1–40. [PubMed] [Google Scholar]
- 2.Hilleman MR. Realities and enigmas of human viral influenza: pathogenesis, epidemiology and control. Vaccine. 2002 Aug 19;20(25–26):3068–3087. doi: 10.1016/s0264-410x(02)00254-2. [DOI] [PubMed] [Google Scholar]
- 3.Belshe RB, Gruber WC. Safety, efficacy and effectiveness of cold-adapted, live, attenuated, trivalent, intranasal influenza vaccine in adults and children. Philos Trans R Soc Lond B Biol Sci. 2001 Dec 29;356(1416):1947–1951. doi: 10.1098/rstb.2001.0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada A, Young JF, Ennis FA. Influenza virus subtype-specific cytotoxic T lymphocytes lyse target cells coated with a protein produced in E. coli. The Journal of experimental medicine. 1985 Nov 1;162(5):1720–1725. doi: 10.1084/jem.162.5.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada YK, Meager A, Yamada A, Ennis FA. Human interferon alpha and gamma production by lymphocytes during the generation of influenza virus-specific cytotoxic T lymphocytes. J Gen Virol. 1986 Nov;67(Pt 11):2325–2334. doi: 10.1099/0022-1317-67-11-2325. [DOI] [PubMed] [Google Scholar]
- 6.Askonas BA, Lin YL. An influenza specific T-killer clone is restricted to H-2Ld and cross-reacts with Dk region. Immunogenetics. 1982;16(1):83–87. doi: 10.1007/BF00364444. [DOI] [PubMed] [Google Scholar]
- 7.Kuwano K, Scott M, Young JF, Ennis FA. HA2 subunit of influenza A H1 and H2 subtype viruses induces a protective cross-reactive cytotoxic T lymphocyte response. J Immunol. 1988 Feb 15;140(4):1264–1268. [PubMed] [Google Scholar]
- 8.Kuwano K, Tamura M, Ennis FA. Cross-reactive protection against influenza A virus infections by an NS1-specific CTL clone. Virology. 1990 Sep;178(1):174–179. doi: 10.1016/0042-6822(90)90391-4. [DOI] [PubMed] [Google Scholar]
- 9.Kuwano K, Braciale TJ, Ennis FA. Cytotoxic T lymphocytes recognize a cross-reactive epitope on the transmembrane region of influenza H1 and H2 hemagglutinins. Viral Immunol. 1989 Fall;2(3):163–173. doi: 10.1089/vim.1989.2.163. [DOI] [PubMed] [Google Scholar]
- 10.Bender BS, Croghan T, Zhang L, Small PA., Jr Transgenic mice lacking class I major histocompatibility complex-restricted T cells have delayed viral clearance and increased mortality after influenza virus challenge. The Journal of experimental medicine. 1992 Apr 1;175(4):1143–1145. doi: 10.1084/jem.175.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham MB, Braciale TJ. Resistance to and recovery from lethal influenza virus infection in B lymphocyte-deficient mice. The Journal of experimental medicine. 1997 Dec 15;186(12):2063–2068. doi: 10.1084/jem.186.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epstein SL, Lo CY, Misplon JA, Bennink JR. Mechanism of protective immunity against influenza virus infection in mice without antibodies. J Immunol. 1998 Jan 1;160(1):322–327. [PubMed] [Google Scholar]
- 13.Mackenzie CD, Taylor PM, Askonas BA. Rapid recovery of lung histology correlates with clearance of influenza virus by specific CD8+ cytotoxic T cells. Immunology. 1989 Jul;67(3):375–381. [PMC free article] [PubMed] [Google Scholar]
- 14.Wells MA, Albrecht P, Ennis FA. Recovery from a viral respiratory infection. I. Influenza pneumonia in normal and T-deficient mice. J Immunol. 1981 Mar;126(3):1036–1041. [PubMed] [Google Scholar]
- 15.Eichelberger M, Allan W, Zijlstra M, Jaenisch R, Doherty PC. Clearance of influenza virus respiratory infection in mice lacking class I major histocompatibility complex-restricted CD8+ T cells. The Journal of experimental medicine. 1991 Oct 1;174(4):875–880. doi: 10.1084/jem.174.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belz GT, Wodarz D, Diaz G, Nowak MA, Doherty PC. Compromised influenza virus-specific CD8(+)-T-cell memory in CD4(+)-T-cell-deficient mice. Journal of virology. 2002 Dec;76(23):12388–12393. doi: 10.1128/JVI.76.23.12388-12393.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas PG, Keating R, Hulse-Post DJ, Doherty PC. Cell-mediated protection in influenza infection. Emerging infectious diseases. 2006 Jan;12(1):48–54. doi: 10.3201/eid1201.051237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ennis FA, Rook AH, Qi YH, Schild GC, Riley D, Pratt R, et al. HLA restricted virus-specific cytotoxic T-lymphocyte responses to live and inactivated influenza vaccines. Lancet. 1981 Oct 24;2(8252):887–891. doi: 10.1016/s0140-6736(81)91389-1. [DOI] [PubMed] [Google Scholar]
- 19.Jameson J, Cruz J, Ennis FA. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. Journal of virology. 1998 Nov;72(11):8682–8689. doi: 10.1128/jvi.72.11.8682-8689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jameson J, Cruz J, Terajima M, Ennis FA. Human CD8+ and CD4+ T lymphocyte memory to influenza A viruses of swine and avian species. J Immunol. 1999 Jun 15;162(12):7578–7583. [PubMed] [Google Scholar]
- 21.Boyam A. Isolation of mononuclear cells and granulocytes from human blood. Scand J Clin Lab Invest. 1968;21 (Suppl.):77. [PubMed] [Google Scholar]
- 22.Ennis FA, Cruz J, Demkowicz WE, Jr, Rothman AL, McClain DJ. Primary induction of human CD8+ cytotoxic T lymphocytes and interferon-gamma-producing T cells after smallpox vaccination. The Journal of infectious diseases. 2002 Jun 1;185(11):1657–1659. doi: 10.1086/340517. [DOI] [PubMed] [Google Scholar]
- 23.Currier JR, Kuta EG, Turk E, Earhart LB, Loomis-Price L, Janetzki S, et al. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. Journal of immunological methods. 2002 Feb 1;260(1–2):157–172. doi: 10.1016/s0022-1759(01)00535-x. [DOI] [PubMed] [Google Scholar]
- 24.Rowe T, Abernathy RA, Hu-Primmer J, Thompson WW, Lu X, Lim W, et al. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. Journal of clinical microbiology. 1999 Apr;37(4):937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki S, Jaimes MC, Holmes TH, Dekker CL, Mahmood K, Kemble GW, et al. Comparison of the influenza virus-specific effector and memory B-cell responses to immunization of children and adults with live attenuated or inactivated influenza virus vaccines. Journal of virology. 2007 Jan;81(1):215–228. doi: 10.1128/JVI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gross PA, Ennis FA, Gaerlan PF, Denson LJ, Denning CR, Schiffman D. A controlled double-blind comparison of reactogenicity, immunogenicity, and protective efficacy of whole-virus and split-product influenza vaccines in children. The Journal of infectious diseases. 1977 Nov;136(5):623–632. doi: 10.1093/infdis/136.5.623. [DOI] [PubMed] [Google Scholar]
- 27.Rastogi S, Gross PA, Bonelli J, Dran S, Levandowski RA, Russo C, et al. Time to peak serum antibody response to influenza vaccine. Clinical and diagnostic laboratory immunology. 1995 Jan;2(1):120–121. doi: 10.1128/cdli.2.1.120-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillespie GM, Wills MR, Appay V, O'Callaghan C, Murphy M, Smith N, et al. Functional heterogeneity and high frequencies of cytomegalovirus-specific CD8(+) T lymphocytes in healthy seropositive donors. J Virol. 2000 Sep;74(17):8140–8150. doi: 10.1128/jvi.74.17.8140-8150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wills MR, Carmichael AJ, Weekes MP, Mynard K, Okecha G, Hicks R, et al. Human virus-specific CD8+ CTL clones revert from CD45ROhigh to CD45RAhigh in vivo: CD45RAhighCD8+ T cells comprise both naive and memory cells. J Immunol. 1999 Jun 15;162(12):7080–7087. [PubMed] [Google Scholar]
- 30.Arlettaz L, Barbey C, Dumont-Girard F, Helg C, Chapuis B, Roux E, et al. CD45 isoform phenotypes of human T cells: CD4(+)CD45RA(−)RO(+) memory T cells reacquire CD45RA without losing CD45RO. Eur J Immunol. 1999 Dec;29(12):3987–3994. doi: 10.1002/(SICI)1521-4141(199912)29:12<3987::AID-IMMU3987>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Catalina MD, Sullivan JL, Brody RM, Luzuriaga K. Phenotypic and functional heterogeneity of EBV epitope-specific CD8+ T cells. J Immunol. 2002 Apr 15;168(8):4184–4191. doi: 10.4049/jimmunol.168.8.4184. [DOI] [PubMed] [Google Scholar]
- 32.Durando P, Fenoglio D, Boschini A, Ansaldi F, Icardi G, Sticchi L, et al. Safety and immunogenicity of two influenza subunit vaccines, with or without MF59 adjuvant, administered to HIV-1 seropositive and seronegative adults. Clin Vaccine Immunol. 2007 Nov 14; doi: 10.1128/CVI.00316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long BR, Michaelsson J, Loo CP, Ballan WM, Vu BA, Hecht FM, et al. Elevated frequency of IFN-{gamma} producing NK cells following Influenza vaccination in healthy adults. Clin Vaccine Immunol. 2007 Nov 14; doi: 10.1128/CVI.00357-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He XS, Holmes TH, Zhang C, Mahmood K, Kemble GW, Lewis DB, et al. Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines. Journal of virology. 2006 Dec;80(23):11756–11766. doi: 10.1128/JVI.01460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mbawuike I, Zang Y, Couch RB. Humoral and cell-mediated immune responses of humans to inactivated influenza vaccine with or without QS21 adjuvant. Vaccine. 2007 Apr 30;25(17):3263–3269. doi: 10.1016/j.vaccine.2007.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rastogi D, Wang C, Mao X, Lendor C, Rothman PB, Miller RL. Antigen-specific immune responses to influenza vaccine in utero. The Journal of clinical investigation. 2007 Jun;117(6):1637–1646. doi: 10.1172/JCI29466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Canas V, Lorbetskie B, Bertrand D, Cyr TD, Girard M. Selective and quantitative detection of influenza virus proteins in commercial vaccines using two-dimensional high-performance liquid chromatography and fluorescence detection. Analytical chemistry. 2007 Apr 15;79(8):3164–3172. doi: 10.1021/ac0621120. [DOI] [PubMed] [Google Scholar]
- 38.McElhaney JE, Hooton JW, Hooton N, Bleackley RC. Comparison of single versus booster dose of influenza vaccination on humoral and cellular immune responses in older adults. Vaccine. 2005 May 9;23(25):3294–3300. doi: 10.1016/j.vaccine.2005.01.080. [DOI] [PubMed] [Google Scholar]
- 39.Deng Y, Jing Y, Campbell AE, Gravenstein S. Age-related impaired type 1 T cell responses to influenza: reduced activation ex vivo, decreased expansion in CTL culture in vitro, and blunted response to influenza vaccination in vivo in the elderly. J Immunol. 2004 Mar 15;172(6):3437–3446. doi: 10.4049/jimmunol.172.6.3437. [DOI] [PubMed] [Google Scholar]
- 40.McElhaney JE, Upshaw CM, Hooton JW, Lechelt KE, Meneilly GS. Responses to influenza vaccination in different T-cell subsets: a comparison of healthy young and older adults. Vaccine. 1998 Nov;16(18):1742–1747. doi: 10.1016/s0264-410x(98)00133-9. [DOI] [PubMed] [Google Scholar]


