Abstract
Formaldehyde is an intermediate formed during the metabolism of methanol or other methylated compounds. Many Gram-negative bacteria generate formaldehyde from methanol via a periplasmic pyrroloquinoline quinone (PQQ)-dependent dehydrogenase in which the α subunit of an α2β2 tetramer has catalytic activity. The genome of the facultative formaldehyde-oxidizing bacterium Rhodobacter sphaeroides encodes XoxF, a homologue of the catalytic subunit of a proposed PQQ-containing dehydrogenase of Paracoccus denitrificans. R. sphaeroides xoxF is part of a gene cluster that encodes periplasmic c-type cytochromes, including CycI, isocytochrome c2 and CycB (a cyt c553i homologue), as well as adhI, a glutathione-dependent formaldehyde dehydrogenase (GSH-FDH), and gfa, a homologue of a glutathione–formaldehyde activating enzyme (Gfa). To test the roles of XoxF, CycB and Gfa in formaldehyde metabolism by R. sphaeroides, we monitored photosynthetic growth with methanol as a source of formaldehyde and whole-cell methanol-dependent oxygen uptake. Our data show that R. sphaeroides cells lacking XoxF or CycB do not exhibit methanol-dependent oxygen uptake and lack the capacity to utilize methanol as a sole photosynthetic carbon source. These results suggest that both proteins are required for formaldehyde metabolism. R. sphaeroides Gfa is not essential to activate formaldehyde, as cells lacking gfa are capable of both methanol-dependent oxygen uptake and growth with methanol as a photosynthetic carbon source.
INTRODUCTION
Formaldehyde is produced at significant levels by abiotic and biological processes (Auerbach et al., 1977; Levy, 1971). Because this compound can inactivate many cellular components, cells have systems to metabolize formaldehyde (Auerbach et al., 1977; Barber & Donohue, 1998). We are studying proteins involved in formaldehyde metabolism by Rhodobacter sphaeroides, a bacterium with an inducible system for the metabolism of this compound (Barber & Donohue, 1998; Hickman et al., 2004).
The study of formaldehyde metabolism in purple non-sulfur photosynthetic bacteria such as R. sphaeroides began when Quayle and co-workers isolated species capable of photosynthetic growth on medium containing methanol and bicarbonate (Quayle & Pfennig, 1975; Sahm et al., 1976). In media supplemented with methanol or methylated compounds such as choline, R. sphaeroides utilizes a GSH-dependent pathway to metabolize formaldehyde (Barber et al., 1996; Barber & Donohue, 1998). Subsequent studies have shown that R. sphaeroides is not able to grow on methylamine (Barber et al., 1996; Barber & Donohue, 1998), and that the genome of this bacterium lacks genes needed for the metabolism of this and other one-carbon (C1) methyl donors (Mackenzie et al., 2001).
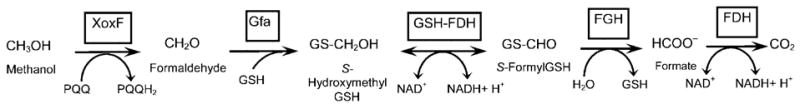
In R. sphaeroides, formaldehyde (CH2O) is added to the thiol of glutathione to form S-hydroxymethylGSH (GS-CH2OH), a reaction which can either occur spontaneously or be facilitated by a glutathione/formaldehyde-activating enzyme (Gfa) (Goenrich et al., 2002). GS-CH2OH is oxidized to S-formylGSH (GS-CHO) by a GSH-dependent formaldehyde dehydrogenase (GSH-FDH), then converted to formate (HCOO−) by S-formylGSH hydrolase (FGH). Finally, formate is oxidized by formate dehydrogenase (FDH) to carbon dioxide (CO2). The R. sphaeroides GSH-FDH structural gene (adhI) is part of a potential formaldehyde metabolism gene cluster (gfa-adhI-cycI-xoxF-cycB-RSP2580-RSP2581) (Mackenzie et al., 2001) that encodes proteins with high amino acid identity to those involved in C1 metabolism by facultative methylotrophs (Fig. 1a) (Chistoserdova, 1996; Harms & van Spanning, 1991; Harms et al., 1996; Ras et al., 1991). In contrast, R. sphaeroides genes encoding FGH and FDH homologues map elsewhere in the genome (Mackenzie et al., 2001).
Fig. 1.
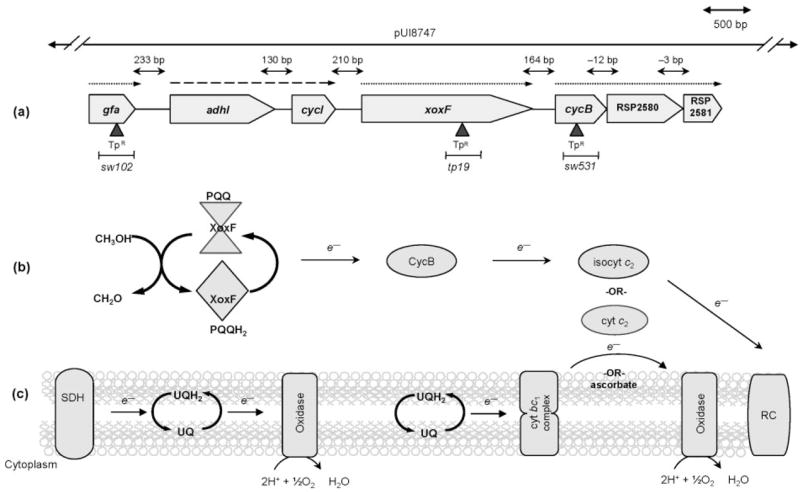
Succinate and methanol oxidation by R. sphaeroides. (a) Shown is the direction of gene transcription (arrowhead) and the intergenic distances within this region of the R. sphaeroides genome. Trimethoprim (TpR) cartridge insertion is indicated by ▲; the extent of wild-type DNA in cosmid pUI8747 is shown above the map. Known operons are shown by a dashed arrow (Barber et al., 1996; Rott et al., 1993), whereas dotted arrows show potential operons (Y. Dufour, personal communication). (b) Periplasmic oxidation of methanol to formaldehyde results in the concomitant transfer of electrons to CycB and to isocyt c2, and the reduction of O2 by a terminal cytochrome oxidase. Alternatively, under photosynthetic conditions, electrons can be transferred to RC complexes in the membrane. (c) During succinate-dependent respiration, succinate dehydrogenase (SDH) reduces ubiquinone (UQH2) via a terminal quinol oxidase. Alternatively, electrons from quinone can pass through cyt bc1, then to periplasmic cytochromes such as isocyt c2, and finally onto membrane-bound cytochrome oxidase to reduce O2.
The periplasmic oxidation of methanol to formaldehyde is often catalysed by proteins that contain pyrroloquinoline quinone (PQQ) as a prosthetic group (Anthony & Williams, 2003; Goodwin & Anthony, 1998). In such PQQ-dependent methanol dehydrogenases, the α subunit (MxaF) has catalytic activity while the role of the β subunit (MxaI) is not known (Anthony & Williams, 2003; Goodwin & Anthony, 1998). R. sphaeroides XoxF has amino acid sequence similarity to MxaF and XoxF of Paracoccus denitrificans. P. denitrificans XoxF has been implicated in formaldehyde formation from methanol, but an XoxF mutant of this bacterium is able to grow with methanol as a sole carbon source, albeit to a reduced extent, presumably because cells also contain MxaFI (Harms et al., 1996; Ras et al., 1991). In Methylobacterium extorquens, the reduction of PQQ is followed by successive one-electron transfers to periplasmic c-type cytochromes which bring reducing power to membrane-bound proteins (Anthony, 1992; Goodwin & Anthony, 1998).
This work takes advantage of features of R. sphaeroides to investigate proteins needed for formaldehyde metabolism. R. sphaeroides is able to co-metabolize methanol and other sources of formaldehyde to CO2 in both the presence and the absence of oxygen. R. sphaeroides does not assimilate the CO2 produced during aerobic respiration, as Calvin cycle enzyme activity is not present (Barber & Donohue, 1998). Under photosynthetic conditions (anaerobic plus light), R. sphaeroides can use methanol as a sole carbon source because it assimilates CO2 via the Calvin cycle (Barber & Donohue, 1998). Since the R. sphaeroides genome lacks mxaFI (MxaFI), the contribution of XoxF to formaldehyde metabolism can be easily studied. In addition, R. sphaeroides encodes isocytochrome (isocyt) c2 and CycB, a homologue of soluble cyt c proteins of M. extorquens (MxaG′) and P. denitrificans (cyt c553i), which could carry electrons from a periplasmic dehydrogenase to the membrane (Anthony, 1992; Harms & van Spanning, 1991; Ras et al., 1991; Rott et al., 1992, 1993). Furthermore, R. sphaeroides Gfa is a homologue of a P. denitrificans protein (Goenrich et al., 2002b; Neculai et al., 2005) which forms the GSH-FDH substrate (Barber et al., 1996; Barber & Donohue, 1998). To analyse the requirements for formaldehyde metabolism by R. sphaeroides, we used growth in the presence of methanol and whole-cell O2 uptake with wild-type cells as well as mutants lacking XoxF, PQQ, one or more c-type cytochromes, or Gfa.
METHODS
Bacterial strains and growth conditions
R. sphaeroides strains (Table 1) were grown in Sistrom’s minimal medium (Sistrom, 1960) containing 35 mM succinate at 30 °C. For testing aerobic methanol metabolism, the medium was supplemented with 100 mM methanol. Monitoring photosynthetic growth by R. sphaeroides utilized either a succinate-based medium or one lacking succinate and amino acids but containing 100 mM methanol and 40 mM sodium bicarbonate. Escherichia coli strains were grown at 37 °C in Luria–Bertani medium. E. coli strain DH5α was used as a plasmid host and S17-1 was used for conjugation of plasmids into R. sphaeroides (Davis et al., 1988). When necessary, the medium was supplemented with 30 μg ml−1 trimethoprim (Tp) or 1 μg ml−1 tetracycline (Tc) for R. sphaeroides, and 100 μg ml−1 ampicillin (Ap), 10 μg Tc ml−1 or 100 μg Tp ml−1 for E. coli.
Table 1.
Plasmids and strains
| Plasmid or strain | Relevant phenotype or genotype | Reference or source |
|---|---|---|
| Plasmids | ||
| pUC19 | ApR | Messing (1979) |
| pSUP202 | ApR, TcR, CmR; Mob+; pBR322 derivative | Simon et al. (1983) |
| Litmus 28i | Multi-purpose cloning vector | New England BioLabs |
| pLA2917 | KnR, TcR; RK2 derivative, cos | Allen & Hanson (1985) |
| p34S-Tp | Source of dhfRII cassette; TpR | Dennis & Zylstra (1998) |
| pWT-P1 | 6.7 kb gfa-adhI-cycI-xoxF-cycB-xoxJ-rsp2581 PstI fragment cloned into pUC19 | Witthuhn, This study |
| pXoxF2 | 1958 bp EcoRI fragment containing xoxF | This study |
| pTP16 | 589 bp deletion and TpR insertion at xoxF codon 260 | This study |
| pTP16-12 | 1.9 kb EcoRI fragment containing ΔxoxF : : Tp from pTP16 cloned into pSUP202 | This study |
| pSW502 | 1.2 kb PstI–EcoRI fragment containing cycB | This study |
| pSW517 | 120 bp deletion and TpR insertion at cycB codon 71 | This study |
| pSW521 | 1.7 kb PstI–EcoRI fragment containing ΔcycB : : Tp from pSW517 cloned into pSUP202 | This study |
| pSW161 | 1.2 kb EcoRI fragment containing gfa | This study |
| pSW161-4 | 147 bp deletion and TpR insertion at gfa codon 74 | This study |
| pSW170 | 1.1 kb EcoRI fragment containing Δgfa : : Tp from pSW161-4 cloned into pSUP202 | This study |
| pUI8747 | TcR; gfa-adhI-cycI-xoxF-cycB-xoxJ DNA in pLA2917 | Rott et al. (1993) |
| Strains | ||
| R. sphaeroides | ||
| Ga | crtD | Laboratory strain |
| BOX9 | crtD ΔadhI : : V SpR box-9 : : Tn5TpR | Ozcan (1996) |
| CCMA1 | ccmA1 : : V | Cox et al. (2001) |
| CYCI1 | cycI : : V | Rott et al. (1993) |
| CYCAI | cycA cycI : : V | Rott et al. (1993) |
| TP39 | crtD cycA spd-7 Tn5 : : I, 2300 | Rott et al. (1993) |
| BC1 | fbcBC : : V TpR | Mouncey et al. (2000) |
| TP19 | crtD ΔxoxF : : Tp | This study |
| SW426 | crtD TP19 with pUI8747 | This study |
| SW531 | crtD ΔcycB : : Tp | This study |
| SW538 | crtD SW531 with pUI8747 | This study |
| SW102 | crtD Δgfa : : Tp | This study |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (Φ80 lacZΔM15) hsdR178 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Laboratories Research (1986) |
| S17-1 | C600 : : RP-4 2-(Tc : : Mu) (Kn : : Tn7) thi pro hsdR hsdM+ recA | Simon et al. (1983) |
Molecular methods and plasmid construction
Standard methods were used for preparation of plasmid DNA from E. coli. Modification enzymes, restriction endonucleases and Taq or Pfu polymerase were used as instructed by the manufacturers. Genomic DNA from wild-type and mutant strains of R. sphaeroides was isolated (Chen & Kuo, 1993) and amplified with Pfu polymerase to facilitate cloning into EcoRV-digested Litmus 28i. Candidate genes were analysed by DNA sequence analysis using the Big Dye Cycle Sequencing protocol (Perkin Elmer). Reaction products were applied to Sephadex G-50 columns and analysed at the University of Wisconsin-Madison DNA Sequencing Facility. Primer sequences used are available from the authors upon request.
Creation of R. sphaeroides mutants
Primers specific to the xoxF upstream and downstream regions, both containing an EcoRI restriction endonuclease site, were used to amplify DNA from pWT-P1. These restriction endonuclease sites facilitated cloning of the 1958 bp PCR product in EcoRI-digested pUC19 (pXoxF2). Divergent primers were used with pXoxF2 to create a 589 bp deletion flanked by 780 bp of xoxF upstream DNA and 520 bp downstream DNA in pUC19. To degrade parental DNA, DpnI was added to the PCR product and incubated for 6 h at 37 °C. The p34S-Tp SmaI-digested TpR cartridge (Dennis & Zylstra, 1998) was ligated into the pXoxF2 PCR product and transformed into E. coli DH5α (pTP16). The EcoRI insert of pTP16 was isolated and cloned into EcoRI-digested pSUP202 (pTP16-12). After mobilization of pTP16-12 into R. sphaeroides, TpR isolates were screened for TcS, to monitor the recombination of ΔxoxF : : dhfRII by an even number of crossover events (strain TP19).
Upstream and downstream cycB primers with PstI restriction endonuclease sites were used to amplify DNA from pWT-P1. The 1210 bp PCR product contained an internal EcoRI site producing a PstI–EcoRI fragment that was cloned into a similarly digested pUC19 (pSW502). Divergent primers with XbaI restriction endonuclease sites were used with pSW502, creating a 120 bp deletion with 524 and 533 bp of upstream and downstream sequence, respectively, in pUC19. This PCR product was treated with DpnI. Following digestion of p34S-Tp with XbaI, a TpR cartridge was inserted into pSW502 (digested with XbaI) and transformed into DH5α (pSW517). The EcoRI–PstI cycB-containing fragment of pSW517 was cloned into pSUP202 to produce pSW521. Following conjugal transfer of pSW521 into R. sphaeroides, strains exhibiting a TpR TcS phenotype were analysed to confirm cycB : : dhfRII gene incorporation into the genome by an even number of crossover events.
A 1.2 kb region that included gfa was amplified with primers containing EcoRI sites and cloned into EcoRI-digested Litmus 28i (pSW161). Using divergent primers with SphI restriction sites, a 147 bp deletion was created in pSW161 and the PCR product was treated with DpnI. Following SphI endonuclease treatment of p34S-Tp, the TpR cartridge was ligated into pSW161 (pSW161-4). The gfa-containing EcoRI fragment of pSW161-4 was cloned into pSUP202 to produce pSW170. After mobilization of pSW170 into R. sphaeroides, TpR strains were screened for TcS to score recombination of the Δgfa : : dhfRII allele into the genome by an even number of crossover events.
Cosmid pUI8747 (Rott et al., 1993), which contains wild-type R. sphaeroides DNA, was mobilized into TP19 (XoxF mutant) or SW531 (CycB mutant) to test for complementation.
Photosynthetic methanol utilization
To test for photosynthetic methanol utilization, aerobically grown cells were harvested and washed with Sistrom’s medium lacking a carbon source. Following dilution into Sistrom’s succinate media (~1.0 × 108 c.f.u. ml−1), cells were placed in filled screw-capped test tubes and exposed to an incandescent light source (10 W m−2; measured with a Yellow-Springs-Kettering model 6.5-A radiometer). After 3 days, cells were harvested, washed twice in media lacking a carbon source and diluted (~1.0 × 108 c.f.u. ml−1) in media containing 100 mM methanol and 40 mM sodium bicarbonate. These tubes were exposed to light and monitored for up to 7 days to score photosynthetic growth.
Whole-cell O2 uptake
Whole-cell O2 uptake was monitored using a YSI model 5300A meter equipped with a Clark-type polarographic O2 probe. For these assays, aerobically grown cultures (~2.5 × 108 c.f.u. ml−1) were exposed to 100 mM methanol for 3 h to induce expression of genes required for methanol metabolism (Hickman et al., 2004). These cells were harvested by centrifugation (10 min at 5000 g) and washed twice in medium lacking succinate. Then, ~5.0 × 108 c.f.u. ml−1 of washed cells were added to the sample chamber for O2-uptake assays. O2-consumption rates were determined in the absence of added carbon sources (endogenous rate), in the presence of 35 mM succinate (succinate-dependent respiration) or in the presence of 25–100 mM methanol (methanol-dependent rate). For each assay, measurements were recorded every 15 s over a 240 s period. By plotting O2 consumption as a function of time, an endogenous, succinate-, or methanol-dependent rate of O2 uptake was calculated [nmol O2 consumed min−1 (mg protein)−1].
Computational prediction of potential operons
To predict co-transcribed ORFs (Fig. 1a), Pearson’s correlation coefficients were calculated based on RNA abundance measurements from public-domain R. sphaeroides global gene expression datasets (Gene Expression Omnibus; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL162) (Y. Dufour, personal communication). Statistical probabilities for co-transcription of ORFs were obtained from available models (www.microbesonline.org) (Bockhorst et al., 2003; Price et al., 2005).
RESULTS
Genes potentially involved in R. sphaeroides formaldehyde metabolism
Previous studies have demonstrated that the structural genes for GSH-FDH and isocyt c2 (adhI–cycI) constitute an inducible operon (Fig. 1a) (Barber et al., 1996; Rott et al., 1993) which is controlled by the presence of formaldehyde or of the metabolic sources of this compound such as methanol (Barber & Donohue, 1998; Hickman et al., 2004). Other products of potential ORFs in this region of the R. sphaeroides genome encode homologues of proteins known or predicted to be involved in the formaldehyde metabolism of other microbes (Fig. 1a).
One of these genes, gfa (Fig. 1a), encodes a protein with 73 % amino acid identity to P. denitrificans Gfa, and stimulates the formation of GS-CH2OH from formaldehyde and GSH, the preferred substrate for GSH-FDH (Goenrich et al., 2002; Neculai et al., 2005). RSP2578 has 80 % amino acid sequence identity to XoxF, a putative PQQ-containing dehydrogenase of P. denitrificans (Harms & van Spanning, 1991; Harms et al., 1996), 75 % amino acid identity to the M. extorquens XoxF homologue (MxaF′) (Chistoserdova & Lidstrom, 1997) and 49 % amino acid identity to the catalytic (α) subunit of methanol dehydrogenase, MxaF. Based on the amino acid identity of RSP2579 (64 %) with the mono-haem cyt c553i (cycB) of P. denitrificans (Harms et al., 1996; Ras et al., 1991), we have designated this ORF as R. sphaeroides CycB (predicted pI of 4.2). A haem-staining protein of a molecular mass predicted for the mature CycB protein (~18 kDa after cleavage of predicted signal peptide) has been observed previously in R. sphaeroides Spd mutants, which have increased expression of some genes in this cluster (Rott et al., 1992). RSP2580 has 63 % amino acid identity to XoxJ of P. denitrificans (Harms et al., 1996) and resembles bacterial periplasmic solute binding proteins (Saier, 1994, 2000). Finally, RSP2581 has 62 % amino acid sequence identity to P. denitrificans XoxI. RSP2581 is predicted to have a rhodanese-like fold (Bordo & Bork, 2002; Spallarossa et al., 2003, 2004), a pI of ~4.8, and a molecular mass of 19 kDa (www.expasy.org). While similar clusters of related genes are found in genomes from several bacteria, the contribution of xoxF-cycB-RSP2580-RSP2581 to formaldehyde metabolism remains unclear. To test the role of these gene products in formaldehyde metabolism, we used enzymic and phenotypic assays to monitor the function of this pathway in wild-type and mutant cells.
Requirements for R. sphaeroides formaldehyde metabolism
One test of R. sphaeroides formaldehyde metabolism was a whole-cell O2-uptake assay. We used this assay because earlier attempts to monitor methanol oxidation by other methods in cell extracts of purple non-sulfur bacteria have been unsuccessful for unknown reasons (Hickman, 2003; Quayle & Pfennig, 1975; Sahm et al., 1976).
To enhance our ability to assay methanol-dependent O2 uptake, cells were grown in the presence of methanol for one doubling prior to analysis. These growth conditions are known to increase the expression of genes required for the metabolism of this carbon source (Hickman et al., 2004) and expression of genes involved in formaldehyde oxidation (Barber & Donohue, 1998). This regimen was preferred over growing cells in the presence of methanol for longer periods because we intended to assay both wild-type and mutant cells that might have contained defects in the formation or metabolism of formaldehyde. Following exposure to methanol, cells were harvested and suspended in medium lacking any carbon source to allow determination of a background (endogenous) O2-uptake rate. O2-uptake activity was then monitored after the addition of methanol or succinate (Fig. 2). The endogenous rate of O2 uptake (O2 consumed per unit time) was subtracted from the rate determined in the presence of succinate or methanol (Fig. 2).
Fig. 2.
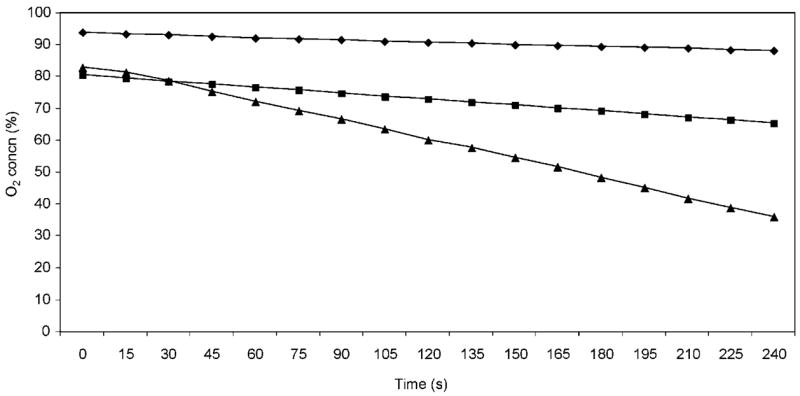
Whole-cell O2 uptake by wild-type cells. Plotted is the percentage O2 concentration as a function of time for each carbon source tested. (◆) Data used to calculate the endogenous rate of O2 uptake (O2 consumption without the addition of any carbon source); (■) samples containing methanol; (▲) samples containing succinate.
R. sphaeroides Ga was used as the parent strain because many of the mutants previously analysed for defects in formaldehyde metabolism have been generated in this background (Barber, 1997; Hickman et al., 2002; Ozcan, 1996; Rott et al., 1993). After subtracting the endogenous rate of O2 uptake, a significant and reproducible rate of methanol-dependent O2 uptake was measured in strain Ga [~5.0 nmol O2 min−1 (mg protein)−1; Table 2]. For comparison, the rate of whole-cell O2 uptake was ~15.8 nmol O2 min−1 (mg protein)−1 when succinate was provided as the electron donor (Table 2). These data indicate that the whole-cell O2 uptake assay is able to report on the use of methanol or succinate as an electron donor. It is not surprising that the rates of O2 uptake are higher with succinate as an electron donor than with methanol, since it is known that wild-type cells have a shorter doubling time in minimal medium that contain this compound as a sole carbon source (Barber et al., 1996; Barber & Donohue, 1998).
Table 2.
Methanol metabolism in PQQ and c-type cyt mutants
| Strain | Relevant genotype | O2 uptake*
|
PS growth†
|
|||
|---|---|---|---|---|---|---|
| Succinate | MeOH | Succinate | Succinate+MeOH | MeOH | ||
| Ga | crtD | 15.8 ± 1.0 | 5.0 ± 0.7 | + | + | + |
| BOX9 | crtD ΔadhI : : ΩSpR box-9 : : Tn5TpR | 13.4 ± 2.0 | ≤ 0.2 | + | + | − |
| CCMA1 | ccmA1 : : ΩSp | 9.7 ± 0.6 | ≤ 0.6 | − | − | − |
| CYCI1 | cycI : : Ω | 17.0 ± 0.4 | 2.2 ± 0.3 | + | + | − |
| CYCAI | cycA cycI : : Ω | 14.1 ± 1.0 | ≤ 0.4 | + | + | − |
Values shown for O2 consumption have been corrected by subtracting the endogenous rate of O2 uptake [~2 nmol O2 min−1 (mg protein)−1].
Photosynthetic (PS) growth of bacteria in liquid medium with succinate, succinate and methanol, or only methanol; (+) growth, (−) no growth.
To further verify the utility of this assay, we monitored whole-cell O2 uptake in mutants with known or predicted defects in formaldehyde metabolism. Based on what is known about the formaldehyde metabolism of other Gram-negative bacteria, we predict that R. sphaeroides would require a PQQ-containing enzyme to generate formaldehyde from methanol. To test this hypothesis, BOX9, a mutant with a loss-of-function mutation in PQQ synthesis, was assayed for methanol oxidation by the whole-cell O2-uptake assay. The rate of O2 consumption in the presence of succinate by R. sphaeroides BOX9 was comparable to that of wild-type cells (Table 2). This was not surprising, since PQQ is not required for electron transport from succinate to O2 (Fig. 1c). In contrast, when methanol was used as an electron donor there was no detectable O2 consumption in the BOX9 strain over background. Results of control experiments indicate that cells lacking AdhI have a rate of methanol-dependent O2 uptake comparable to that of wild-type cells (data not shown). Thus, the deficiency in methanol-dependent O2 uptake exhibited by BOX9 demonstrates the requirement for the PQQ prosthetic group for the conversion of methanol to formaldehyde by R. sphaeroides (Table 2).
Methanol metabolism by Gram-negative bacteria often involves periplasmic c-type cytochromes to transfer electrons from a PQQ-dependent dehydrogenase to membrane-bound enzymes of the respiratory chain (Anthony, 1982, 1992) (Fig. 1b). As predicted, in CCMA1 cells, which contain a mutation that blocks assembly of many periplasmic and membrane-associated cyt c proteins, there is no detectable methanol-dependent O2 uptake compared to background (Table 2). In contrast, succinate-dependent O2 uptake is not abolished in CCMA1 cells (Table 2), presumably because R. sphaeroides contains a quinol oxidase which allows cyt c-independent electron transfer from succinate via quinol to O2 (Fig. 1c). These results demonstrate that methanol-dependent respiration in R. sphaeroides requires one or more cyt c proteins, presumably to transfer electrons from a periplasmic PQQ-dependent enzyme to O2. Indeed, a reduced methanol-dependent O2-uptake rate [~2.2 nmol O2 min−1 (mg protein)−1] is observed in strain CYCI1 (lacks CycI; Table 2), suggesting a potential role for isocyt c2 in electron transfer during formaldehyde generation by this periplasmic protein. On the other hand, no detectable methanol-dependent O2 uptake was measured in CYCAI, which has mutations that inactivate both cycA and the gene that encodes its isoform, cycI (Table 2).
Additional proteins required for R. sphaeroides
Using the O2-uptake assay, we also analysed the properties of R. sphaeroides cells containing defined mutations in xoxF, cycB or gfa (Fig. 1a). Based on the direction of transcription of xoxF and gfa as well as their predicted positions in potential operons, we do not expect the properties of these mutants to be complicated by polarity on downstream genes (Fig. 1a).
Methanol-dependent O2 uptake of R. sphaeroides cells that lacked XoxF (TP19) was not detectable over background (Table 3). As expected, cells lacking XoxF exhibited normal rates of O2 uptake when using succinate as an electron donor (Table 3). Also, the ability to complement the defect of TP19 with cosmid pUI8747 (strain SW426) demonstrates that XoxF is required for methanol-dependent O2 uptake by R. sphaeroides (Table 3). Additionally, cells that lacked CycB (SW531) were incapable of methanol-dependent O2 consumption (Table 3). When SW531 cells containing pUI8747 (strain SW538) were analysed, rates of methanol-dependent O2 uptake were restored (Table 3) to levels observed in wild-type cells. This suggests that CycB is also required for methanol-dependent O2 uptake by R. sphaeroides, presumably since it carries electrons from the enzyme that generates formaldehyde to the respiratory chain (Fig. 1b). Thus, it appears that inhibiting electron transfer from the site of formaldehyde formation is sufficient to prevent methanol-dependent O2 uptake. In the case of both TP19 and SW531, the addition of ascorbate was able to support O2 uptake (data not shown). Ascorbate donates electrons directly to cytochrome oxidases; therefore, both of these strains contain a functional terminal cytochrome oxidase(s) (Fig. 1b). Thus, the failure of TP19 and SW531 to exhibit detectable methanol-dependent O2 uptake is not due to lack of this terminal enzyme in the respiratory chain.
Table 3.
Properties of methanol metabolism mutants
| Strain | Relevant genotype | O2 uptake*
|
PS growth†
|
|||
|---|---|---|---|---|---|---|
| Succinate | MeOH | Succinate | Succinate+MeOH | MeOH | ||
| Ga | crtD | 15.8 ± 1.0 | 5.0 ± 0.7 | + | + | + |
| TP19 | crtD ΔxoxF : : Tp | 21.5 ± 1.0 | ≤ 0.2 | + | + | − |
| SW426 | crtD TP19 with cosmid pUI8747 | 12.6 ± 0.5 | 6.2 ± 1.0 | + | + | + |
| SW531 | crtD ΔcycB : : Tp | 16.1 ± 2.0 | ≤ 0.2 | + | + | − |
| SW538 | crtD SW531 with cosmid pUI8747 | 13.0 ± 0.5 | 6.6 ± 0.7 | + | + | + |
| SW102 | crtD Δgfa : : Tp | 18.0 ± 1.4 | 3.6 ± 0.9 | + | + | + |
Values shown for O2 consumption have been corrected by subtracting the endogenous rate of O2 uptake [~2 nmol O2 min−1 (mg protein)−1].
Photosynthetic (PS) growth of bacteria in liquid medium with succinate, succinate and methanol, or only methanol; (+) growth, (−) no growth.
To analyse the role of R. sphaeroides Gfa in formaldehyde metabolism (Goenrich et al., 2002) we tested O2 uptake by cells that lacked Gfa (SW102). Cells lacking Gfa (SW102) were capable of using succinate or methanol as an electron donor to support O2 uptake; however, the rate of methanol-dependent O2 uptake was slightly lower than that of wild-type cells (Table 3). Based on the observed activity of SW102, we conclude that loss of Gfa is not sufficient to prevent methanol-dependent O2 uptake by R. sphaeroides.
Methanol metabolism during photosynthetic growth
R. sphaeroides requires a GSH-FDH-dependent pathway to use methanol as a sole photosynthetic carbon source. Under these conditions, formaldehyde is oxidized to CO2 and subsequently assimilated by the Calvin cycle (Barber & Donohue, 1998). To further test the role of these proteins in formaldehyde metabolism, we also analysed the ability of wild-type and mutant strains to grow photosynthetically using either succinate or methanol as a carbon source. Based on the pivotal role of c-type cytochromes in photosynthetic and methanol-dependent electron transport (Davis et al., 1988; Rott et al., 1993; Zannoni et al., 1976), it is not surprising that CCMA1, which lacks all c-type cytochromes, is photosynthetically incompetent when using either succinate or methanol as a carbon source (Table 2). Also, it is not surprising that cells lacking isocyt c2 (CYCI1) are able to grow photosynthetically in media containing either succinate or methanol (Table 2), since previous results have shown that cyt c2 can support photosynthetic electron transport under both conditions (Fig. 1b) (Davis et al., 1988; Rott et al., 1992, 1993).
Both PQQ (BOX9) and XoxF (TP19) mutants grew photosynthetically in the presence of succinate. However, BOX9 and TP19 were photosynthetically incompetent when methanol was the sole carbon source (Tables 2 and 3). Presumably, the defect in using methanol during photosynthetic growth for both the PQQ and XoxF mutants is a consequence of the requirement for a PQQ-dependent enzyme such as XoxF. In addition, the CycB mutant (SW531) was able to grow photosynthetically in the presence of succinate but not when methanol was used as the sole carbon source (Table 3). Based on this we predict that CycB is needed to bring electrons from the PQQ-dependent dehydrogenase XoxF to the photosynthetic electron transport chain (Fig. 1b). As expected, the photosynthetic growth defects in the presence of methanol of the XoxF and CycB mutants were each complemented by a cosmid (pUI8747, Fig. 1a) that carries this entire region of the genome (Table 3). Finally, photosynthetic growth of cells that lack XoxF, CycB or PQQ occurred when the media contained both succinate and methanol (Tables 2 and 3), indicating that these mutants are not simply sensitive to methanol. Rather, it appears that the loss of PQQ, XoxF or CycB causes the growth defect by blocking the pathway needed to generate formaldehyde from methanol under photosynthetic conditions.
In contrast, cells lacking Gfa (SW102) grew photosynthetically when using either succinate or methanol (Table 3). These data, plus the ability to measure methanol-dependent O2 uptake in SW102 cells, support the notion that Gfa is not absolutely required for formaldehyde activation and its subsequent metabolism by GSH-FDH in R. sphaeroides.
DISCUSSION
In this work, we analysed products from a region of the R. sphaeroides genome, gfa-adhI-cycI-xoxF-cycB-RSP2580-RSP2581, which encodes proteins implicated in the metabolism of formaldehyde (Fig. 1). To address the function of these gene products in R. sphaeroides, mutants lacking the PQQ-dependent dehydrogenase (XoxF), a homologue of the periplasmic cyt c553i (CycB) or the glutathione formaldehyde activating enzyme (Gfa), were monitored for methanol-dependent O2 uptake or photosynthetic growth using methanol as a carbon source (Table 3). We used methanol as a carbon source for these studies, as previous work has shown that growth under these conditions requires the activity of GSH-FDH to metabolize the formaldehyde that is produced (Barber et al., 1996; Hickman et al., 2002). Also, by analysing strains that lacked periplasmic c-type cytochromes (CYCI1, CYCAI and CCMA1) or possessed a defect in PQQ synthesis, we were able to study other requirements for formaldehyde metabolism (Table 2). Our results show that formaldehyde generation from methanol by R. sphaeroides requires XoxF, PQQ and periplasmic c-type cytochromes, including CycB. However, R. sphaeroides does not require Gfa for the metabolism of formaldehyde.
Whole-cell methanol-dependent O2 uptake and the use of methanol as a sole photosynthetic carbon source require either XoxF or PQQ biosynthesis (Table 3). Because R. sphaeroides BOX9 contains a lesion in PqqE, this finding suggests that XoxF requires PQQ for formaldehyde metabolism (Ozcan, 1996). The properties of the R. sphaeroides XoxF mutant (TP19) and the absence of a gene related to mxaI predict that XoxF might function as an α2 homodimer. In R. sphaeroides and other bacteria that contain homologues of XoxF (Fig. 3), these proteins lack a C-terminal domain of MxaF which interacts with MxaI (Ghosh et al., 1995). This also suggests that active XoxF might not need a β subunit to form an active enzyme. Phylogenetic and bioinformatic analysis found high amino acid identity among other XoxF homologues (Fig. 3), even though the genomes of P. denitrificans, M. extorquens and Rhodopseudomonas palustris BisA53 contain both mxaFI and a homologue of xoxF.
Fig. 3.
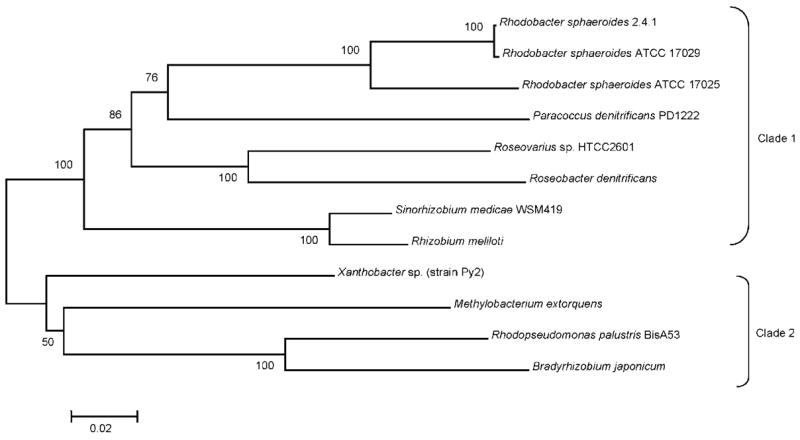
Phylogenetic analysis of XoxF orthologues. Amino acid sequence identities among α-proteobacteria XoxF homologues are represented. The phylogenetic tree shows all three wild-type strains of R. sphaeroides grouping in one of two clades based on evolutionary distance. This analysis was generated with MEGA (Kumar et al., 2004), and bootstrap values above 50 % from 500 replicates are shown. Scale bar, 0.02 substitutions per site.
Interestingly, R. sphaeroides lacks detectable activity using the dye-linked assay that is commonly employed with α2β2 methanol dehydrogenases (Fig. 3) (Hickman, 2003; Quayle & Pfennig, 1975; Sahm et al., 1976). The inability to detect dye-linked methanol dehydrogenase activity in extracts may be due to the absence of a β subunit which can stabilize the enzyme (Ghosh et al., 1995).
It is accepted that periplasmic c-type cytochromes are involved in electron transfer to the membrane following the oxidation of methanol (Anthony, 1992). Since R. sphaeroides possesses well-characterized periplasmic electron transport carriers, we also tested the role of these proteins in methanol metabolism. One ORF in the proposed R. sphaeroides formaldehyde metabolism gene cluster encodes a c-type cyt with high amino acid identity to cyt c553i, and mutational analysis of cycB showed that the protein is required for growth and O2 uptake when R. sphaeroides is placed in formaldehyde-generating conditions. Although cyt c553i homologues of P. denitrificans or M. extorquens (MxaG′) have been suggested to be involved in C1 metabolism, earlier biochemical or genetic attempts to demonstrate a role in this pathway have been unsuccessful (Chistoserdova & Lidstrom, 1997; Harms et al., 1996; Ras et al., 1991). Also, studies in P. denitrificans have demonstrated that the primary electron acceptor of MxaFI is cyt c551i and not cyt c553i, while in M. extorquens cyt cL directly accepts electrons from methanol dehydrogenase (Anthony, 1992; Long & Anthony, 1991). While we cannot distinguish whether CycB is a direct electron acceptor from XoxF or an intermediate carrier of electrons to the membrane, we have shown that this protein is required by R. sphaeroides when methanol is oxidized to formaldehyde. The intergenic distance between R. sphaeroides cycB and adjacent ORFs (RSP2580 and RSP2581) combined with computational analysis of global gene-expression studies suggests that these three genes might be co-transcribed (Y. Dufour, personal communication). Thus, our findings could also reflect the effect of the cycB mutation on the expression of RSP2580 and RSP2581. If this was the case, then it would also provide the first evidence for a specific role of these uncharacterized proteins in formaldehyde metabolism.
Our data predict that one or more class I c-type cyt proteins, isocyt c2 or cyt c2 is involved in electron transport from the periplasmic site of formaldehyde formation to the membrane. We propose that one or both of the class I c-type cytochromes transfers electrons to the membrane, especially since previous studies have shown that each cyt c2 isoform can transfer electrons to reaction centre (RC) complexes (Fig. 1b) (Rott et al., 1993).
Purified Gfa of P. denitrificans catalyses the condensation of formaldehyde and glutathione to generate the hydroxymethylGSH adduct used as a substrate by GSH-FDH (Goenrich et al., 2002). We conclude that R. sphaeroides does not absolutely require Gfa for formaldehyde activation because cells lacking Gfa (SW102) were capable of electron transfer from methanol to O2 and able to use methanol as a sole photosynthetic carbon source. Cells that lack GSH-FDH accumulate formaldehyde and are killed by methanol (Barber et al., 1996; Rott et al., 1993). Thus, the properties of SW102 suggest that formaldehyde can spontaneously condense with GSH at a sufficient rate in vivo to prevent accumulation of formaldehyde. In M. extorquens the formaldehyde activating enzyme (Fae) catalyses the condensation of formaldehyde and tetrahydromethanopterin (Vorholt et al., 2000). Unlike Gfa of R. sphaeroides, Fae is necessary for growth on methanol (Vorholt et al., 2000).
In summary, these experiments have identified additional proteins that are required by cells to generate formaldehyde from methylated compounds and to participate in other steps of the metabolism of this compound. Our results indicate that formaldehyde metabolism by R. sphaeroides requires XoxF, PQQ and periplasmic c-type cyt proteins, including CycB, isocyt c2 or cyt c2. However, Gfa is not absolutely required by R. sphaeroides to condense formaldehyde with glutathione. Based on these findings, we predict that the PQQ-dependent dehydrogenase XoxF oxidizes methanol to formaldehyde, which stimulates electron transfer to CycB and isocyt c2, and then onto membrane-associated electron acceptors (Fig. 1b). These findings make specific predictions for further analysis of this pathway in R. sphaeroides and other bacteria which contain a similar putative formaldehyde gene cluster.
Acknowledgments
This study was supported by grants GM020609-01 (National Institute of General Medical Sciences) to S. M. W. and DE-FG02-05ER15653 (Department of Energy) to T. J. D.
Abbreviations
- Ap
ampicillin
- cyt
cytochrome
- GS-CH2OH
S-hydroxymethylGSH
- GSH-FDH
glutathione-dependent formaldehyde dehydrogenase
- isocyt
isocytochrome
- PQQ
pyrroloquinoline quinone
- RC
reaction centre
- Tc
tetracycline
- Tp
trimethoprim
References
- Allen LN, Hanson RS. Construction of broad-host-range cosmid cloning vectors: identification of genes necessary for growth of Methylobacterium organophilum on methanol. J Bacteriol. 1985;161:955–962. doi: 10.1128/jb.161.3.955-962.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C. The Biochemistry of Methylotrophs. London: Academic Press; 1982. [Google Scholar]
- Anthony C. The c type cytochromes of methylotrophic bacteria. Biochim Biophys Acta. 1992;1099:1–15. [PubMed] [Google Scholar]
- Anthony C, Williams P. The structure and mechanism of methanol dehydrogenase. Biochim Biophys Acta. 2003;1647:18–23. doi: 10.1016/s1570-9639(03)00042-6. [DOI] [PubMed] [Google Scholar]
- Auerbach C, Moutschen-Dahmen M, Moutschen J. Genetic and cytogenetical effects of formaldehyde and related compounds. Mutat Res. 1977;39:317–361. doi: 10.1016/0165-1110(77)90011-2. [DOI] [PubMed] [Google Scholar]
- Barber RD. Formaldehyde sensing and metabolism in Rhodobacter sphaeroides. University of Wisconsin-Madison; 1997. PhD dissertation. [Google Scholar]
- Barber RD, Donohue TJ. Pathways for transcriptional activation of a glutathione-dependent formaldehyde dehydrogenase gene. J Mol Biol. 1998;280:775–784. doi: 10.1006/jmbi.1998.1900. [DOI] [PubMed] [Google Scholar]
- Barber RD, Rott MA, Donohue TJ. Characterization of a glutathione-dependent formaldehyde dehydrogenase from Rhodobacter sphaeroides. J Bacteriol. 1996;178:1386–1393. doi: 10.1128/jb.178.5.1386-1393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethesda Laboratories Research. BRL pUC host: E. coli DH5α™ competent cells. Bethesda Res Lab Focus. 1986;8:9–12. [Google Scholar]
- Bockhorst J, Qiu Y, Glasner J, Liu M, Blattner F, Craven M. Predicting bacterial transcription units using sequence and expression data. Bioinformatics. 2003;19:i34–i43. doi: 10.1093/bioinformatics/btg1003. [DOI] [PubMed] [Google Scholar]
- Bordo D, Bork P. The rhodanese/Cdc25 phosphatase superfamily. Sequence–structure–function relations. EMBO Rep. 2002;3:741–746. doi: 10.1093/embo-reports/kvf150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WP, Kuo TT. A simple and rapid method for the preparation of Gram-negative bacterial genomic DNA. Nucleic Acids Res. 1993;21:2260. doi: 10.1093/nar/21.9.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. Metabolism of formaldehyde in M. extorquens AM1. In: Lidstrom ME, Tabita FR, editors. Microbial Growth on C1 Compounds. Dordrecht: Kluwer Academic Publishers; 1996. pp. 16–24. [Google Scholar]
- Chistoserdova L, Lidstrom ME. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology. 1997;143:1729–1736. doi: 10.1099/00221287-143-5-1729. [DOI] [PubMed] [Google Scholar]
- Cox RL, Patterson C, Donohue TJ. Roles for the Rhodobacter sphaeroides CcmA and CcmG proteins. J Bacteriol. 2001;183:4643–4647. doi: 10.1128/JB.183.15.4643-4647.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J, Donohue TJ, Kaplan S. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J Bacteriol. 1988;170:320–329. doi: 10.1128/jb.170.1.320-329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis JJ, Zylstra GJ. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of Gram-negative bacterial genomes. Appl Environ Microbiol. 1998;64:2710–2715. doi: 10.1128/aem.64.7.2710-2715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Anthony C, Harlos K, Goodwin MG, Blake C. The refined structure of the quinoprotein methanol dehydrogenase from Methylobacterium extorquens at 1.94 A. Structure. 1995;3:177–187. doi: 10.1016/s0969-2126(01)00148-4. [DOI] [PubMed] [Google Scholar]
- Goenrich M, Bartoschek S, Hagemeier CH, Griesinger C, Vorholt JA. A glutathione-dependent formaldehyde activating enzyme (Gfa) from Paracoccus denitrificans detected and purified via 2D proton exchange NMR spectroscopy. J Biol Chem. 2002;277:3069–3072. doi: 10.1074/jbc.C100579200. [DOI] [PubMed] [Google Scholar]
- Goodwin PM, Anthony C. The biochemistry, physiology and genetics of PQQ and PQQ-containing enzymes. Adv Microb Physiol. 1998;40:1–80. doi: 10.1016/s0065-2911(08)60129-0. [DOI] [PubMed] [Google Scholar]
- Harms N, van Spanning RJ. C1 metabolism in Paracoccus denitrificans: genetics of Paracoccus denitrificans. J Bioenerg Biomembr. 1991;23:187–210. doi: 10.1007/BF00762217. [DOI] [PubMed] [Google Scholar]
- Harms N, Ras J, Koning S, Reijnders WNM, Stouthamer AH, van Spanning RJM. Genetics of C1 metabolism regulation in Paracoccus denitrificans. In: Lidstrom ME, Tabita FR, editors. Microbial Growth on C1 Compounds. Dordrecht: Kluwer Academic Publishers; 1996. pp. 126–132. [Google Scholar]
- Hickman J. Physiology and regulation of glutathione-dependent formaldehyde metabolism. University of Wisconsin-Madison; 2003. PhD dissertation. [Google Scholar]
- Hickman JW, Barber RD, Skaar EP, Donohue TJ. Link between the membrane-bound pyridine nucleotide transhydrogenase and glutathione-dependent processes in Rhodobacter sphaeroides. J Bacteriol. 2002;184:400–409. doi: 10.1128/JB.184.2.400-409.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JW, Witthuhn VC, Jr, Dominguez M, Donohue TJ. Positive and negative transcriptional regulators of glutathione-dependent formaldehyde metabolism. J Bacteriol. 2004;186:7914–7925. doi: 10.1128/JB.186.23.7914-7925.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Levy H. Normal atmosphere: large radical and formaldehyde concentrations predicted. Science. 1971;173:141–143. doi: 10.1126/science.173.3992.141. [DOI] [PubMed] [Google Scholar]
- Long AR, Anthony C. Characterization of the periplasmic cytochromes c of Paracoccus denitrificans: identification of the electron acceptor for methanol dehydrogenase, and description of a novel cytochrome c heterodimer. J Gen Microbiol. 1991;137:415–425. doi: 10.1099/00221287-137-10-2353. [DOI] [PubMed] [Google Scholar]
- Mackenzie C, Choudhary M, Larimer FW, Predki PF, Stilwagen S, Armitage JP, Barber RD, Donohue TJ, Hosler JP, et al. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth Res. 2001;70:19–41. doi: 10.1023/A:1013831823701. [DOI] [PubMed] [Google Scholar]
- Messing J. A multipurpose cloning system based on the single-standed DNA bacteriophage, M13. Recomb DNA Tech Bull. 1979;2:43–48. [Google Scholar]
- Mouncey NJ, Gak E, Choudhary M, Oh J, Kaplan S. Respiratory pathways of Rhodobacter sphaeroides 2.4.1T: identification and characterization of genes encoding quinol oxidases. FEMS Microbiol Lett. 2000;192:205–210. doi: 10.1111/j.1574-6968.2000.tb09383.x. [DOI] [PubMed] [Google Scholar]
- Neculai AM, Neculai D, Griesinger C, Vorholt JA, Becker S. A dynamic zinc redox switch. J Biol Chem. 2005;280:2826–2830. doi: 10.1074/jbc.C400517200. [DOI] [PubMed] [Google Scholar]
- Ozcan S. Pyrroloquinoline quinone (PQQ) synthesis is required for butanol metabolism. University of Wisconsin-La Crosse; 1996. MS thesis. [Google Scholar]
- Price MN, Huang KH, Alm EJ, Arkin AP. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 2005;33:880–892. doi: 10.1093/nar/gki232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quayle JR, Pfennig N. Utilization of methanol by Rhodospirillaceae. Arch Microbiol. 1975;102:193–198. doi: 10.1007/BF00428368. [DOI] [PubMed] [Google Scholar]
- Ras J, Reijnders WN, van Spanning RJ, Harms N, Oltmann LF, Stouthamer AH. Isolation, sequencing, and mutagenesis of the gene encoding cytochrome c553i of Paracoccus denitrificans and characterization of the mutant strain. J Bacteriol. 1991;173:6971–6979. doi: 10.1128/jb.173.21.6971-6979.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott MA, Fitch J, Meyer TE, Donohue TJ. Regulation of a cytochrome c2 isoform in wild-type and cytochrome c2 mutant strains of Rhodobacter sphaeroides. Arch Biochem Biophys. 1992;292:576–582. doi: 10.1016/0003-9861(92)90033-s. [DOI] [PubMed] [Google Scholar]
- Rott MA, Witthuhn VC, Schilke BA, Soranno M, Ali A, Donohue TJ. Genetic evidence for the role of isocytochrome c2 in photosynthetic growth of Rhodobacter sphaeroides Spd mutants. J Bacteriol. 1993;175:358–366. doi: 10.1128/jb.175.2.358-366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahm H, Cox RB, Quayle JR. Metabolism of methanol by Rhodopseudomonas acidophila. J Gen Microbiol. 1976;94:313–322. doi: 10.1099/00221287-94-2-313. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- Sistrom WR. A requirement for sodium in the growth of Rhodopseudomonas sphaeroides. J Gen Microbiol. 1960;22:778–785. doi: 10.1099/00221287-22-3-778. [DOI] [PubMed] [Google Scholar]
- Spallarossa A, Forlani F, Pagani S, Salvati L, Visca P, Ascenzi P, Bolognesi M, Bordo D. Inhibition of Azotobacter vinelandii rhodanese by NO-donors. Biochem Biophys Res Commun. 2003;306:1002–1007. doi: 10.1016/s0006-291x(03)01067-2. [DOI] [PubMed] [Google Scholar]
- Spallarossa A, Forlani F, Carpen A, Armirotti A, Pagani S, Bolognesi M, Bordo D. The “rhodanese” fold and catalytic mechanism of 3-mercaptopyruvate sulfurtransferases: crystal structure of SseA from Escherichia coli. J Mol Biol. 2004;335:583–593. doi: 10.1016/j.jmb.2003.10.072. [DOI] [PubMed] [Google Scholar]
- Vorholt JA, Marx CJ, Lidstrom ME, Thauer RK. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J Bacteriol. 2000;182:6645–6650. doi: 10.1128/jb.182.23.6645-6650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannoni D, Melandri BA, Baccarini-Melandri A. Energy transduction in photosynthetic bacteria. X. Composition and function of the branched oxidase system in wild type and respiration deficient mutants of Rhodopseudomonas capsulata. Biochim Biophys Acta. 1976;423:413–430. doi: 10.1016/0005-2728(76)90197-3. [DOI] [PubMed] [Google Scholar]


