Abstract
The genomes of two closely related lytic Thermus thermophilus siphoviruses with exceptionally long (~800 nm) tails, bacteriophages P23-45 and P74-26, were completely sequenced. The P23-45 genome consists of 84,201 bp with 117 putative ORFs (Open Reading Frames), and the P74-26 genome has 83,319 bp and 116 putative ORFs. The two genomes are 92% identical with 113 ORFs shared. Only 25% of phage gene product functions can be predicted from similarities to proteins and protein domains with known functions. The structural genes of P23-45, most of which have no similarity to sequences from public databases, were identified by mass-spectrometric analysis of virions. An unusual feature of the P23-45 and P74-26 genomes is the presence, in their largest intergenic regions, of long polypurine-polypyrimidine (R-Y) sequences with mirror repeat symmetry. Such sequences, abundant in eukaryotic genomes but rare in prokaryotes, are known to form stable triple helices that block replication and transcription and induce genetic instability. Comparative analysis of the two phage genomes shows that the area around the triplex-forming elements is enriched in mutational variations. In vitro, phage R-Y sequences form triplexes and block DNA synthesis by Taq DNA polymerase in orientation-dependent manner, suggesting that they may play a regulatory role during P23-45 and P74-26 development.
Keywords: Thermus thermophilus, thermophages, virion proteomics, bioinformatics, triplex-forming sequence
Introduction
Bacteriophages are the most abundant and diverse form of life on Earth and may exert major influence over the microbial world 1; 2. Comparative viral genomics has already provided important insights into diversity and evolution of phage genomes and the functions of various phage genes. To date, more than 400 phage genomes have been completely sequenced (NCBI, National Center for Biotechnology Information – July 2007). However, phages that infect thermophilic eubacteria have remained mostly unexplored. Genomes of only a few of such phages have been completely sequenced 3; NC_004735; NC_004462). Available genomic data indicate that phages infecting thermophilic bacteria may be substantially different from other known phages. The lack of similarity complicates functional annotations of thermophilic phage genomes. Very little is also known about strategies used by these phages to regulate their gene expression as well as gene expression of their hosts during the infection process. The only phage infecting thermophilic bacteria that has been characterized on molecular level is ϕYS40, a myovirus that infects Thermus thermophilus 3; 4. The functional analysis of the ϕYS40 genome and its gene expression strategy has revealed a wealth of new data about transcription and translation regulation in Thermus 5, indicating that further studies of phages infecting thermophilic bacteria are warranted.
A collection of phages infecting Thermus was reported recently 6. Here, these phages will be referred to as thermophages. Among the 115 thermophage isolates in the collection, four major morphological types of bacteriophages were observed. However, no molecular characterization of these phages was undertaken. The thermophage collection contained 11 siphoviruses (phages with long non-contractile tails). While siphoviruses form the most abundant 7 and, in terms of genomic sequences (NCBI, July 2007) and functionally annotated proteins 8, best characterized morphological group of phages, no genome of phages from the Siphoviridae family that infect the Thermus group of bacteria has been reported. Two thermophages, P23-45 and P74-26, stand out from other siphoviruses because of their extremely long (~800 nm) tails. These phages were isolated from hot springs in geographically close, yet distinct (~30 km apart) areas of the Kamchatka peninsula in Far East, the Dolina Geyser valley (P23-45) and the Uzon valley (P74-26) 6. Limited restriction digest patterns of their DNA indicated that the two phages are closely related to each other 6. To learn more about siphoviruses infecting Thermus and to get evolutionary insight through comparative genomic analysis, we here determined the complete genome sequences of P23-45 and P74-26 and analyzed the two genomes.
Results
Comparison of the P23-45 and P74-26 genomes
The sequences of the P23-45 and P74-26 genomes (84,201 bp and 83,319 bp, respectively) were determined using Fimer technology and assembled using the phredPhrap package (see Materials and Methods). The sequences of the genomes appeared to be almost identical (92.2% identity in global alignment using the Stretcher program from EMBOSS package 9; 10. The C+G content for both phages (57.8%) is significantly lower than that of their host, T. thermophilus (69.4%), but is still in stark contrast with ϕYS40, a Thermus myophage that is AT-rich (a C+G content of 32.6%) 3. While both P23-45 and P74-26 genomes encode putative DNA methylases (ORF14, see below), they are susceptible to several methylation-sensitive restriction endonucleases (data not shown), suggesting that phage DNA is not extensively methylated. No phage DNA ends were identified during genome sequencing and assembly stages.
A total of 117 ORFs longer than 30 codons were predicted in the P23-45 genome using the GeneMark.hmm 2.0/VIOLIN program 11 and 116 ORFs were predicted in the P74-26 genome. The non-coding regions were scanned for the presence of additional genes by searching GenBank, GenPept, and the database of unfinished microbial genomes at NCBI but no additional ORFs were found. Search by the tRNAscan-SE program 12 did not reveal any tRNA-encoding genes.
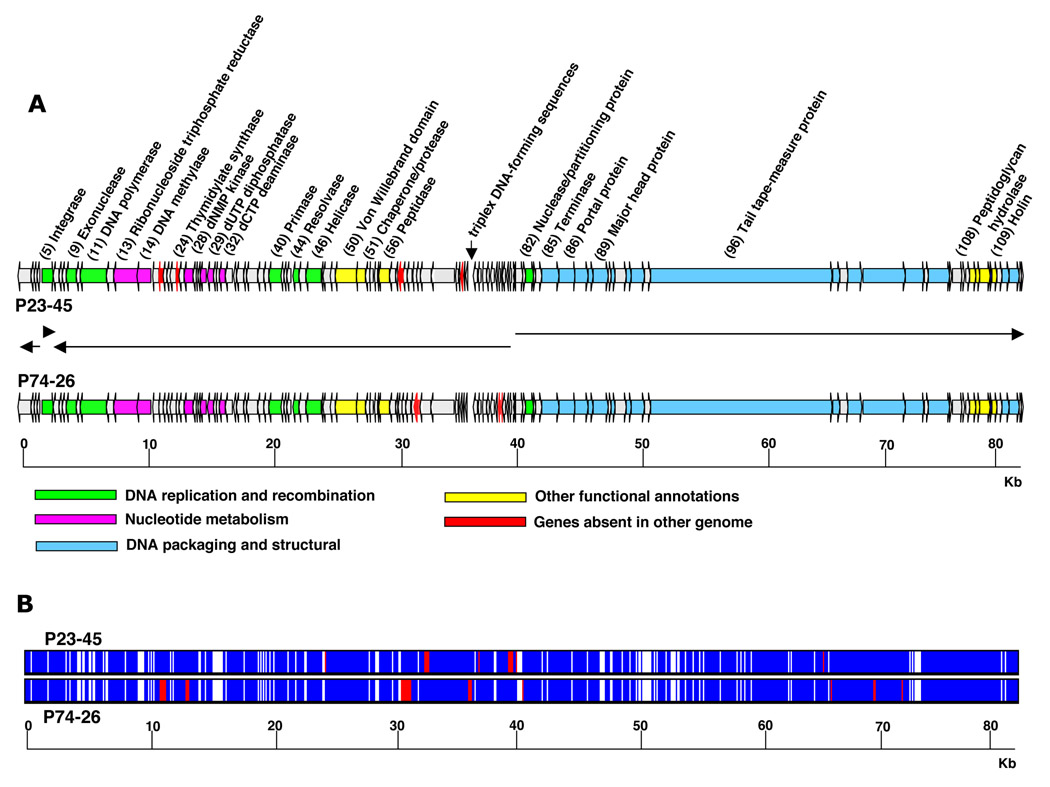
In each phage, almost half of the genome is transcribed in the same direction (leftward in Fig. 1 and in the rest of the paper) and form a single cluster at the left arm of the genome; the remaining genes are transcribed in the rightward direction and also form a cluster at the right arm of the genome. Also, one ORF from the left arm, ORF5, is transcribed in the rightward direction. The P23-45 genome lacks ORFs corresponding to P74-26 ORFs 60, 72, and 73. Conversely, the P74-26 genome has no equivalents corresponding to P23-45 ORFs 16, 22, 59, and 67 (Supplementary Table 1 andFig. 1). None of these ORFs code for proteins with known functions and/or structural motifs, except for P74-26 ORF60 that encodes a protein with WD40-like repeats (Supplementary Table 1). The products of these ORFs must be dispensable for phage development, though they may contribute to different host ranges exhibited by the two thermophages 6.
Figure 1.
A. Schematic representation of the P23-45 and P74-26 phage genomes. Predicted ORFs are indicated by arrows. The end points of the genomes are selected to separate large groups of genes transcribed from opposite DNA strands. The direction of the arrows indicates the direction of transcription. Green, magenta, blue and yellow colors show functions assigned from amino acid similarity with proteins or protein domains in the database; red color shows ORFs that are absent in the other genome (See Supplementary Table 1 for details). The numbers in parentheses next to the predicted gene products represent their numbers for P23-45 shown in Supplementary Table 1.
B. Overview of global P23-45 and P74-26 genomic alignment. White, identical regions; blue, regions with substitutions; red, deletions.
Similarly to other bacteriophage genomes, the coding density of the P23-45 and P74-26 genomes is high. Potentially coding sequences comprise ~96% of their genomes. The length of predicted P23-45 ORFs varies between 30 and 5002 codons and that of P74-26 between 33 and 5006 codons. A 102 bp-long ORF79 (ORF78 in P74-26) is located within ORF78 (ORF77 in P74-26). ORF79 does not have a good SD sequence and may not be a real ORF. Other general features of the two genomes are summarized in Table 1. The longest non-coding regions are located between ORF68 and ORF69 (P23-45) and ORF65 and ORF66 (P74-26), and contain unusual sequences that potentially can form DNA triplexes (see below).
Table 1.
General features of the P23-45 and P74-26 genomes.
| Feature | P23-45 | P74-26 |
|---|---|---|
| Total number of predicted ORFs | 117 | 116 |
| Smallest ORF length (codons) | 30 | 33 |
| Longest ORF length (codons) | 5002 | 5006 |
| Longest non-coding region (bp) | 847 | 887 |
| Number of overlaps between ORFs | 47 | 41 |
| Length of overlaps (bp) | 1–46 | 1–44 |
| Start codon usage | AUG(96) | AUG(93) |
| GUG(13) | GUG(13) | |
| UUG(8) | UUG(10) | |
| Stop codon usage | TGA(36) | TGA(37) |
| TAG(40) | TAG(40) | |
| TAA(41) | TAA(39) |
Comparative sequence analysis of P23-45 and P74-26 ORFs
P23-45 and P74-26 share 113 ORFs. The average rates of nonsynonymous and synonymous nucleotide substitutions over the 113 genes are low (, for nonsynonymous and synonymous nucleotide substitutions, respectively). There are 13 orthologs that have a Ka >0.1; two of them have more than 50% difference at the amino acid level (ORFs 74 and 71 in P23-45 and ORFs 75 and 74 in P74-26, correspondingly). Almost all ORFs with a Ka >0.1 have no homologs in public databases. Interestingly, 9 out of these 13 divergent ORFs are clustered in the genome (ORFs 62, 65, 66, 68, 70, 72-75 in P23-45). This region of the genome also contains a long intergenic spacer with triplex-forming sequences as well as the largest number of indels between the two genomes (Fig. 1, see also below).
In order to study codon usage variations between the phages and the host, we calculated the average RSCU (Relative Synonymous Codon Usage) in P23-45, P74-26, and T. thermophilus. RSCU is defined as a ratio of observed codon frequency to codon frequency expected if all the synonymous codons for each amino acid were used with equal frequency 13. RSCU values would be close to 1 in the absence of codon usage bias. It is known that codon usage patterns in some phages are phage- but not host-specific 14; 15, a property that was proposed to correlate with the presence of phage-encoded DNA polymerases 14. Though P23-45 and P74-26 code for their own DNA polymerase (ORF11 in both genomes, see below) the RSCU values for both phages are similar to those of T. thermophilus (Supplementary Table 2).
The P23-45 and P74-26 genomes encode four and three ORFs, respectively, that are absent from the other genome (Supplementary Table 1). Recently acquired ORFs are expected to have atypical codon usage and compositional bias (reviewed in 16). We calculated individual Nc (effective number of codons) values for 117 P23-45 ORFs and 116 P74-26 ORFs, using CodonW software (www.molbiol.ox.ac.uk/cu) (data not shown). In extremely biased ORFs, Nc can approach 20, while in unbiased ORFs it will approach 61. The average Nc was about 46.1±7 in both genomes. Among four “orphan” genes of the P23-45 phage and three “orphans” of P74-26, only ORF67 of P23-45 exhibited an extreme Nc value of 31.6 (Nc values for other “orphans” were close to 50). This may be an indication that most of the “orphan” ORFs were generated through the loss of their counterparts in another genome rather than by gene acquisition.
Predicted P23-45 and P74-26 proteins
The deduced protein sequences of predicted ORFs were compared to proteins in the non-redundant NCBI database using the PSI-BLAST program with a slightly relaxed cut-off for profile inclusion (-h parameter set at 0.01). Because of the high degree of similarity between the two phages, the analysis below focuses on phage P23-45 (see Supplementary Table 1 for comparison with P74-26, where corresponding genes of both phages are listed). About 25% of P23-45 gene products exhibit sequence similarity to proteins of known function or contain predicted conserved domains, which is somewhat lower than the average number for the large sample of phages in public databases 8. A similar situation is found in the case of unrelated Thermus phage ϕYS40 that we have analyzed earlier 3.
Three predicted ORFs with unknown functions (ORFs 103, 105, and 106) have significant sequence similarities with hypothetical proteins from a small (19,603 bp-size genome) T. aquaticus phage IN93 (NC_004462) and one deduced P23-45 protein, the product of ORF112, has an ORF from ϕYS40 as its BLAST best match (see Supplementary Table 1). The result suggests that diverse Thermus phages have access to a common gene pool, a situation also observed for other phages 17; 18; 19. Since three deduced P23-45 gene products are most similar to proteins from mesophilic phages (Supplementary Table 1), the gene pool accessible to thermophages is not limited to thermophilic organisms. Six more ORFs have proteins from other thermophilic bacteria as their best BLAST matches, including two gene products from T. thermophilus. Thus, at least 10 of 34 functionally annotated P23-45 ORFs (~30%) have proteins from thermophilic organisms as their nearest database neighbors, which is a much larger than the corresponding number for the ϕYS40 phage (less than 10% 3).
The predicted P23-45 ORFs show limited gene order conservation: for example, ORFs 103, 105, and 106 match three adjacent ORFs (22, 23, and 24) of phage IN93, while ORFs 108 and 111 show similarities with adjacent IN93 ORFs 9 and 10. Such conservation of gene order is consistent with modular mode of phage evolution 19; 20.
Proteins involved in DNA metabolism
Like many other large bacteriophages, P23-45 and P74-26 encode a group of proteins that appear to be involved in nucleotide metabolism, including ribonucleoside-triphosphate reductase (ORF13), thymidilate synthase (ORF24), dNMP kinase (ORF28), dUTP diphosphatase (ORF29), and dCTP deaminase (ORF32). Remarkably, these proteins share stronger sequence similarity with homologs from bacteria (both Gram-positive and Gram-negative) and Archaea rather than with phage proteins. The corresponding genes are clustered in the left arm of the genome (Fig. 1 and Supplementary Table 1) and expression of some, or perhaps all of them, may be co-regulated.
Proteins involved in DNA replication, recombination, and transcription
P23-45 and P74-26 encode a set of proteins required for DNA replication, namely, a type A DNA polymerase (ORF11), a putative small subunit of eukaryotic-type DNA primase (ORF40), and replicative helicase DnaB (ORF46). They also encode at least three recombination proteins, an integrase (ORF5), a RecB family exonuclease (ORF9), and a protein homologous to Holliday junction resolvase of the archaeal type (ORF44). The ORF5 integrase is related to site-specific tyrosine recombinases XerC and XerD that are common in eubacteria and are involved in chromosome segregation as well as in integration/excision of temperate phage genomes into and out of host chromosomes 21. This may be an indication that P23-45 and P74-26 are able to enter a lysogenic state.
Replication, repair, and recombination genes are scattered over the left arm in both genomes (Fig. 1 and Supplementary Table 1). No ORFs coding for a DNA ligase, a large subunit of DNA primase, single-strand DNA-binding protein, or RNase H was found, suggesting that the corresponding functions are provided by the host. Notably, T. thermophilus encodes only bacterial DnaG-type DNA primase. It is possible that phages use the host enzyme to prime the replication of their genomes, while phage-encoded small subunit of eukaryotic-type DNA primase may have another role in phage development.
P23-45 and P74-26 do not encode an RNA polymerase (RNAP) or any recognizable σ factors and must therefore rely on host RNAP to transcribe all of their genes. Interestingly, no phage-encoded transcription factors could be predicted, similarly to the case of ϕYS40, where only transcription regulator, ORF18, was predicted 3.
Structural proteins and protein composition of P23-45 virions
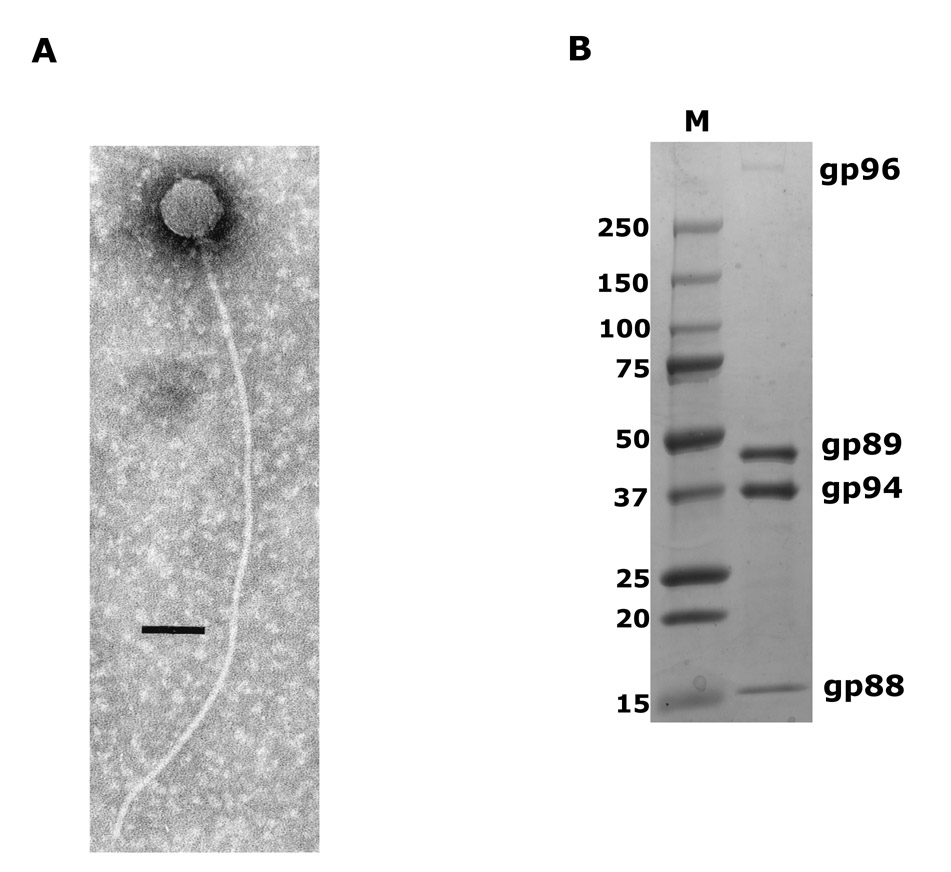
Analysis of amino acid sequences of predicted proteins did not reveal similarities with known bacteriophage structural proteins from public databases except for a putative portal protein ORF86, which is homologous to a putative protein in Corynebacterium (a likely remnant of an integrated prophage, with a putative terminase gene next to it, data not shown). To identify protein composition of phage virions, P23-45 particles (Fig. 2A) were purified by two-step centrifugation in a CsCl gradient and subjected to SDS-PAGE (Fig. 2B). Three major bands of ~17, ~40, and ~50 kDa, as well as one band migrating significantly slower than the largest molecular weight marker (250 kDa), were observed (Fig. 2B).
Figure 2.
A. Electron micrograph of a P23-45 virion showing its extremely long flexible tail and icosahedral capsid. Scale bar, 100 nm.
B. Protein composition of phage P23-45 in SDS-PAGE. The three major structural proteins, gp88, 89 and 94, and the tail-tape measure protein, gp96, are indicated. Lane M, protein molecular weight marker.
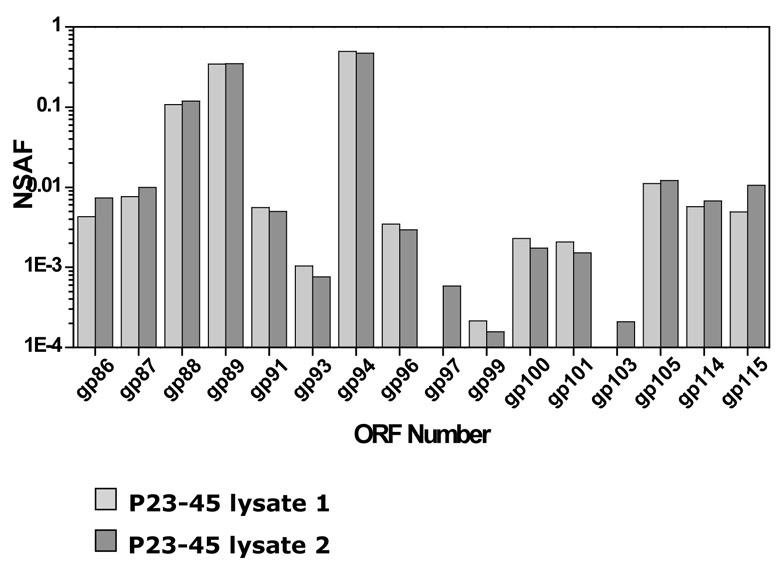
The preparation of P23-45 virions was analyzed by MudPIT (Multidimensional Protein Identification Technology), a shotgun proteomics approach where proteolytic peptides of a protein complex under study (in our case, phage virions) are generated, loaded onto triphasic microcapillary HPLC columns, eluted over several chromatography steps, and analyzed directly by tandem mass spectrometry 22; 23. The results are presented in Fig. 3. Peptides from 16 predicted phage proteins were found in infectious virion preparation (shown in red in Supplementary Table 1). The virion sample was very pure as evidenced by the fact that low levels of only three T. thermophilus proteins were detected. The structural proteins of the phage included gp86, a putative portal protein, six proteins with homology to conserved hypothetical proteins and/or conserved domains from different prokaryotes and phages (gp87, gp89, gp94, gp96, gp103, and gp105), and nine additional proteins without any significant similarity to sequences in public databases. Almost all P23-45 virion proteins (14 of the 16) are encoded by adjacent ORFs 86–105 clustered in the right arm of the genome. These ORFs must form the cluster of phage late genes. The cluster of structural genes appears to be discontinuous, since the products of genes 90, 92, 95, 98, 102, and 104 were not detected by MudPIT (even after lowering the selection criteria for spectrum/peptides matches). The corresponding proteins may either be present in the virions in very low amounts that prevent their detection by MudPIT or may indeed not be part of the virion. One ORF in the cluster, ORF96, encodes a protein of 5002 amino acid residues (the corresponding protein in P74-26, the product of ORF95, is 5006 amino acid residues-long). To our knowledge, these proteins are among the longest known bacteriophage or bacterial proteins; gp96 may correspond to the high-molecular weight band seen on the SDS gel shown in Fig. 2B.
Figure 3.
MudPIT analysis of P23-45 lysates. Normalized spectral abundance factor (NSAF) values are shown for P23-45 structural proteins detected in two replicate runs (white and grey bars). The proteins gp88, gp89, gp94 and gp96 are also shown in Fig. 2B.
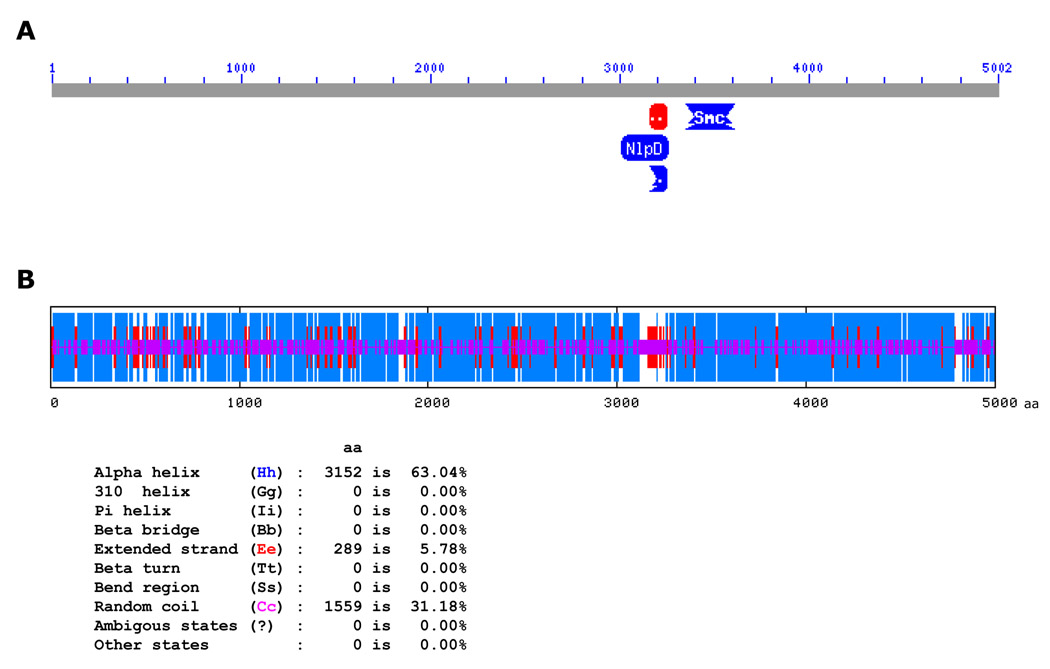
Both phages have extremely long tails (over 800 nm; 6;Fig. 2A). The N-terminal and C-terminal parts of gp96 are reminiscent of fibrillar regions present in tail-tape measure proteins (TMPs), a diverse group of structural proteins that determine the length of phage tails. The two fibrillar regions of gp96 are separated by a metallopeptidase-like domain (Fig. 4A). Some known TMPs, for example, one from Staphylococcus aureus phage phiNM3 (NC_008617), also have a metallopeptidase domain in a similar location. We therefore hypothesize that gp96 is a TMP. Though the primary sequences of TMPs are poorly conserved, these proteins have similar secondary structures and location of their genes on the genome is conserved between different phages 19; 24. TMPs are long and their lengths are linearly correlated with the lengths of cognate virions tails 25; 26. TMPs are also largely α-helical. The analysis of gp96 secondary structure by a metaserver 27 that combines prediction of secondary structure by several algorithms, suggests that 65.7% of gp96 amino acids are within α-helices, 5.8% are involved in extended strands, and 25.7% are involved in random coils (Fig. 4B). These proportions are close to predicted values for a well-characterized TMP from phage λ, gpH (64.3%, 6.2%, and 27.1%, respectively). Using the equation of Katsura and Hendrix25, a tail length regulated by gp96 should be ~700 nm 6. The ~15% discrepancy of the calculated (700 nm) and observed (800 nm) numbers for P23-45 tail length maybe due to the presence of extended strands in gp96, since the calculation assumes a TMP to be completely α-helical.
Figure 4.
P23-45 putative tape measure protein gp96.
A. PSI-BLAST search for putative conserved domains in gp96 revealed a domain with strong similarity to proteins of the metallopeptidase family (shown as red and blue segments between amino acid residues 3000 and 3200).
B. gp96 protein secondary structure predicted by a HNN (Hierarchical Neural Network) method available at NPS@ server. Blue, amino acid residues involved in α-helices; red, amino acid residues involved in extended strands; purple, amino acid residues involved in random coils.
Spectral counting derived from MudPIT allows one to make inferences on relative abundance of proteins in mixtures analyzed 28; 29. Three P23-45 virion proteins, gp88, gp89, and gp94, were detected with highest NSAF values (Normalized Spectral Abundance Factor; see Materials and Methods) (Fig. 3). The calculated molecular weights of these proteins (16.6 kDa, 46.6 kDa, and 37.9 kDa, respectively) correspond well to apparent molecular weights of three major bands observed after SDS-PAGE separation of proteins from phage virions (Fig. 2B). According to the NSAF values, an approximate stoichiometry of 1 gp88 to 3 gp89 to 5 gp94 in phage virions can be estimated. Other virion proteins were detected in lower abundances and must correspond to minor components of phage virions.
Unusual triplex-forming sequences in the longest non-coding regions of both phage genomes
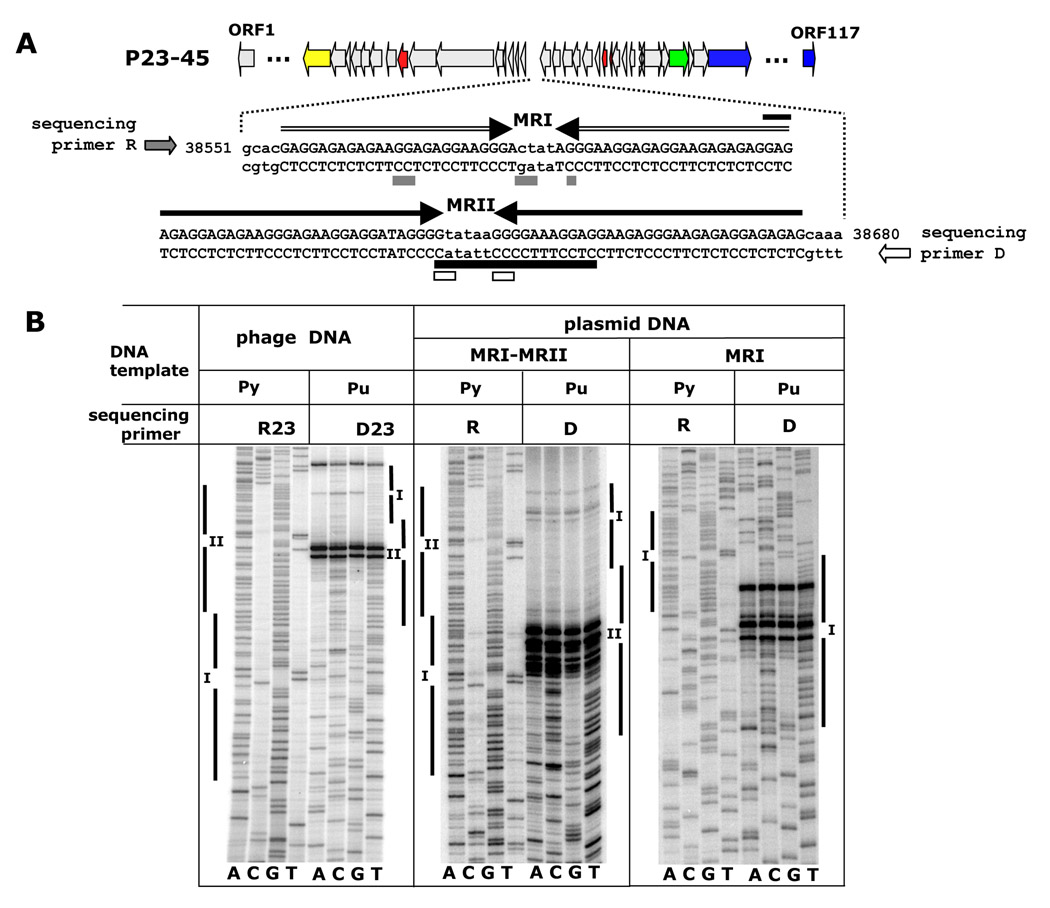
An intriguing feature of the P23-45 and P74-26 genomes is the presence of homopurine-homopyrimidine (R-Y) mirror repeats (MRs) between ORFs 68 and 69 (P23-45) and ORFs 65 and 66 (P74-26) (Fig. 1 and Fig 5A). Each genome contains two nonidentical sets of tandemly located and partially overlapping repeats, MRI and MRII. MRI consists of two 25-nucleotide R-Y tracts with perfect axis symmetry; MRII consists of two 33-nucleotide R-Y tracts with nearly perfect axis symmetry (one mismatch). MRI and MRII overlap by 3 nucleotides (Fig. 5A). MRI and MRII of P23-45 and P74-26 are identical.
Figure 5.
Triplex-forming mirror repeats MRI and MRII located in the longest non-coding region of the P23-45 genome.
A. P23-45 genome is schematically represented. Sequences of partially overlapping regions MRI and MRII are shown in capital letters and marked by horizontal arrows. Replication blockage sites for different templates are marked by boxes: empty, phage DNA; black, plasmid-encoded MRI-MRII; grey, plasmid-encoded MRI.
B. DNA polymerization on the triplex-forming templates MRI and MRI-II of P23-45. The blockage of DNA polymerization reaction by Taq DNA polymerase depends on template nucleotide content, polypurine (Pu) or polypyrimidine (Py). D23 and R23, phage DNA-specific direct and reverse sequencing primers, respectively; D and R, universal T7 promoter and M13 reverse sequencing primers, respectively; vertical bars indicate positions of MRs I and II on the sequencing gels.
Homopurine-homopyrimidine MRs are relatively abundant in eukaryotes, but are only rarely found in prokaryotic genomes 30; 31. To our knowledge, the MRs of P23-45 and P74-26 are the longest observed to date in bacteria or their viruses. The R-Y sequences organized in an MR have special structural properties and may adopt unusual triplex DNA conformations in vitro 32; 33 and in vivo 35 under appropriate conditions. Two different types of DNA triplexes are known. At physiological pH and in the presence of bivalent cations, a triplex can be built from a pyrimidine strand and a hairpin formed by a purine strand (YR.R conformation) 33. Under acidic conditions, a triplex can be built from a purine strand and a hairpin formed by a pyrimidine strand (YR.Y+ conformation) 32. At our experimental conditions (pH 9.0 at 25 °C and the presence of Mg2+ in the sequencing mixture, see Materials and Methods), the YR.R conformation appears to be more favorable than the alternative YR.Y+ conformation.
Triplexes affect DNA synthesis in vitro 35; 36; 37; 38 and in vivo 39; 40; 41. We used P23-45 phage DNA as a template for a thermal cycle sequencing reaction catalyzed by Taq DNA polymerase using oligonucleotides that annealed at either side of MRs as primers (the 3’ ends of the primers were directed towards the repeats, Fig. 5A, sequencing primers D and R). As can be seen from data presented in Fig. 5B, the MRI-II region acted as a unidirectional replication block: the arrest was only observed when Taq polymerase approached the center of the repeat in the purine-containing template strand at 72°C (Fig. 5A). Similar result was obtained when the MRI-II region was cloned into a E. coli plasmid: robust DNA polymerization blockage was evident in both supercoiled (Fig. 5A, B,) and linearized (data not shown) plasmid templates and the sites of blockage mostly coincided with those observed in phage DNA fragments. We believe that polymerization arrests in the middle of MR occur through the formation of a triplex in which a purine template strand and a portion of a growing pyrimidine strand are involved (YR.R conformation, Fig. 5B and Fig. 6B). While we did not observe any arrests when a pyrimidine strand served as a template for Taq DNA polymerase (Fig. 5A and Fig. 6A), strong DNA polymerization arrests were observed at both purine and pyrimidine denatured templates at 37 °C, when Sequenase (modified T7 DNA polymerase) was used (data not shown). A similar behavior was observed for triplex-forming DNA sequences earlier 35; 36. We conclude that the MRI-II sequences form triplexes and that these triplexes affect DNA synthesis in an orientation-dependent manner. In order to test whether the MRI and MRII can act separately or they function as a single element, the MRI region was cloned into the same plasmid. During polymerization reaction, multiple stop sites were also present at the center of this mirror repeat, though the block was less efficient (compare polymerization efficiency on the MRI-MRII and MRI templates in plasmid DNA in Fig. 5B). We conclude that both MRI and MRII can form triplexes and block replication in vitro.
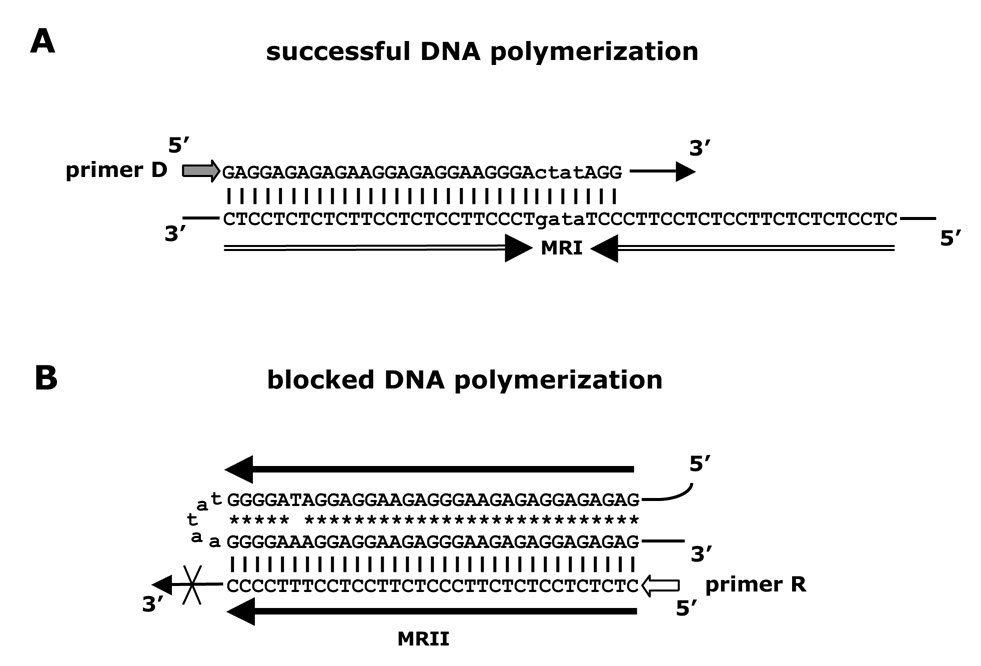
Figure 6.
Schematic models of DNA polymerization reaction and arrest at polypurine and polypirimidine DNA templates.
A. DNA polymerization is successful when a pyrimidine strand served as a template for Taq DNA polymerase.
B. DNA polymerization is blocked in the middle of MR through the formation of a triplex in which a purine template strand and a portion of a growing pyrimidine strand are involved. |, Watson-Crick hydrogen bonds; *, Reverse Hoogsteen hydrogen bonds; →, 3’-end of growing DNA chain.
The comparison of P23-45 and P74-26 genome sequences revealed increased levels of point mutations, deletions, and insertions around the triplex-forming sequences (Fig. 1B). Notably, 4 of 7 ORFs that are absent from one of the two phage genomes are located close to MRs, at a distance less than 4 kbp. Recently, it has been shown that triplexes formed by R-Y tracts cause genomic instabilities: point mutations, amplifications, deletions and translocations 42; 43; 44 and references therein). Thus, it seems plausible that phage MRs could be responsible for increased variability of their surrounding genomic regions.
Discussion
Here, we report a comparative study of complete genomes for two T. thermophilus dsDNA phages with siphoviral morphology, P23-45 and P74-26, and proteomic characterization for one of them (P23-45). These thermophages were isolated from geographically close but distinct regions of the Kamchatka peninsula 6. As suggested by similar physical properties and close, though not identical restriction digest patterns 6, the P23-45 and P74-26 genomes are very similar, showing 92% overall nucleotide identity and ~95% common ORFs.
P23-45 and P74-26 nucleotide sequence data were used to check the possible presence of these phages and their close relatives in several distinct hot water sources with different temperature and pH conditions in the Uzon and Dolina Geyser valleys. PCR analysis with pairs of primers specific to ORF11 encoding putative DNA polymerase and ORF85 (ORF84 in P74-26) encoding putative terminase large subunit revealed the presence of this group of phages in every water sample collected in Kamchatka hot springs. In contrast, similar PCR analysis with primers specific to several genes of ϕYS40 revealed the absence of this phage in the samples (ZB and LM, data not shown).
The high level of similarity and the ubiquitous presence of long-tailed Thermus phages suggests that viral (and by extension, bacterial) populations of hot springs in the Uzon and Dolina Geyser valleys may be experiencing genetic exchange, either through common underground aquifers or through air. Additional comparative studies of microbial diversity in these locations should shed light on this issue.
As in T. thermophilus phage ϕYS40 3, 25% of predicted P23-45 and P74-proteins show sequence similarity to proteins from a broad phylogenetic range of microorganisms, including Gram-negative, Gram-positive bacteria, and Archaea, as well as from bacteriophages infecting diverse bacteria, such as Thermus, Bacillus, and Pseudomonas groups.
P23-45 and P74-26 encode exceptionally long tail tape-measure proteins (TMPs) (ORF96 of 5002 codons and ORF95 of 5006 codons, respectively), which are likely responsible for extreme tail lengths of both phages 6. Though phage-like particles with 2,800 nm long tails have been reported in ocean water 2 and references therein), they have not been characterized. Thus, at present P23-45 and P74-26 have the longest tails amongst the characterized phages. The advantages of such extremely long tails are not clear. In addition to determining tail length, TMPs were shown to be important for stability and assembly of phage tails 24; 45. Putative P23-45 and P74-26 TMPs contain 80–120 amino acids regions with strong similarity to endopeptidases of the M23/M37 metallopeptidase family. This peptidase domain may be part of membrane-puncturing device 46. TMPs are thought to be partially ejected from the tail into the bacterial cytoplasm ahead of phage DNA 47. Alternatively, this region of TMP may be involved into proteolytic processes necessary for tail assembly.
An unusual feature of the P23-45 and P74-26 genomes is the presence of long polypurine-polypyrimidine (R-Y) mirror repeats (MRs) in their longest non-coding regions. Such triplex-forming sequences, extremely rare in prokaryotes but abundant in non-coding regions, regulatory elements, and introns of eukaryotic genomes 30; 31; 48, are known to form stable triple helices in vitro and in vivo that block replication and transcription 32; 33; 38; 49; 50. Since the coding density of phage genomes is known to be very high, long non-coding regions may be expected to play a regulatory role. We therefore hypothesize that in both phages MRs are involved in control of replication or gene expression. While the exact molecular mechanisms of this control remain to be investigated, several hypotheses appear to be plausible. The triplex-forming sequences are located between two convergently transcribed genes in the orientation that inhibits transcription of the downstream gene from an upstream promoter (LM, unpublished observations). Thus, MRs may attenuate transcription of downstream genes either by causing transcription termination or arrest/pausing. MRs may also downregulate replication of phage DNA in transcription-dependent manner due to a collision between the replication fork and an RNA polymerase stopped at the triplex-forming sequence 50. Finally, as revealed by the MRs’ in vitro behavior, they may provide a unidirectional block for phage DNA replication, which may be important for generation of unit-length copies of the genome. The biological role of these unusual sequences and the necessity of tandem MRs for phage development is currently being investigated in our laboratory.
Recently, it has been revealed that triplex-forming sequences are frequently found near mutation hotspots and at breakpoints of genetic rearrangements in both E. coli and mammalian cell cultures 42; 43; 51. Moreover, several human diseases and disorders are found to be associated with mutations and rearrangements promoted by triplex-forming sequences 51; 52; 53. The results of comparative analysis of the two thermophage genomes presented here are consistent with a similar role for these triplex-forming sequences. Thus, MRs may contribute to genetic instability and genome plasticity of P23-45 and P74-26 and may therefore play an important role in the evolution of these thermophages and their relatives.
Experimental procedures
Bacterial growth and phage infection
Bacteriophages P23-45 and P74-26 were generously provided by Dr. Michael Slater, Promega Corporation (Madison, WI). To isolate individual P23-45 or P74-26 plaques, 150 µl of freshly grown Thermus thermophilus HB8 culture (OD600~0.4) in the TB medium [0.8% (w/v) Tryptone, 0.4%(w/v) yeast extract, 0.3%(w/v) NaCl, 1 mM MgCl2, and 0.5 mM CaCl2] was combined with 100 µl dilutions of phage stock, incubated for 10 min at 65°C, plated in soft TB agar (0.75 %), and incubated overnight at 65°C. To prepare a phage stock suspension, an individual plaque was picked up and subjected to two more rounds of plaque purification. To prepare the phage lysate, a single plaque was resuspended in a small volume of the TB medium and mixed with 0.1 ml of freshly grown HB8 culture. The mixture was incubated for 10 minutes at 65 °C to allow phage absorption, 20 ml of fresh TB medium was added and the culture was incubated on a rotary shaker at 65 °C until complete lysis occurred (usually after 6–10 hours). Cell debris was removed from the lysate by centrifugation at 6,000g for 10 minutes. The resultant phage stock (~109 pfu/ml) was saturated with chloroform and stored at 4 °C. The P23-45 and P74-26 stocks were used to prepare larger volumes of phage lysates using a scale-up of the procedure described above.
Purification of P23-45 virions
P23-45 virion purification was done as described 3. Briefly, DNase I and RNase A (Sigma-Aldrich, St. Louis, MO, final concentration of 3 units/ml and 1 µg/ ml, respectively) were added to P23-45 lysate and the mixture was incubated for 30 min at 30°C. Solid NaCl was added a final concentration of 1 M and dissolved by swirling. The lysed culture was left on ice for 1 h and centrifuged at 11,000 g for 10 min at 4°C. To precipitate P23-45, PEG 8000 was added to the supernatant to the final concentration of 10% (w/v) followed by 1 – 4 h incubation on ice. Precipitated P23-45 particles were recovered by centrifugation at 11,000g for 10 min at 4 °C. The phage pellet was resuspended in 2 ml of SM buffer (100 mM NaCl, 1 mM MgSO4, 50 mM Tris-HCl pH7.5, 2% gelatin). PEG 8000 and cell debris were extracted from the phage suspension by adding an equal volume of chloroform and centrifuged at 3,000g for 15 min at 4°C. A quantity of 0.5 g of solid CsCl per milliliter of phage suspension was added to the aqueous phase, which contained the phage particles, and dissolved by gentle mixing. CsCl step gradients (three steps of 1.45, 1.50, and 1.70 g/l density) were prepared in Beckman SW41 polypropylene centrifuge tubes and centrifuged at 22,000 rpm for 2 hrs at 4 °C and at 38.000 rpm for 24 hrs at 4°C (Beckman SW50.1 rotor, Beckman Coulter, Fullerton, CA). Purified bacteriophage suspension was dialyzed twice at 4°C overnight against a 1000-fold volume of 10 mM NaCl, 50 mM Tris-HCl pH 8.0, 10 mM MgCl2. Purified virions were mixed with loading SDS-buffer, boiled for 5 min, and loaded onto a 4–20% gradient Tris-HCl polyacrylamide gel containing SDS (BioRad, Hercules, CA). The gel was stained with Coomassie Blue R-250 (BioRad). The approximate molecular weights of the protein bands were determined by comparison to a protein standard (Precision Plus Protein, BioRad).
Electron microscopy
CsCl density-gradient purified viruses were washed once in 0.1 M ammonium acetate (pH 7.0), using a Beckman (Palo Alto, CA) J2-21 centrifuge and a JA-18.1 fixed angle rotor operated for 60 min at 25,000 g. Phage particles were deposited on carbon-coated copper grids, stained with 2% potassium phosphotungstate (pH 7.0) or uranyl acetate (pH 4.5) and examined in a Philips EM 300 electron microscope. Magnification was monitored using T4 phage tails, which measure 113 nm in the extended state 6.
Extraction of phage DNA and genome sequencing
P23-45 and P74-26 phage DNA were extracted with Lambda Midi kit (Qiagen, Valencia, CA) following a procedure recommended by the manufacturer.
Initial sequence data were obtained using mini shotgun libraries of P23-45 and P74-26 DNA. For that, purified phage DNA was digested with HindIII, and the resulting fragments were ligated into HindIII-linearized pTZ19R (Fermentas, Glen Burnie, MD). The recombinant clones were subjected to sequence analysis, and on the basis of the sequencing results obtained, specific primers were designed. Several rounds of sequencing reactions were performed directly on phage DNA using ThermoFidelase and Fimer technology 54; 55. Trace assembly was done with phredPhrap package (http://www.phrap.com) 11. The final round of sequencing resulted in one pseudocircular contig. All the restriction enzymes, T4 DNA ligase, and T4 polynucleotide kinase used in this work were purchased from New England Biolabs (Ipswich, MA).
Molecular cloning and DNA polymerization reaction
A 113 bp and 257 bp DNA fragments of P23-45 genome comprising MRI and both MRI and MRII sequences, respectively, were amplified with primers containing engineered BamHI and PstI sites. The PCR fragments were digested with the appropriate restriction enzymes and ligated into BamHI-PstI-digested Bluescript SK(-)(Stratagene, La Jolla, CA). The resultant plasmids, pBsktriplex1 and pBsktriplex1-2, were used as templates for sequencing reactions with 32P end-labeled T7 promoter direct and M13 reverse universal sequencing primers. P23-45 genome DNA was also used as a template for sequencing reactions with 32P (PerkinElmer, Waltham, MA) end-labeled D and R phage DNA-specific sequencing primers. The sequencing reactions carried out with the fmol DNA Cycle Sequencing kit (Promega Corp.) and with the Sequenase version 2.0 DNA sequencing kit (USB Corporation, Cleveland, OH) were performed according to the manufacturer’s procedure. The reaction products were resolved on 6% (w/v) sequencing gels and visualized using a PhosphorImager Storm840 (Molecular Dynamics, GE Healthcare, Piscataway, NJ). The sequences of the phage DNA-specific primers are available from the authors upon request.
Sequence analysis
ORFs of the P23-45 and P74-26 genomes were predicted using the GeneMark server (http://opal.biology.gatech.edu/GeneMark/heuristic_hmm2.cgi). The PSI-BLAST program 57 was used to detect the homologs of the P23-45 and P74-26 genes in DNA and protein databases, with profile inclusion cutoff E-value in PSI-BLAST (-h parameter) set at 0.01. Both options for low-complexity filtering (-F parameter) and composition-based statistics (-t parameter) were sometime adjusted for better detection of sequence similarities. HHpred 57 comparison to the CDD database at NCBI was also used to predict structural and functional protein similarities.
tRNA genes were searched by using the tRNAscan-SE program 12 Searches for the presence of the transmembrane helices and coiled coil regions were done with the aid of the SEALS package 58.
Pairwise global alignment of two phage genomes was generated using the Stretcher program from EMBOSS package 9; 10. Alignment visualization and calculation of percent of identical bases in aligned genomes was done using the Base-By-Base (BBB) global alignment editor 59. Orthologous genes were aligned using ClustalW 60. SeqinR package for the R statistical computing environment 61; SeqinR is available at http://pbil.univ-lyon1.fr/software/SeqinR/seqinr_home.php) was used for the analysis of synonymous and nonsynonymous substitutions and RSCU biases. Protein secondary structures were predicted using a Web server NPS@ (Network Protein Sequence Analysis, the server is available at http://pbil.ibcp.fr/NPSA) 27.
MudPIT
P23-45 lysate was purified by double sedimentation in CsCl gradients and had phage titer of 2×109 pfu/ml. Subsequent steps were done as previously described 3. Briefly, the phage lysate was treated for 30 minutes at 37°C with 0.1U of benzonase (Sigma-Aldrich), then precipitated in 20% trichloroacetic acid, 100mM Tris-HCl, pH 8.5, overnight at 4°C. The dried protein pellet was denatured, reduced, alkylated and digested with endoproteinase LysC (Roche Applied Science, Indianapolis, IN) and trypsin (Promega Corp.). The peptide mixture was split in half and each aliquot was pressure-loaded onto a triphasic microcapillary column, installed in-line with a Quaternary Agilent 1100 series HPLC pump coupled to Deca-XP ion trap tandem mass spectrometer (ThermoElectron, San José, CA) and analyzed via ten-step chromatography 23.
The MS/MS datasets were searched using SEQUEST 62 against a database of 117 P23-45 predicted gene products, combined with 2238 protein sequences from T. thermophilus HB8 (including chromosome and large plasmids), as well as usual contaminants such as human keratins, IgGs, and proteases. Additionally, to estimate background correlations, each sequence in the database was randomized (keeping the same amino acid composition and length) and the resulting "shuffled" sequences were concatenated to the "forward" sequences and searched at the same time. The total number of non-redundant sequences searched was 5038.
The DTASelect/CONTRAST program 63 was used to select spectra/peptide matches with normalized difference in cross-correlation score (DeltCn) of at least 0.08, a minimum cross-correlation score (XCorr) of 1.8 for singly-, 2.0 for doubly-, and 3.0 for triply-charged spectra, a maximum Sp rank of 10, and a minimal length of 7 amino acids. Combining both replicate runs, proteins were identified by at least two peptides or one peptide with two independent spectra. Only one peptide matching shuffled protein sequences passed this criteria set, leading to a false discovery rate of 0.09% for both replicate runs. Spectral counts are considered to be a good estimation of absolute protein abundance 64. To account for the fact that larger proteins tend to contribute more peptide/spectra, spectral counts were divided by protein length defining a Spectral Abundance Factor (SAF) 65. SAF values are normalized against the sum of all SAFs for each run, allowing us to compare protein levels across different runs using the Normalized Spectral Abundance Factor (NSAF) value 28; 29.
GenBank accession code
The coordinates of the new sequences have been deposited at the GenBank under accession codes EU100883 and EU100884 for P23-45 and P74-26, respectively.
Supplementary Material
Acknowledgements
Bacteriophages P23-45 and P74-26 were generously provided by Dr. Michael Slater from Promega Corporation. We are grateful to Dr. S. Mirkin (Tufts University, Medford, MA) for critical reading of the manuscript and helpful suggestions. This work was supported by NIH grant RO1 GM59295 (to KS), and partially by NIH R21 grant AI074769-01 (to LM) and by National Center for Biotechnology of Republic of Kazakhstan (to ZB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl Acad. Sci. USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naryshkina T, Liu J, Florens L, Swanson SK, Pavlov AR, Pavlova NV, Inman R, Minakhin L, Kozyavkin SA, Washburn M, Mushegian A, Severinov K. Thermus thermophilus bacteriophage phiYS40 genome and proteomic characterization of virions. J. Mol. Biol. 2006;364:667–677. doi: 10.1016/j.jmb.2006.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakaki Y, Oshima T. Isolation and characterization of a bacteriophage infectious to an extreme thermophile, Thermus thermophilus HB8. J. Virol. 1975;15:1449–1453. doi: 10.1128/jvi.15.6.1449-1453.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sevostyanova A, Djordjevic M, Kuznedelov K, Naryshkina T, Gelfand M, Severinov S, Minakhin L. Temporal regulation of viral transcription during development of Thermus thermophilus bacteriophage phiYS40. J. Mol. Biol. 2007;366:420–435. doi: 10.1016/j.jmb.2006.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu MX, Slater MR, Ackermann HW. Isolation and characterization of Thermus bacteriophages. Arch. Virol. 2006;151:663–679. doi: 10.1007/s00705-005-0667-x. [DOI] [PubMed] [Google Scholar]
- 7.Ackermann HW. 5500 Phages examined in the electron microscope. Arch. Virol. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Glazko G, Mushegian A. Protein repertoire of double-stranded DNA bacteriophages. Virus Res. 2006;117:68–80. doi: 10.1016/j.virusres.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Myers EW, Miller W. Optimal alignments in linear space. Comput. Appl. Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 10.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 11.Besemer J, Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunisawa T, Kanaya S, Kutter E. Comparison of synonymous codon distribution patterns of bacteriophage and host genomes. DNA Res. 1998;5:319–326. doi: 10.1093/dnares/5.6.319. [DOI] [PubMed] [Google Scholar]
- 15.Sau K, Gupta SK, Sau S, Ghosh TC. Synonymous codon usage bias in 16 Staphylococcus aureus phages: implication in phage therapy. Virus Res. 2005;113:123–131. doi: 10.1016/j.virusres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence JG. Catalyzing bacterial speciation: correlating lateral transfer with genetic headroom. Syst. Biol. 2001;50:479–496. doi: 10.1080/10635150120286. [DOI] [PubMed] [Google Scholar]
- 17.Ravin V, Ravin N, Casjens S, Ford ME, Hatfull GF, Hendrix RW. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 2000;299:53–73. doi: 10.1006/jmbi.2000.3731. [DOI] [PubMed] [Google Scholar]
- 18.Yuzenkova J, Nechaev S, Berlin J, Rogulja D, Kuznedelov K, Inman R, Mushegian A, Severinov K. Genome of Xanthomonas oryzae bacteriophage Xp10: an odd T-odd phage. J. Mol. Biol. 2003;330:735–748. doi: 10.1016/s0022-2836(03)00634-x. [DOI] [PubMed] [Google Scholar]
- 19.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker NR, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR, Jr, Hendrix RW, Hatfull GF. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 20.Susskind MM, Botstein D. Molecular genetics of bacteriophage P22. Microbiol. Rev. 1978;42:385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber KE, Waldor MK. Filamentous phage integration requires the host recombinases XerC and XerD. Nature. 2002;417:656–659. doi: 10.1038/nature00782. [DOI] [PubMed] [Google Scholar]
- 22.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 23.Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol. Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 24.Pedersen M, Ostergaard S, Bresciani J, Vogensen FK. Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology. 2000;276:315–328. doi: 10.1006/viro.2000.0497. [DOI] [PubMed] [Google Scholar]
- 25.Katsura I, Hendrix RW. Length determination in bacteriophage lambda tails. Cell. 1984;39:691–698. doi: 10.1016/0092-8674(84)90476-8. [DOI] [PubMed] [Google Scholar]
- 26.Katsura I. Determination of bacteriophage lambda tail length by a protein ruler. Nature. 1987;327:73–75. doi: 10.1038/327073a0. [DOI] [PubMed] [Google Scholar]
- 27.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 28.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl Acad. Sci. USA. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 30.Schroth GP, Ho PS. Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucleic Acids Res. 1995;23:1977–1983. doi: 10.1093/nar/23.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox R, Mirkin SM. Characteristic enrichment of DNA repeats in different genomes. Proc. Natl Acad. Sci. USA. 1997;94:5237–5242. doi: 10.1073/pnas.94.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirkin SM, Lyamichev VI, Drushlyak KN, Dobrynin VN, Filippov SA, Frank-Kamenetskii MD. DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature. 1987;330:495–497. doi: 10.1038/330495a0. [DOI] [PubMed] [Google Scholar]
- 33.Kohwi Y, Kohwi-Shigematsu T. Magnesium ion-dependent triple-helix structure formed by homopurine-homopyrimidine sequences in supercoiled plasmid DNA. Proc. Natl Acad. Sci. USA. 1988;85:3781–3785. doi: 10.1073/pnas.85.11.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohwi Y, Panchenko Y. Transcription-dependent recombination induced by triple-helix formation. Genes Dev. 1993;7:1766–1778. doi: 10.1101/gad.7.9.1766. [DOI] [PubMed] [Google Scholar]
- 35.Baran N, Lapidot A, Manor H. Formation of DNA triplexes accounts for arrests of DNA synthesis at d(TC)n and d(GA)n tracts. Proc. Natl Acad. Sci. USA. 1991;88:507–511. doi: 10.1073/pnas.88.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dayn A, Samadashwily GM, Mirkin SM. Intramolecular DNA triplexes: unusual sequence requirements and influence on DNA polymerization. Proc. Natl Acad. Sci. USA. 1992;89:11406–11410. doi: 10.1073/pnas.89.23.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samadashwily GM, Dayn A, Mirkin SM. Suicidal nucleotide sequences for DNA polymerization. EMBO J. 1993;12:4975–4983. doi: 10.1002/j.1460-2075.1993.tb06191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krasilnikov AS, Panyutin IG, Samadashwily GM, Cox R, Lazurkin YS, Mirkin SM. Mechanisms of triplex-caused polymerization arrest. Nucleic Acids Res. 1997;25:1339–1346. doi: 10.1093/nar/25.7.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohshima K, Montermini L, Wells RD, Pandolfo M. Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J. Biol. Chem. 1998;273:14588–14595. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 40.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol. Cell Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollard LM, Sharma R, Gomez M, Shah S, Delatycki MB, Pianese L, Monticelli A, Keats BJ, Bidichandani SI. Replication-mediated instability of the GAA triplet repeat mutation in Friedreich ataxia. Nucleic Acids Res. 2004;32:5962–5971. doi: 10.1093/nar/gkh933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faruqi AF, Datta HJ, Carroll D, Seidman MM, Glazer PM. Triple-helix formation induces recombination in mammalian cells via a nucleotide excision repair-dependent pathway. Mol. Cell Biol. 2000;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasquez KM, Narayanan L, Glazer PM. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 44.Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutat. Res. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 45.Abuladze NK, Gingery M, Tsai J, Eiserling FA. Tail length determination in bacteriophage T4. Virology. 1994;199:301–310. doi: 10.1006/viro.1994.1128. [DOI] [PubMed] [Google Scholar]
- 46.Piuri M, Hatfull GF. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol. Microbiol. 2006;62:1569–1585. doi: 10.1111/j.1365-2958.2006.05473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roessner CA, Ihler GM. Proteinase sensitivity of bacteriophage lambda tail proteins gpJ and pH in complexes with the lambda receptor. J. Bacteriol. 1984;157:165–170. doi: 10.1128/jb.157.1.165-170.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bacolla A, Collins JR, Gold B, Chuzhanova N, Yi M, Stephens RM, Stefanov S, Olsh A, Jakupciak JP, Dean M, Lempicki RA, Cooper DN, Wells RD. Long homopurine*homopyrimidine sequences are characteristic of genes expressed in brain and the pseudoautosomal region. Nucleic Acids Res. 2006;34:2663–2675. doi: 10.1093/nar/gkl354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grabczyk E, Fishman MC. A long purine-pyrimidine homopolymer acts as a transcriptional diode. J. Biol. Chem. 1995;270:1791–1797. doi: 10.1074/jbc.270.4.1791. [DOI] [PubMed] [Google Scholar]
- 50.Krasilnikova MM, Samadashwily GM, Krasilnikov AS, Mirkin SM. Transcription through a simple DNA repeat blocks replication elongation. EMBO J. 1998;17:5095–5102. doi: 10.1093/emboj/17.17.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bacolla A, Jaworski A, Larson JE, Jakupciak JP, Chuzhanova N, Abeysinghe SS, O'Connell CD, Cooper DN, Wells RD. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl Acad. Sci. USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 53.Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr. Opin. Struct. Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 54.Polushin N, Malykh A, Morocho AM, Slesarev A, Kozyavkin S. High-throughput production of optimized primers (fimers) for whole-genome direct sequencing. Methods Mol. Biol. 2005;288:291–304. doi: 10.1385/1-59259-823-4:291. [DOI] [PubMed] [Google Scholar]
- 55.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 56.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walker DR, Koonin EV. SEALS: a system for easy analysis of lots of sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:333–539. [PubMed] [Google Scholar]
- 59.Brodie R, Smith AJ, Roper RL, Tcherepanov V, Upton C. Base-By-Base: single nucleotide-level analysis of whole viral genome alignments. BMC Bioinformatics. 2004;5:96. doi: 10.1186/1471-2105-5-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W:improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J. Comp. Graph. Stat. 1996;5:299–314. [Google Scholar]
- 62.Eng J, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 63.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 65.Powell DW, Weaver CM, Jennings JL, McAfee KJ, He Y, Weil PA, Link AJ. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol. Cell Biol. 2004;24:7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.