Abstract
To understand how nuclear processes involving DNA are regulated, knowledge of the determinants of chromatin condensation is required. From recent structural studies it has been concluded that the formation of the 30-nm chromatin fiber does not require the linker histone. Here, by comparing the linker histone-dependent compaction of long, reconstituted nucleosome arrays with different nucleosome repeat lengths (NRLs), 167 and 197 bp, we establish that the compaction behavior is both NRL- and linker histone-dependent. Only the 197-bp NRL array can form 30-nm higher-order chromatin structure. Importantly for understanding the regulation of compaction, this array shows a cooperative linker histone-dependent compaction. The 167-bp NRL array displays a limited linker histone-dependent compaction, resulting in a thinner and topologically different fiber. These observations provide an explanation for the distribution of NRLs found in nature.
Keywords: 30-nm fiber, electron microscopy, heterochromatin, nucleosome array reconstitution, sedimentation velocity analysis
During the past decade it has emerged that the packaging of eukaryotic DNA by histones into chromatin is a key regulator of nuclear processes involving DNA, such as transcription and replication. Although the structure of the first level of DNA folding, the nucleosome core, is known at atomic resolution (1, 2), the structure of the second level of folding, whereby a string of nucleosomes folds into a fiber with an approximate diameter of 30 nm (the 30-nm chromatin fiber) remains undetermined (3). Early evidence for the presence of a 30-nm chromatin fiber in vivo came from EM analysis of Balbiani ring genes in Chironomus tentans (4) and x-ray diffraction studies of nuclei that show spacings of 30–40 nm (5).
The structure of the 30-nm chromatin fiber is controversial (reviewed in ref. 3). Recent structural analyses using in vitro-reconstituted model nucleosome arrays based on the strong 601 DNA nucleosome positioning sequence (6) have led to the proposal of two models for the 30-nm chromatin fiber that differ in topology, dimension, and nucleosome packing density. The first model was constructed by using the crystal structure at 9 Å of a tetra-nucleosome core array and is of a two-start helix type (7). It is based on a zigzag arrangement of nucleosome cores that stack on top of each other and has a 24- to 25-nm diameter with a packing density of five to six nucleosomes per 11 nm. The second model was derived from tight constraints obtained from measurements of the physical dimensions of long nucleosome arrays visualized by EM (8). It is of the one-start helix type in which nucleosomes from adjacent gyres are interdigitated. It has a diameter of 34 nm with a packing density of 11 nucleosomes per 11 nm.
The key difference between the two 30-nm chromatin fiber models is that the interdigitated model was derived from nucleosome arrays saturated with linker histone, whereas the two-start helix model was derived from a tetra-nucleosome core array in the absence of linker histone. Whereas early studies on native chromatin had suggested an essential role for the linker histone in 30-nm chromatin fiber formation (9, 10), recent studies using reconstituted short nucleosome arrays have concluded that the linker histone is dispensable for compaction (7, 11). The linker histone, on binding to the nucleosome core, not only constrains an additional 20 bp of DNA (12, 13), but also determines the trajectory of the DNA entering and exiting the nucleosome (14, 15), which in turn will direct the relative positioning of successive nucleosomes in an array. Consequently, the linker histone is likely to have a pivotal role in the formation of chromatin higher-order structures (16). Besides a structural role, it is also emerging that the linker histone and its subtypes have diverse biologically important roles (reviewed in refs. 3 and 17). A gene knockout of three of the six murine H1 gene variants is embryonic lethal (18), whereas single knockouts modulate gene expression and affect chromatin compaction (19, 20). Additionally, in vivo experiments show linker histone variants control chromatin dynamics during early embryogenesis (19, 20).
A second difference between the two 30-nm chromatin fiber models is that they were derived from arrays with different nucleosome repeat lengths (NRLs). Adjacent nucleosomes are joined by linker DNA that varies in length from 0 to 80 bp in a tissue- and species-dependent manner, giving rise to different NRLs (21). The interdigitated one-start helix model was based on the constant diameter and nucleosome packing ratio of fibers containing NRLs ranging from 177 to 207 bp (8). By contrast, the two-start helix model was derived from a tetra-nucleosome core array with a NRL of only 167 bp (7). Although all of these NRLs are found in nature, they have a very distinct distribution of occurrence: NRLs centered on 188 and 196 bp are by far the most common, whereas short NRLs such as 165 bp, found in Saccharomyces cerevisiae, are comparatively rare (21). The reason, structural or functional, for the differential occurrence of linker DNA length in nature is not understood.
Results and Discussion
Reconstitution of Nucleosome Arrays with Different NRLs.
To define the roles of the NRL and linker histone in nucleosome array compaction and higher-order structure formation, we compared the compaction properties of two nucleosome arrays containing different NRLs (167 or 197 bp). Both arrays were based on the strong nucleosome positioning 601 DNA sequence onto which the histone octamer positions uniquely (6). To minimize end effects and better reflect the folding behavior of native chromatin, we constructed DNA arrays containing 80 and 61 tandem repeats of the 601 DNA sequence with 167- and 197-bp NRLs, respectively. The two DNA arrays were reconstituted with histone octamer and linker histone as described (22).
The Linker Histone Produces a Dramatic Increase in the Compaction of the 197-bp NRL Array.
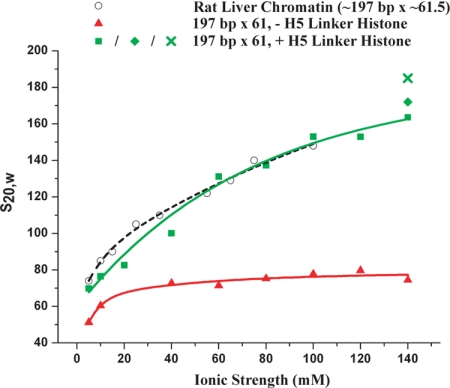
First, to define the folding properties of a reconstituted model nucleosome array and compare it with those of native chromatin, we analyzed the salt-dependent compaction of the 197-bp NRL array, which is the most commonly found NRL in nature (21). The 197-bp NRL array was reconstituted in the presence and absence of a saturating concentration of linker histone. For this experiment we used the linker histone H1 variant, H5, found in chicken erythrocytes. H5 binds with greater affinity (23), but has very similar compaction behavior to H1 [supporting information (SI) Fig. S1]. The two samples were then dialyzed into folding buffers of increasing ionic strengths (0–140 mM NaCl), and the resulting salt-dependent compaction was measured in solution by sedimentation velocity analysis (Fig. 1 and Fig. S2). These salt conditions were chosen because they were those classically used to investigate the folding behavior of native chromatin (24). Although the array devoid of H5 shows a small degree of salt-dependent compaction, the array saturated with H5 has a considerably steeper compaction curve. In 140 mM NaCl, the sedimentation coefficient (S20,w) of the 197-bp NRL array saturated with H5 is 164 S, whereas in the absence of linker histone it is a value more than twice that of the same array lacking the linker histone, 74.5 S. The doubling of the sedimentation coefficient provides unambiguous evidence for the important role of the linker histone in nucleosome array compaction. This effect is much higher than previously reported from studies using short and poorly defined nucleosome arrays (25). Importantly, for establishing the relevance of using reconstituted model nucleosome arrays to study chromatin structure and function, the S20,w values for the 197-bp NRL arrays saturated with H5 were compared with those of native chromatin fragments isolated from rat liver (24) and were found to be essentially identical (Fig. 1). The native arrays had a similar NRL (≈197 bp) and stoichiometric H1 content and were of a similar length (an average of 61.5 nucleosomes). Inclusion of Mg2+ ions in the folding buffers increases the compaction by 5–10%: 172 S in 5 mM MgCl2, 10 mM KCl, and 100 mM NaCl and 182 S in 1.6 mM MgCl2 alone (Fig. 1). In summary, this experiment shows that the linker histone gives rise to a compaction that is approximately twice that obtained in its absence, resulting in a reconstituted model nucleosome array that has the same folding behavior and reaches the same compaction as chromatin isolated from native sources.
Fig. 1.
The linker histone is essential for obtaining a native-like compaction. Sedimentation velocity analysis of the 197-bp NRL array (197 bp × 61) reconstituted without (red triangles) or with (green squares) stoichiometric concentrations of linker histone H5 and folded in increasing concentrations of NaCl and 10 mM TEA, pH 7.4 is shown. For comparison, the sedimentation coefficients (S20,w) are plotted against those of rat liver chromatin fragments containing a full complement of H1 and an average size of 61.5 nucleosomes (black circles) (24). The raw data from which the sedimentation coefficients were derived (35) are shown in Fig. S2 and are those of the monomer peaks. The sedimentation coefficients for the array folded in 5 mM MgCl2, 100 mM KCl, and 10 mM NaCl (green diamond) and folded in 1.6 mM MgCl2 alone (green cross) are also shown.
The 167- and 197-bp NRL Arrays Have Different Linker Histone Binding and Compaction Behaviors.
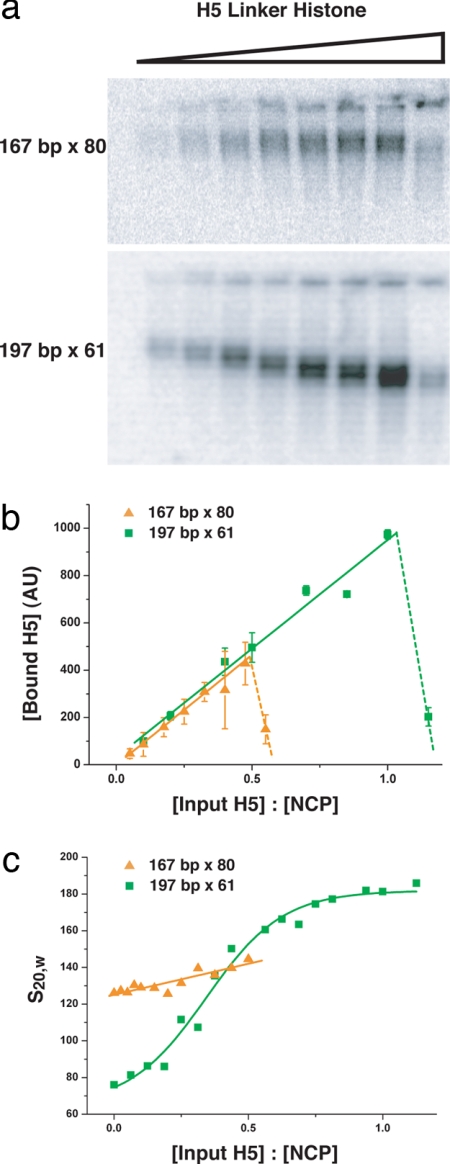
Having established that the linker histone is an essential determinant of compaction, we next investigated whether the NRL affects the linker histone-dependent compaction. To enable us to obtain insights into the mechanism of compaction it is necessary to experimentally separate the binding of the linker histone from the effect of the linker histone on compaction. The 167- and 197-bp NRL arrays were reconstituted with increasing concentrations of H5, the samples were fixed with 0.1% (vol/vol) glutaraldehyde, and the analysis was carried out by native gel electrophoresis (Fig. 2a). To be able to quantify the amount of linker histone bound to the two nucleosome arrays, H5 was P32-labeled by exploiting the phosphorylation sites present in the C-terminal tail of H5. The labeling was restricted to phosphorylation of less than one site per 1,500 linker histones, which does not affect linker histone binding to any detectable level (data not shown). Quantification of the radioactivity present in the bands in the gels, corresponding to the nucleosome arrays, reveals a linear increase in bound linker histone as a function of input linker histone until the saturation point is reached, after which the nucleosome arrays precipitate. Comparison of the binding plots (Fig. 2b) for the 167- and the 197-bp NRL arrays shows a striking difference: whereas saturation of linker histone binding for the 197-bp NRL array occurs as expected at a stoichiometry of about one H5 molecule per nucleosome core (22, 26), the 167-bp NRL array reaches saturation at ≈0.5 H5 molecules per nucleosome core. Because the linker histone extends protection of the DNA from nucleases by ≈20 bp from the 147 bp bound by the histone octamer to 167 bp (12), a likely explanation for the limited binding is that there is not sufficient linker DNA between adjacent nucleosome cores for the linker histone to bind to. It is nevertheless striking that saturation is reached at exactly half of stoichiometry, or one linker histone per two nucleosomes.
Fig. 2.
Differential linker histone-dependent binding and compaction for the 167- and 197-bp NRL arrays. (a) Gel electrophoretic analysis of the amount of linker histone H5 bound the 167- and 197-bp NRL arrays. The 167-bp × 80 and 197-bp × 61 nucleosome arrays were reconstituted with histone octamer and increasing concentrations of H5 and fixed in 0.1% (vol/vol) glutaraldehyde. To permit the direct visualization of array-bound linker histone, H5 was labeled with 32P to trace levels. Analysis was by electrophoresis in native 0.8% agarose gels. (b) Quantification of the amount of H5 bound to the nucleosome arrays in each band in the gel in a. The quantification was by densitometer tracing. Plots for the H5 saturation of the 167-bp NRL arrays (orange triangles) and the 197-bp NRL arrays (green squares) are shown. After H5 saturation, the arrays precipitate (dotted lines). (c) Sedimentation velocity analysis of the 167- and 197-bp NRL arrays reconstituted with histone octamer and increasing concentration of H5 as in a and all samples folded in 1.6 mM MgCl2 and 20 mM TEA pH 7.4 is shown. The plots show the sedimentation coefficients (S20,w) for the 167-bp (orange triangles) 197-bp NRL arrays (green squares). The raw data from which the sedimentation coefficients were derived (35) are shown in Fig. S3.
To obtain a quantitative measure of how the concentration of bound linker histone affects compaction, the 167- and 197-bp NRL arrays reconstituted with increasing concentrations of H5 were folded in 1.6 mM MgCl2 and analyzed by sedimentation velocity analysis. The MgCl2 concentration used gives maximal compaction (22) and results in predominantly monodispersed fibers. The S20,w values plotted are those from the monomer peaks (Fig. S3). Fig. 2c shows that the sedimentation behavior of the two nucleosome arrays in the presence of increasing linker histone concentrations is strikingly different. For the 197-bp NRL array, the S20,w values more than double upon saturation with linker histone H5, reaching maximal compaction at 182 S. The same effect was observed with linker histone H1 (Fig. S1). Significantly, the S20,w values increase sigmoidally as a function of linker histone concentration, indicating that the linker histone acts cooperatively in the compaction of the 197-bp NRL array. Because linker histone binding increases linearly with linker histone concentration (Fig. 2b), the observed cooperativity in the compaction of the 197-bp NRL array suggests that chromatin folding requires that contiguous nucleosomes in an array have linker histones bound. Indeed, it has previously been concluded that the linker histone must be bound to five to seven contiguous nucleosomes to allow the formation of higher-order chromatin structures (27). Additionally, the binding of linker histone to adjacent nucleosomes may be required for the bending of the linker DNA between them, which may be necessary for the formation of the 30-nm chromatin fiber (8, 28). In contrast, the 167-bp NRL array displays only a limited linker histone-dependent increase in S20,w values, from 125 to 142 S (Fig. 2c), suggesting a compaction mechanism that is much less dependent on the linker histone than that of the 197-bp NRL array. This result is partly in agreement with previous observations (11) and provides an explanation for why the role of the linker histone in nucleosome array compaction was missed. Together, the binding and sedimentation results provide direct evidence for different linker histone-dependent compaction mechanisms for the 167- and 197-bp NRL arrays.
The 167- and 197-bp NRL Arrays Have Different Linker Histone-Dependent Compaction Pathways and Different Structures.
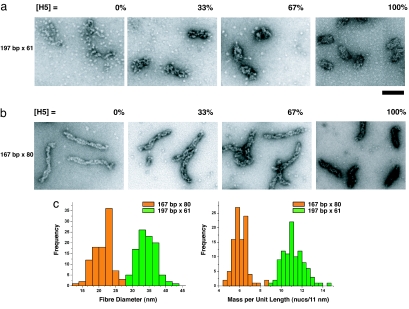
To visualize the folding pathways and rationalize the observed differences in the compaction behavior of the 167- and 197-bp NRL arrays, chromatin fibers containing increasing linker histone concentrations (0%, 33%, 67%, and 100% linker histone saturation) were visualized by EM. The negatively stained images show that the 197-bp NRL array in the absence of H5 (Fig. 3a, [H5] = 0%) forms “puddles” of nucleosomes. So although the charge neutralization by Mg2+ ions present in the folding buffer promotes nucleosome–nucleosome interactions, they are disordered. As the H5 content in the array is increased (Fig. 3a, [H5] = 33% and 67%), the fibers become increasingly compact and regular. Importantly, fibers with a diameter of 33–35 nm are only observed once the arrays are saturated with H5 (Fig. 3a, [H5] = 100%; corresponding to one bound linker histone per nucleosome core), consistent with our previous observations (22). The 167-bp NRL array in the absence of linker histone (Fig. 3b, [H5] = 0%) forms a highly ordered “ladder”-like structure consisting of two parallel columns of stacked nucleosomes consistent with a two-start helix topology. This stacking is reminiscent of the nucleosome stacking seen in the crystal structure of the four-nucleosome core array also lacking the linker histone (7). The formation of such well ordered structures accounts for the relatively high S20,w values of these samples in the absence of linker histone (Fig. 2c). The two columns of nucleosome cores twist around each other at irregular intervals. Upon addition of H5, the frequency of this twisting increases and the fibers shorten in length, forming increasingly compact fibers (Fig. 3b, [H5] = 33% and 67%). Upon H5 saturation (Fig. 3b, [H5] = 100%; corresponding to 0.5 bound linker histones per nucleosome core), the 167-bp NRL array reaches maximal compaction forming thin, regular fibers. Comparison of the dimensions of these fully folded fibers with those formed by the 197-bp NRL array reveals striking differences. Diameter and length measurements (Fig. 3c) of 99 fully folded fibers from negatively stained EM images of each of the two types of fibers ([H5] = 100%) show that the 197-bp NRL array has an average diameter of 34.3 nm (SD = 2.7) and a packing ratio of 11.2 nucleosomes per 11 nm (SD = 1.0). This finding is in agreement with our previous analysis showing that nucleosome arrays with NRLs of 177, 187, 197, and 207 bp form fibers with a constant 34-nm diameter (8). The 167-bp NRL fibers have an average diameter of 21.3 nm (SD = 3.0) and only 6.1 nucleosomes per 11 nm (SD = 0.74) and hence represent a significantly less compact structure. Therefore, not only do the 167- and 197-bp NRL arrays have very different linker histone-dependent folding pathways, but they result in fully compacted fibers that are very different. Organization of the 197-bp NRL array into a higher-order structure is clearly driven by the binding of linker histone, whereas that of the 167-bp NRL is driven primarily by nucleosome–nucleosome interactions. The reason for the difference in nucleosome arrangements in the two fibers must reside in the different lengths of their linker DNA, which is 20 bp in the 167-bp NRL array and 50 bp in the 197-bp NRL array (147 bp are bound by the histone octamer). It would therefore seem that the short linker DNA constrains adjacent nucleosomes, forcing them to stack on top of each other in a zigzag arrangement, thus determining the topology of the fiber, whereas for nucleosome arrays containing longer linker DNA the nucleosome arrangement is determined by the linker histone.
Fig. 3.
The 167- and 197-bp NRL arrays have different linker histone-dependent compaction pathways and different structures. (a and b) For EM analysis, the 167-bp × 80 and 197-bp × 61 nucleosome arrays were reconstituted with different concentrations of linker histone as in Fig. 2 and folded in 1.6 mM MgCl2. The percentage of linker histone H5 saturation (as derived from Fig. 2b) is indicated above each panel. The particles seen in the background of the EM micrographs are individual nucleosomes resulting from excess histone octamer bound to competitor DNA. (a) EM micrographs showing the folding pathway of the 197-bp NRL array from disordered puddles in [H5] = 0%, to the formation of regular 30-nm chromatin fibers at saturation of H5 binding ([H5] = 100%). (b) EM micrographs showing the folding pathway of the 167-bp NRL arrays from thin ladder-like structures formed by the stacking of nucleosome cores in [H5] = 0%, to the formation of thin, more twisted, fibers at saturation of H5 binding ([H5 = 100%]). (c) Histogram representations of the diameter and mass per unit length for the 197-bp × 61 (green) and 167-bp × 80 (orange) fully folded chromatin fibers saturated with H5. Ninety-nine measurements were taken from EM micrographs for each array. The average diameter and mass per unit length for the 167-bp NRL fibers is 21.3 nm (SD = 3.0) and 6.1 nucleosomes per 11 nm (SD = 0.74), and the average diameter and mass per unit length for the 197-bp NRL fibers is 34.3 nm (SD = 2.7) and 11.2 nucleosomes per 11 nm (SD = 1.0). (Scale bar: 100 nm.)
Conclusions
The results presented here define how the linker histone together with the NRL determines nucleosome array compaction leading to the formation of chromatin higher structures with different topologies, dimension, and nucleosome packing ratios. We show that the formation of a 30-nm chromatin fiber with similar compaction properties to native chromatin requires a sufficiently long linker DNA to permit stoichiometric linker histone binding. For long NRL arrays, binding of the linker histone drives the folding of a nucleosome array with irregular nucleosome–nucleosome interactions into a highly regular and compact fiber with a diameter of 33–35 nm (Fig. 4). By contrast, a short NRL gives rise to a highly organized nucleosome–nucleosome stacking that is much less affected by linker histone binding, resulting in the formation of a less compact fiber with small diameter of 21 nm (Fig. 4). These results not only provide an understanding of the mechanism of nucleosome array compaction, but also have an impact on future structural analyses of chromatin fibers.
Fig. 4.
The NRL and the linker histone determine chromatin higher-order structure. (Upper) Without linker histone. (Lower) With linker histone. Selected regions of the EM micrographs shown in Fig. 3 a and b are shown next to schematic representations to illustrate the folding pathway. The schematic representations representing different folding states are taken from Monte Carlo simulations performed by Kepper et al. (37) on long nucleosome arrays to explore their conformation dependence on nucleosome geometry and internucleosomal interactions. (Upper Left) Unfolded 167-bp NRL fiber showing the two-start helix arrangement typified by the stacking of nucleosome cores in the absence of linker histone. This nucleosome arrangement is very similar to that seen by Richmond and colleagues (7) in the crystal structure of the 167-bp NRL array containing four nucleosome cores. (Lower Left) Fully folded 167-bp NRL fiber in the presence of saturating linker histone concentrations. (Upper Right) Unfolded 197-bp NRL array showing the formation of puddles in the absence of linker histone. (Lower Right) Folded 197-bp NRL fibers in the presence of saturating linker histone concentrations. This nucleosome packing is in agreement with the model for the 30-nm chromatin fiber proposed by Rhodes and colleagues (8) for NRLs centered on 197 bp with stoichiometric concentrations of linker histone. (Scale bar: 50 nm.)
The demonstration that the formation of the 30-nm chromatin fiber requires a sufficiently long linker DNA to permit stoichiometric linker histone binding suggests that studies of nucleosome arrays lacking the linker histone or with short NRLs (7, 29) cannot determine the structure or compaction/decompaction properties of the 30-nm fiber. Additionally, our results suggest an explanation for the distribution of NRL found in nature: 188- and 196-bp NRLs are the most abundant (21) because they favor the formation of the 30-nm chromatin fiber (3).
Because the linker histone content in different cell types is close to one per nucleosome core (26, 30), it would seem likely that the fiber resulting from stoichiometric linker histone binding represents the dominant structure in higher eukaryotes whose genomes are largely transcriptionally inactive and packaged into heterochromatin. The question is how heterochromatin is rendered active. The observed cooperative linker histone-dependent compaction for the 197-bp NRL array suggests an elegant mechanism for the decompaction of the 30-nm chromatin fiber, whereby depletion of a few linker histone molecules could destabilize a large region of chromatin. This result is consistent with findings that transcriptionally active chromatin regions have a low linker histone content (30, 31).
Materials and Methods
601 DNA Arrays.
The DNA arrays are based on the Widom 601 DNA nucleosome positioning sequence (6). For these experiments we constructed DNA arrays with NRLs of 167 and 197 bp containing 80 and 61 repeats, respectively. In each case, the monomer DNA was designed so that the dyad of the nucleosome was fixed in the same position at the center of the 601 DNA sequence. Monomeric DNA fragments were ligated together in a tandem arrangement to form multimers and then cloned into pUC18 as described (8, 22). Plasmids were grown in DH5α Escherichia coli cells and purified. For blunt-ended release, multimer arrays (13.4 or 12 kbp) were excised by digestion with EcoRV. To purify the 601 DNA arrays from the linear vector, the vector was digested into short fragments (<1 kbp) by using HaeII and DraI. The array was then precipitated with 5–8% PEG 6000 in 0.5 M NaCl, which allows the selective precipitation of long DNA fragments.
Mixed sequence competitor DNA (crDNA) ≈147 bp in length was obtained from nucleosome core preparations. Nucleosome cores were produced from chicken erythrocyte nuclei by limited micrococcal nuclease digestion of long chromatin (32). The DNA was extracted by using phenol/chloroform and precipitated with ethanol.
Histone Purification.
The histone octamer and linker histone proteins were purified from chicken erythrocyte nuclei essentially as described (33, 34). The histone octamer was stored in 2 M NaCl, 10 mM triethanolamine (TEA) (pH 7.4), and 0.1 EDTA, and the linker histones H1 and H5 were stored in 10 mM sodium phosphate buffer, pH 7.4 at 4°C.
Reconstitution and Folding of Nucleosome Arrays.
Nucleosome arrays were reconstituted at 25 μg/ml DNA by using our in vitro reconstitution method (22). The molar input ratio of histone octamer required to obtain saturation was empirically determined. For binding and compaction studies, the linker histone (H5 or H1) was added to the reconstitution in increasing concentrations. Mixed sequence crDNA (≈147 bp) was added in all reconstitutions at a crDNA/601 DNA array mass ratio of 1:2 to prevent supersaturation of the 601 DNA arrays with excess histone octamer, ensuring that one histone octamer was bound per 601 DNA repeat. After reconstitution, chromatin arrays were dialyzed into various folding buffers, all containing 10–20 mM TEA, pH 7.4. The reconstitution and folding of nucleosome arrays was monitored by electrophoresis in native agarose gels (22).
Electrophoretic Gel-Shift Analysis.
Gel mobility-shift assays were carried out in 19- × 17-cm 0.9% agarose gels equilibrated with 0.2× Tris/borate buffer (18 mM Tris, 18 mM boric acid). Before analysis, samples were fixed with 0.1% (vol/vol) glutaraldehyde on ice for 30 min. Gels were run at 30 mA. For visualization by standard phosphorimaging techniques, the H5 linker histone was phosphorylated at very low levels by using CDK2-CyclinA (New England Biolabs) and [γ-32P]-ATP at 30°C for 15 min. This process yields approximately one [32P]phosphate label per 1,500 H5 linker histone molecules.
Analytical Ultracentrifugation.
Sedimentation coefficients were determined by using a Beckman XL-A analytical ultracentrifuge equipped with scanner optics. The initial sample absorbance at A260 was between 0.8 and 1.2. Sedimentation runs were carried out for 2 h at 5°C at speeds ranging between 15,000 and 22,000 rpm in 12-mm double-sector cells and a Beckman AN60 rotor. Sedimentation coefficients were determined by the time-derivative method described by Stafford (35). Differential apparent sedimentation coefficient distribution g(s*) was calculated by using John Philo's Dcdt+ data analysis program (version 2.05) (36). Sedimentation coefficients were corrected to S20,w by using the partial specific volume of each nucleosome array, calculated by assuming values of 0.725 and 0.55 for the protein and DNA content, respectively. g(s*) values were not mass-corrected according to linker histone composition because of their relatively small contribution to mass (never >9%) and because the frictional coefficient is unpredictable due to the possible disordering of histone tails. Solvent viscosity and solvent density were corrected according to buffer composition.
Electron Microscopy.
For visualization in negative stain, chromatin samples at a concentration of 50 μg/ml were gently fixed on ice in 0.1% (vol/vol) glutaraldehyde for 30 min. Carbon-coated electron microscope grids were glow-discharged. A 4-μl droplet of the chromatin suspension was deposited onto a grid, rinsed with 40 μl of 2% uranyl acetate, blotted, and air-dried. Images were recorded at −1- to −2-μm defocus and ×56,000 magnification with a Philips 208S microscope operating at 80 kV (Fig. 3) or a FEI Tecnai F30 microscope operating at 200 kV (Fig. 4).
Supplementary Material
Acknowledgments.
We thank Philip Robinson, Tony Crowther, and Lynda Chapman for critical reading of the manuscript and helpful advice and discussions; Jo Bulter for advice on analytical ultracentrifugation; John Widom (Northwestern University, Evanston, IL) for providing the 601 DNA sequence; Jean Thomas (Cambridge University, Cambridge, U.K.) for providing H1 linker histone; and Nick Kepper and Karsten Rippe for the production of the schematic models in Fig. 4. S.S. is supported by a European Molecular Biology Organization long-term fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802336105/DCSupplemental.
References
- 1.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent-mediated interactions in the structure of the nucleosome core particle at 1.9-Å resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8-Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Robinson PJ, Rhodes D. Structure of the 30-nm chromatin fiber: A key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Andersson K, Mahr R, Bjorkroth B, Daneholt B. Rapid reformation of the thick chromosome fiber upon completion of RNA synthesis at the Balbiani ring genes in Chironomus tentans. Chromosoma. 1982;87:33–48. doi: 10.1007/BF00333508. [DOI] [PubMed] [Google Scholar]
- 5.Langmore JP, Schutt C. The higher-order structure of chicken erythrocyte chromosomes in vivo. Nature. 1980;288:620–622. doi: 10.1038/288620a0. [DOI] [PubMed] [Google Scholar]
- 6.Lowary PT, Widom J. New DNA sequence rules for high-affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 7.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fiber. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 8.Robinson PJJ, Fairall L, Huynh VAT, Rhodes D. EM measurements define the dimensions of the 30-nm chromatin fiber: Evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorigo B, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 12.Noll M, Kornberg RD. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977;109:393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- 13.Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 14.Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A. Linker histone-dependent DNA structure in linear mononucleosomes. J Mol Biol. 1996;257:30–42. doi: 10.1006/jmbi.1996.0144. [DOI] [PubMed] [Google Scholar]
- 15.Bednar J, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hizume K, Yoshimura SH, Takeyasu K. Linker histone H1 per se can induce three-dimensional folding of chromatin fiber. Biochemistry. 2005;44:12978–12989. doi: 10.1021/bi050623v. [DOI] [PubMed] [Google Scholar]
- 17.Izzo A, Kamieniarz K, Schneider R. The histone H1 family: Specific members, specific functions? Biol Chem. 2008;389:333–343. doi: 10.1515/BC.2008.037. [DOI] [PubMed] [Google Scholar]
- 18.Fan Y, et al. Histone h1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 19.Saeki H, et al. Linker histone variants control chromatin dynamics during early embryogenesis. Proc Natl Acad Sci USA. 2005;102:5697–5702. doi: 10.1073/pnas.0409824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alami R, et al. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc Natl Acad Sci USA. 2003;100:5920–5925. doi: 10.1073/pnas.0736105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Widom J. A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc Natl Acad Sci USA. 1992;89:1095–1099. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huynh VA, Robinson PJ, Rhodes D. A method for the in vitro reconstitution of a defined 30-nm chromatin fiber containing stoichiometric amounts of the linker histone. J Mol Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 23.Thomas JO, Rees C. Exchange of histones H1 and H5 between chromatin fragments. A preference of H5 for higher-order structures. Eur J Biochem. 1983;134:109–115. doi: 10.1111/j.1432-1033.1983.tb07538.x. [DOI] [PubMed] [Google Scholar]
- 24.Bates DL, Butler PJ, Pearson EC, Thomas JO. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur J Biochem. 1981;119:469–476. doi: 10.1111/j.1432-1033.1981.tb05631.x. [DOI] [PubMed] [Google Scholar]
- 25.Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: Mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 26.Bates DL, Thomas JO. Histones H1 and H5: One or two molecules per nucleosome? Nucleic Acids Res. 1981;9:5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graziano V, Gerchman SE, Ramakrishnan V. Reconstitution of chromatin higher-order structure from histone H5 and depleted chromatin. J Mol Biol. 1988;203:997–1007. doi: 10.1016/0022-2836(88)90124-6. [DOI] [PubMed] [Google Scholar]
- 28.Yao J, Lowary PT, Widom J. Direct detection of linker DNA bending in defined-length oligomers of chromatin. Proc Natl Acad Sci USA. 1990;87:7603–7607. doi: 10.1073/pnas.87.19.7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shogren-Knaak M, et al. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 30.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 31.Krishnakumar R, et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 32.Finch JT, et al. Structure of nucleosome core particles of chromatin. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- 33.Thomas JO, Butler PJ. Size dependence of a stable higher-order structure of chromatin. J Mol Biol. 1980;144:89–93. doi: 10.1016/0022-2836(80)90215-6. [DOI] [PubMed] [Google Scholar]
- 34.Clark DJ, Thomas JO. Salt-dependent cooperative interaction of histone H1 with linear DNA. J Mol Biol. 1986;187:569–580. doi: 10.1016/0022-2836(86)90335-9. [DOI] [PubMed] [Google Scholar]
- 35.Stafford WF., 3rd Boundary analysis in sedimentation transport experiments: A procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 36.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 37.Kepper N, Foethke D, Stehr R, Wedemann G, Rippe K. Nucleosome geometry and internucleosomal interactions control the chromatin fiber conformation. Biophys J. 2008 doi: 10.1529/biophysj.107.121079. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.