Abstract
Almost all immuno-biosensors are inherently limited by the quality of antibodies available for the target molecule, and obtaining a highly sensitive antibody for a given target molecule is a challenge. We describe a highly efficient and flexible way to enhance immunoassay detection sensitivity and binding kinetics using a nanofluidic based electrokinetic preconcentrator. The device is a microfluidic integration of charge-based biomolecule concentrator and a bead-based immunoassay. Because the preconcentrator can increase the local biomolecule concentration by many orders of magnitude, it gives the immuno-sensor better sensitivity and faster binding kinetics. With a 30 min preconcentration, we were able to enhance the immunoassay sensitivity (with molecular background) by more than 500 fold from higher 50 pM to the sub 100 fM range. Moreover, by adjusting the preconcentration time, we can switch the detection range of the given bead-based assay (from 10–10,000 ng/ml to 0.01–10,000 ng/ml) to have a broader dynamic range of detection. As the system can enhance both detection sensitivity and dynamic range, it can be used to address the most critical detection issues in the detection of common disease biomarkers.
Detecting low abundance biomolecules from either bloodstream or environmental samples has been a major challenge in proteomic studies and disease/pharmacokinetic biomarker monitoring. While immunoassays are the method of choice due to its high sensitivity, detection at ultra low concentration is still challenging. Recently, many significant advances have been reported for improving detection sensitivity of immunoassays. These include surface plasma resonance (SPR)1, cantilever based sensors2, nanowire-based sensors3, nanoparticle-based assays4, optical microcavity sensors5, and immuno-PCR techniques6, with impressive detection sensitivities. Many of these novel immunoassays, however, still rely on the primary immunobinding between the low-concentration target and the antibody. The process is usually diffusion-limited7–9, especially when the target concentration is significantly below the Kd (dissociation constant, typically ranges from 10−8 to 10−12 M) of the antibody-antigen pair. Therefore, longer incubation (binding time) is required10, and only a small fraction of targets are bound at binding equilibrium, leading to statistical noise in sensing. In addition, a good quality antibody (with low Kd) would be required to enable low-concentration sensing, which is not always feasible. Mirkin and coworkers4 improved binding kinetics by making much more antibodies (through the use of beads) available for the primary immuno-binding. Still, this technique would require chemical processing and relatively large sample volumes. The other critical issue of immunoassays is the dynamic range of detection, typically only ~103, while biomarker concentrations could vary over many orders of magnitude. When multiplexed detection of multiple biomarkers is desired, tracking various targets with large concentration differences posts formidable challenges.11,12
In this communication, we introduce a novel strategy using electrokinetic preconcentration for pre-binding concentration enhancement. Instead of chemically amplifying the signal after the primary immuno-reaction, we seek to increase the concentration of the target molecule before the reaction, using a unique molecular preconcentration device. In this way, one would be able to drive the kinetics of primary immuno-reaction toward the bound state, improving both the sensitivity and the speed of detection. This is demonstrated by integrating a standard bead-based immunoassay with a nanofluidic preconcentrator in a microfluidic device format. While pre-binding enhancement can be a powerful tool for immuno-sensing, it has only been realized in microchip electrophoretic immunoassay format with limited sensitivity improvement.13 Our recently developed nanofluidic preconcentrator14 can precisely locate the collected molecule at a pre-determined spot with high preconcentration factors, therefore, is ideally suited for integration with bead/surface-based immunoassays. This strategy can be applied to most charged biomolecues by balancing the ion depletion force with counter flows (electrokinetic or pressure driven). Because it requires no physical confinement and complex buffer reagents, it has no interfacing issues with any other detectors, post amplification chemistries or multiplexing techniques.
The nanofluidic protein preconcentration device is shown in Figure 1(a). Biomolecules are trapped by the depletion force coming from the nanofluidic concentration polarization effect. The details of the concentration polarization mechanism have been extensively studied in membrane science15.
Fig. 1.
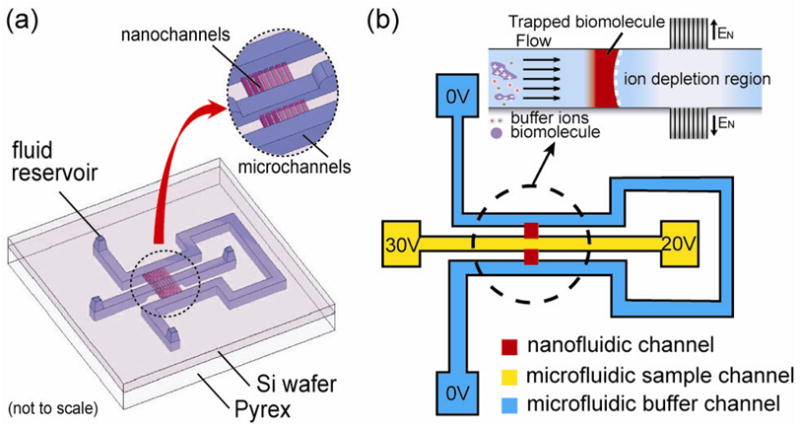
(a) Schematics of the nanofluidic preconcentration device. The middle sample channel is connected to the U shaped buffer channel by a nanochannel array with a depth of 40 nm; (b) Voltage scheme used for the biomolecule preconcentration and the electrokinetic trapping mechanism.
When an electric field is applied across the nanochannel array (behaves as charge selective membrane due to electrical double layer overlapping), according to the classic theory of concentration polarization, co-ions will be prohibited from entering the charge selective membrane. To maintain the charge neutrality in the vicinity of the membrane after selective positive ion transfer, the concentration of both positive and negative ions in the anodic side of the charge selective membrane will decrease. By balancing this depletion force with an external flow (pressure or electrokinetic driven), one can preconcentrate biomolecules as illustrated in Figure 1(b) with high efficiency.
To integrate the preconcentrator with an immunoassay, bead-based antibody chemistry was chosen due to its flexibility and robustness. Streptavidin coated polystyrene beads (diameter 6–8 micrometer) were first labeled with biotinylated Green Fluorescent Protein (GFP) antibodies or R-phycoerythrin (R-PE) antibodies and then loaded into the device. To locate the assay adjacent to the preconcentrator, we used a two-step wet etching process to form a dam-like structure around the nanochannels with shallow region smaller than microbeads. The deep region was etched to a depth of 12 μm while the shallow region had only a 5.5 μm depth. The specificity of the bead assay was tested with R-PE on GFP antibodies labeled beads or vice versa, and no cross reactions between the two targets were observed. By coupling the immunoassay with the nanofluidic preconcentrator using this simple bead loading and preconcentration procedure, as shown in Figure 2, one can locally increase the concentration and facilitate the binding kinetics and the sensitivity without changing the binding pair or detection system. The open geometry of this device makes it compatible to post amplification steps, various detectors and different antibody pairs for further enhancing the detection sensitivity.
Fig. 2.
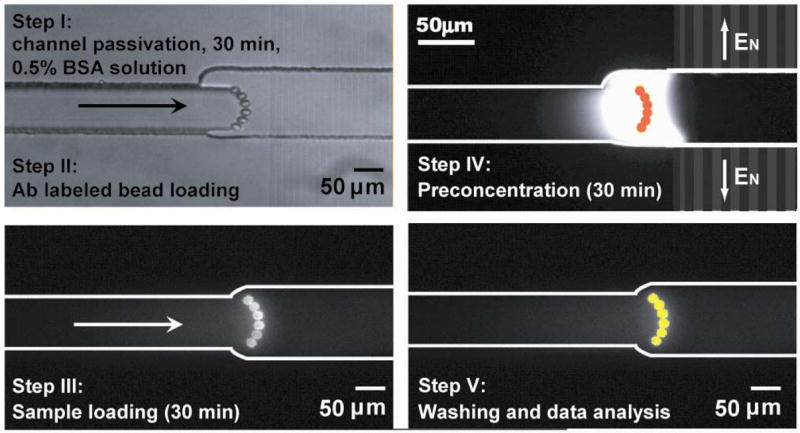
Bead loading, immunosensing and preconcentration procedure.
With this device, bead-based immuno-sensing can be realized with or without preconcentration. Due to analyte depletion and/or the diffusive constraint, Kd can be 10- to 100-fold smaller than a saturated situation at low target concentration environment.7 Therefore, as the antigen concentration decreases, the time required for binding equilibrium will increase dramatically. Such kinetic behavior is clearly shown in Figure 3(a), where the equilibrium time of R-PE binding on primary antibodies (on beads) increases dramatically with decreasing target concentrations. As a result, when low concentration antigen diagnosis is needed, incubations from several hrs to overnight are usually used.10 Such a long incubation is a common problem in immuno-biosensing once the analyte concentration goes below the scaling limit.7,9,16
Fig. 3.
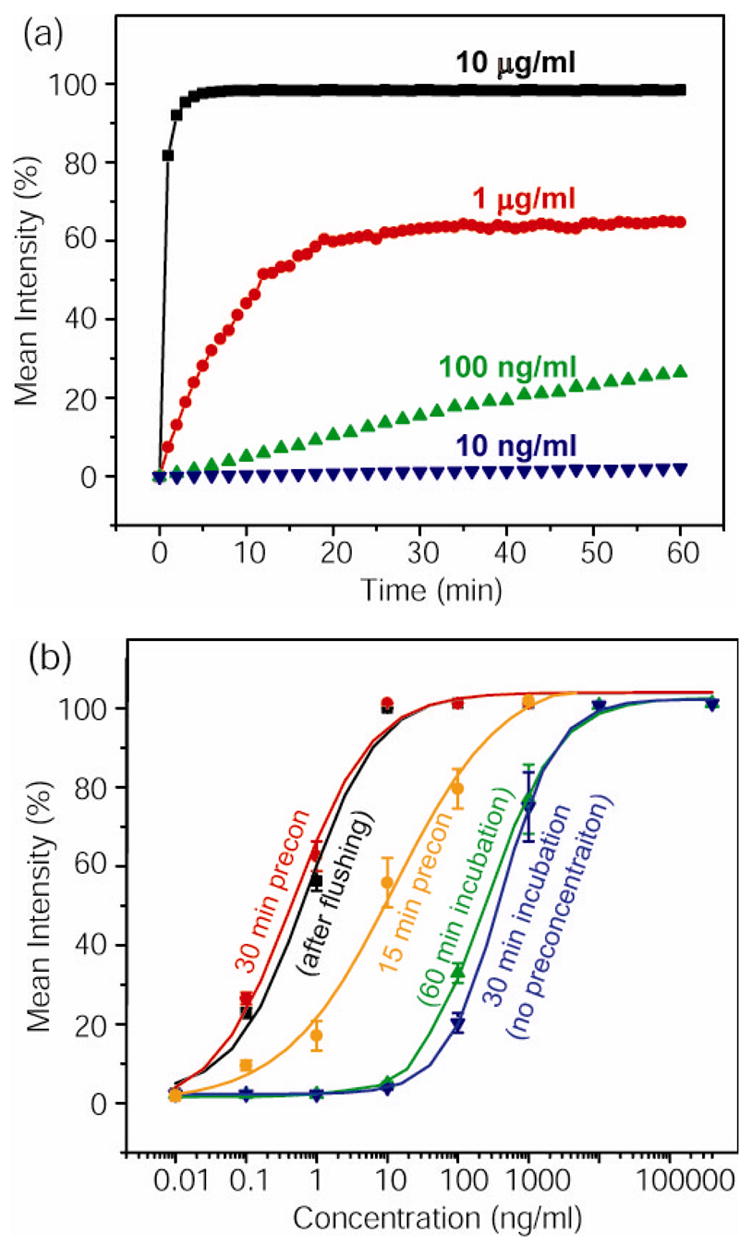
(a)Binding kinetics of R-PE samples with various concentrations; (b) Plots showing the dose response of the immunoassay without preconcentration, and with 15 min or 30 min preconcentration. Through maintaining a 30 min on-site preconcentration, this approach can lower the sensitivity limit by about 500 fold from 50 pM to sub 100 fM range. (with 10 μg/ml GFP as simulated molecular background)
As described above, by coupling the nanofluidic preconcentrator with the bead-based assay, we can achieve a continuous on-site sample preconcentration on various charged biomolecules. Therefore, the presented strategy can be applied to different biomolecules with very little or no adaptation. Figure 3(b) shows the dose response (binding curve) of this bead based immunoassay with the presence of background analytes (in this case, higher concentration of GFP). As we increase the reaction time from 30 min to 1 hr, the response signal is marginally improved, without the significant changes in the lower detection limit maintained at around ~10 ng/ml (~42 pM) level for the given monoclonal antibody-antigen (R-PE) pair. Nevertheless, by doing a 30 min on-site preconcentration, this system can lower the sensitivity limit by ~500 fold from the 50 pM to 100 fM range. In addition, the preconcentration and immuno-binding response is not affected by adding 10 μg/ml GFP (370 nM) as the background molecules, demonstrating that the operation of the device was unaffected even when much higher concentration background molecules were presented and co-concentrated along with the target molecule. As shown in Figure 3(b), the enhanced signals (bound R-PE on the bead) were not affected by the additional washing/flushing step, clearly demonstrating the enhanced binding made available by the preconcentration step.
In this communication, we have demonstrated that pre-binding preconcentration can solve many of the challenges in the early screening of common diseases biomarkers (dynamic range of detection, sensitivity, binding kinetics) without using ultra-sensitive biosensors, or high-quality antibodies. With a given antibody-antigen pair, the device can increase the dynamic range of detection, as well as the sensitivity (by at least 500 fold). This is of special importance when it comes to detecting low abundance markers after multiple sample preparation steps. Furthermore, through increasing the concentration of target antigen around the immunoassay site, the binding kinetics is significantly improved. This microfluidic device requires only a small amount of sample and reagent for detection, therefore, is ideal for miniaturized, point-of-care biosensing applications. In additions, multiplexing for many different targets is highly feasible due to the possibility of massive parallelization of lab-on-a-chip systems and the use of commercialized bead-based sensors (flow cytometry assays like Bio-Plex from BioRad or xMAP from Luminex Corp.). Because of the simplicity of the presented method, the adaptation of this strategy to sandwich assays (using both primary and secondary antibodies) could be done easily in the future. Meanwhile, any existing novel biosensing strategies that utilize post-binding amplification can also be coupled to further enhance the detection sensitivity and reliability with wider range of concentration variation.
Supplementary Material
Electronic Supplementary Information (ESI) available: [device fabrication, protein sample preparation, antibody surface immobilization, fluorescent imaging, and immunoassay response measurement.].
Acknowledgments
This work was supported by NIH grants including: CDP center (GM68762), NIBIB program (EB005743), and NCI program (CA119402).
Notes and references
- 1.Kurita R, Yokota Y, Sato Y, Mizutani F, Niwa O. Analytical Chemistry. 2006;78:5525–5531. doi: 10.1021/ac060480y. [DOI] [PubMed] [Google Scholar]
- 2.Wu G, Datar RH, Hansen KM, Thundat T, Cote RJ, Majumdar A. Nat Biotechnol. 2001;19:856–860. doi: 10.1038/nbt0901-856. [DOI] [PubMed] [Google Scholar]
- 3.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 4.Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 5.Armani AM, Kulkarni RP, Fraser SE, Flagan RC, Vahala KJ. Science. 2007;317:783–787. doi: 10.1126/science.1145002. [DOI] [PubMed] [Google Scholar]
- 6.Schweitzer B, Wiltshire S, Lambert J, O’Malley S, Kukanskis K, Zhu Z, Kingsmore SF, Lizardi PM, Ward DC. Proc Natl Acad Sci U S A. 2000;97:10113–10119. doi: 10.1073/pnas.170237197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mol NJd, Plomp E, Fischer MJE, Ruijtenbeek R. Analytical Biochemistry. 2000;279:61–70. doi: 10.1006/abio.1999.4464. [DOI] [PubMed] [Google Scholar]
- 8.Nair PR, Alam MA. Appl Phys Lett. 2006;88:233120. [Google Scholar]
- 9.Kusnezow W, Syagailo YV, Ruffer S, Klenin K, Sebald W, Hoheisel JD, Gauer C, Goychuk L. Proteomics. 2006;6:794–803. doi: 10.1002/pmic.200500149. [DOI] [PubMed] [Google Scholar]
- 10.Cesaro-Tadic S, Dernick G, Juncker D, Buurman G, Kropshofer H, Michel B, Fattinger C, Delamarche E. Lab on a Chip. 2004;4:563–569. doi: 10.1039/b408964b. [DOI] [PubMed] [Google Scholar]
- 11.Anderson NL, Anderson NG. Molecular & Cellular Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov YD, Govorun VM, Bykov VA, Archakov AI. Proteomics. 2006;6:1399–1414. doi: 10.1002/pmic.200402087. [DOI] [PubMed] [Google Scholar]
- 13.Herr AE, Hatch AV, Throckmorton DJ, Tran HM, Brennan JS, Giannobile WV, Singh AK. PNAS. 2007;104:5268–5273. doi: 10.1073/pnas.0607254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang YC, Stevens AL, Han J. Analytical Chemistry. 2005;77:4293–4299. doi: 10.1021/ac050321z. [DOI] [PubMed] [Google Scholar]
- 15.Yaroslavtsev AB, Nikonenko VV, Zabolotsky VI. Russian Chemical Reviews. 2003;72:393–421. [Google Scholar]
- 16.Shim HW, Lee JH, Hwang TS, Rhee YW, Bae YM, Choi JS, Han J, Lee CS. Biosensors and Bioelectronics. 2007;22:3188–3195. doi: 10.1016/j.bios.2007.02.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electronic Supplementary Information (ESI) available: [device fabrication, protein sample preparation, antibody surface immobilization, fluorescent imaging, and immunoassay response measurement.].


