Abstract
There is a growing belief that the metabolic program of breast tumor cells could be a therapeutic target. Yet, without detailed information on central carbon metabolism in breast tumors it is impossible to know which metabolic pathways to target, and how their inhibition might influence different stages of breast tumor progression. Here we perform the first comprehensive profiling of central metabolism in the MCF10 model of mammary carcinoma, where the steps of breast tumor progression (transformation, tumorigenicity and metastasis) can all be examined in the context of the same genetic background. The metabolism of [U-13C]-glucose by a series of progressively more aggressive MCF10 cell lines was tracked by 2D NMR and mass spectrometry. From this analysis the flux of carbon through distinct metabolic reactions was quantified by isotopomer modeling. The results indicate widespread changes to central metabolism upon cellular transformation including increased carbon flux through the pentose phosphate pathway (PPP), the TCA cycle, as well as increased synthesis of glutamate, glutathione and fatty acids (including elongation and desaturation). The de novo synthesis of glycine increased upon transformation as well as at each subsequent step of breast tumor cell progression. Interestingly, the major metabolic shift in metastatic cells is a large increase in the de novo synthesis of proline. This work provides the first comprehensive view of changes to central metabolism as a result of breast tumor progression.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-007-9732-3) contains supplementary material, which is available to authorized users.
Keywords: Cellular metabolism, Flux modeling, Glucose, Glycolysis, Metastasis, Tumor progression
Introduction
More than 80 years ago Otto Warburg observed that slices of tumor tissue took up high levels of glucose and produced high levels of lactate, even under aerobic conditions [1]. His findings were striking because aerobic conditions normally support the complete metabolism of glucose to CO2, not to the production of lactate, leading Warburg to postulate that tumor cells make use of “aerobic glycolysis.” This metabolic shift is often called the Warburg effect (or Warburg hypothesis). Many recent observations have been taken as support for the Warburg hypothesis, and there is a common perception that tumor cells have a “sweet tooth” because of their increased glycolytic activity [2]. Another reason for this perception is that tumors in patients often uptake a large amount of glucose. This fact is supported by the clinical use of PET scanning with the tracer 2-[18F]fluoro-2-deoxy-d-glucose, the most sensitive and the most accurate method with which to diagnose solid tumors, including those of the breast [3]. However, glucose uptake is biochemically distinct from the process of glycolysis. Although the evidence points to an increase of glucose metabolism in tumors, it does not definitively identify glycolysis as the process that is involved.
The metabolic fate of this glucose is not well understood and there are reasons to question the universality of the Warburg effect. In 2004, Zu and Guppy [4] systematically analyzed the literature on the topic and concluded that “there is no evidence that tumor cells are inherently glycolytic,” that both normal and tumor cells exhibit broad ranges of glycolytic dependence, and that the ranges of values have significant overlap. There are also reasons to be more vigilant about dissecting the individual steps of glucose metabolism, which were certainly not apparent at the time of Warburg. His observation spoke only to the increased lactate production of tumors. It is now clear that lactate production can be brought about not only by glycolysis but also by the pentose phosphate pathway (PPP). Furthermore, little work has been done on measuring the flux of glucose into the downstream reactions of central carbon metabolism. This information is vitally important because it will reveal how the tumor cell allocates the available energy. If such comparisons were available for tumor and normal cells, or even across different stages of tumor progression, it may be possible to design anti-tumor therapies targeting appropriate metabolic pathways.
The purpose of the present study was to perform the first comprehensive analysis of carbon flux through central metabolism in a cellular model of breast tumor progression. For this purpose we used the MCF10 model of mammary carcinoma in which progressively more malignant mammary epithelial lines were obtained by transformation and subsequent cycling as tumors in nude mice [5]. All cells in the model are derived from the spontaneously immortalized MCF10-A cell line [6] and therefore have essentially the same genetic background. The model includes the spontaneously immortalized MCF10 cells; the MCF10-AT cells, which are a derivative of these cells transformed with H-ras and that are mildly hyperplastic in vivo [7]; the MCF10-AT1 cells that yield atypical hyperplasia and carcinoma in situ [8], and the metastatic MCF10-CA1a cells which have the highest tumorigenic potential [9]. The genetics [10] and proteomics [11] of this model have been investigated but the metabolic program has not.
A major reason for the paucity of details regarding breast tumor metabolism has been the lack of appropriate experimental techniques. Metabolic modeling and flux analysis has thus far been largely confined to bacterial and plant systems due to these systems relative simplicity and scalability. However, with recent advances in spectroscopy, molecular biology and mammalian tissue culture techniques the direct investigation of the human cellular metabolism is becoming possible. We used 2D HSQC-NMR, GC-MS and enzymatic assays to quantify the activity of 15 metabolic reactions or pathways, and to measure the relative abundance of 21 metabolites. From these analyses, we were able to develop an isotopomer model of carbon flux through several key metabolic pathways in each of the four MCF10 cell lines. The results show that the first step toward tumorigenesis, transformation, is accompanied by several transitions in central metabolism. These include of catabolism of glucose through the pentose phosphate pathway (both the oxidative and non-oxidative branches), increased synthesis of glycine and glutathione, increased flux through the TCA cycle and a substantial increase in fatty acid synthesis and desaturation. The study also reveals a dramatic increase in the biosynthesis of proline that coincides with the acquisition of the metastatic phenotype, the first reported link between the acquisition of metastatic capability, and a shift in central metabolism. To our knowledge, this is also the first study to report on metabolic shifts that accompany distinct stages of breast tumor progression.
Experimental procedures
Cell culture and sample collection
The MCF10-A cell line was purchased from ATCC (Manassas, VA). This cell line was deposited there at an early passage (#39) by the former Michigan Cancer Foundation (now The Barbara Ann Karmanos Cancer Institute) as CRL#10317, the lot used in this study. The MCF10-AT, MCF10-AT1 and MCF10-CA1a cells were purchased directly from The Barbara Ann Karmanos Cancer Institute (Detroit, MI) at passage number 29, 19 and 25, respectively. Cells were maintained in supplemented MEM media (10% v/v Fetal Bovine Serum, 1% v/v antibiotic/antimycotic solution (Omega), 1% v/v MEM non-essential amino acids (Cellgro), 1% v/v MEM vitamins (Irvine Scientific), 1 mM sodium pyruvate (Sigma), 2 mM l-glutamine (Sigma), and 100 ng/ml human EGF (Sigma). Cells were labeled with [U-13C]-glucose for NMR analysis by seeding cells into 150 mm dishes at a density of approximately 4 × 106 cells per dish. At time (t) = 0 h, the complete MEM was replaced with MEM supplemented with 1 g/l glucose (Sigma), 0.5 g/l [U-13C] glucose (Cambridge Isotope Laboratories), 10% v/v Fetal Bovine Serum, 1% v/v antibiotic/antimycotic solution (Omega) and 1% v/v MEM non-essential amino acids (Cellgro), 1% v/v MEM vitamins (Irvine Scientific) and 100 ng/ml human EGF (Sigma). The total glucose concentration was 2.5 g/l with 20% [U-13C] glucose. Sodium pyruvate and l-glutamine were not included in the labeling media to minimize the alternative carbon sources for cells. All cell lines were passaged less than 10 times between thawing and the completion of [U-13C] glucose labeling.
A sample of the labeling media was taken at t = 0 and stored as a reference for analysis. Additional samples of the growth media were collected at 24 and 48 h. Following the 48 h labeling period cells were rinsed with PBS:EDTA, detached with trypsin and subjected to centrifugation at 500g for 5 min. This procedure yielded pellets containing approximately 300 × 106 cells, which were split into fractions of 250 × 106 cells and 50 × 106 cells for NMR and GCMS analysis, respectively. Cell pellets were stored at −80°C for subsequent analysis.
Quantification of total protein and growth rates
The amount of protein present in each tissue culture sample was determined using the DC Protein Assay Kit (Bio-Rad). The assay was performed generally according to the manufacturer’s protocol in microplate format. Briefly, 2.5 μl of cell lysate was added to 225 μl of reaction mixture and incubated at room temperature for 30 min. The reaction was via UV absorbance at 650 nm using a Bio-Rad 3550-UV microplate reader. Each measurement was performed in triplicate. The protein concentration was calculated via a Bovine Serum Albumin (BSA) standard curve (0.25–5 mg/ml; R2 = 0.9966) and the rate of growth for each cell line was determined by growth rate = (ln(g proteint = 48 h) − ln(g proteint = 0 h))/48 h.
Quantification of glucose consumption and lactate production
The amount of glucose present in the media samples was determined using the Glucose Assay Kit (GAGO-20) from Sigma. The assay was performed generally according to the manufacturer’s protocol but scaled down to microplate format. Briefly, 2.5 μl of media was added to 100 μl of enzyme mixture and incubated at 37°C for 30 min. The reaction was quenched with 100 μl 12 N H2SO4 and the reaction was observed via UV absorbance at 490 nm using a Bio-Rad 3550-UV microplate reader. Each measurement was performed in triplicate. The glucose concentration was calculated via a standard curve (0.25–2.5 mg/ml; R2 = 0.9973). The rate of glucose consumption for each cell line was determined by ((mmol glucose = 0 h) − (mmol glucoset = 48 h))/(48 h·((g proteint = 48 h + g proteint = 0 h)/2)).
The amount of lactate present in the media samples was determined by generally following the Sigma Diagnostics Procedure No. 826-UV. All components were purchased from Sigma. Nicotinamide adenine dinucleotide (10 mg) was dissolved in 2 ml glycine buffer, 4 ml water and 100 μl lactate dehydrogenase (1000 U/ml). A media sample (2.5 μl) was added to 145 μl of this enzyme mixture and incubated at room temperature for 30 min. The level of lactate was calculated from the observed decrease in absorbance at 340 nm. Each measurement was performed in triplicate. The lactate concentration was calculated via a standard curve (0.25–1.25 mg/ml; R2 = 0.9997). The rate of lactate production for each cell line was determined by ((mmol lactate = 48 h) − (mmol lactatet = 0 h))/(48 h·((g proteint = 48 h + g proteint = 0 h)/2)).
Extraction of cellular metabolites
After the experimental incubations, approximately 2.5 × 108 cells were harvested and cell metabolites were extracted using the cold methanol extraction method. It has been reported that the cold methanol extraction is superior to other procedures commonly used for metabolome analysis, which include acid or alkaline treatments, high-temperature extraction in the presence of ethanol or methanol, and extraction with chloroform–methanol [12]. Cell pellets were suspended in phosphate buffered saline, and l-methionine in an amount of 5 × 10−6 mol was added for quantification of metabolite concentration and calibration of chemical shift in the NMR spectra. The cells were extracted with an equal volume of cold (−20°C) absolute methanol. After rapid mixing, the tube was transferred into a dry ice bath for 30 min and subsequently thawed in an ice bath for 10 min. Cells were subjected to centrifugation (18,000g, 10 min) and the supernatant was collected. The cell pellet was subjected to extraction another time with 0.5 ml of 50% v/v cold (−20°C) methanol, and the first and second extracts were combined. The cell extract was evaporated to dryness, and then dissolved in 650 μl of D2O (99.9% enriched; Cambridge Isotope Laboratories). The sample was filtered through a 0.2-μm-pore-size filter and used for NMR measurements.
Analysis of fatty acids by GCMS
The GCMS samples were prepared from about 5 × 107 cells as described previously [13]. Briefly, the cell pellet was saponified with 1 ml of 30% KOH:ethanol (1:1 v/v) at 70°C overnight. Heptadecanoic acid (C17) in an amount of 1 mg was added as an internal standard. Neutral lipids were removed with petroleum ether extraction. The aqueous layer was acidified by adding 1.25 ml of 2.5 N sulfuric acid, and then the fatty acids were extracted with 2 ml petroleum ether. The petroleum ether layer was washed with Milli-Q water and evaporated to dryness. The fatty acids were methylated with 0.5 N methanolic HCl (Supelco, Bellfonte, PA) and incubated at 65°C for 30 min.
GC-MS measurements were performed on a Trace GC/Trace MS Plus system (Thermo Electron Corporation, Waltham, MA). The settings are as follows: flow rate of carrier gas (helium), 2 ml/min; source temperature, 200°C; interface temperature, 250°C. The column of Rtx-5MS (fused silica, 15 m × 0.25 mm × 0.25 μm) (Restek, Bellefonte, PA) was used for GC-MS analysis. Electron impact (EI) spectra were obtained at −70 eV. The GC oven temperature was programmed from 180°C (1 min) to 210°C at 3°C/min. Palmitate, palmitoleate, stearate and oleate were monitored at m/z 270, 268, 298 and 296, respectively.
Mass isotopomer distribution was determined after correcting the contribution of labeling arising from natural abundances of carbon (13C), oxygen (17O, 18O), and hydrogen (2H) [14]. The 13C enrichment of acetyl units and the de novo synthesis of fatty acids were determined from the distribution of mass isotopomers of fatty acids. De novo synthesis produces fatty acids with 2, 4, or 6 13C atoms (m2, m4, and m6). Thus the enrichment of acetyl units was calculated from the m4/m2 or m6/m4 ratio using the formula m4/m2 = (n − 1)/2 · (p/q) and m6/m4 = (n − 2)/3 · (p/q), where n is the number of acetyl units in fatty acids (n = 8 for palmitate and palmitoleate; n = 9 for stearate and oleate), p is the labeled fraction, and q is the unlabeled fraction (p + q = 1). The de novo synthesis was then calculated by dividing the observed to the predicted mass isotopomer fraction, i.e., m2/(8 · p · q7) or m4/(28 · p2 · q6) for palmitate and palmitoleate, m2/(9 · p · q8) or m4/(36 · p2 · q7) for stearate and oleate.
Identification and quantification of metabolites by NMR spectroscopy
NMR experiments were performed at 30°C and 500 MHz on a Bruker Avance 500 spectrometer (Bruker, Karlsruhe, Germany). One-dimensional 1H spectra were acquired using a spectral width of 5000 Hz and 32 K data points. Two-dimensional [13C, 1H] HSQC spectra were obtained using a standard gradient-based sequence. The acquisition parameters were t1max = 183 ms, t2max = 157 ms; data size was 3,072 points in t1 and 1,024 points in t2. Sweep width was 80 ppm and the carrier position was 50 ppm for 13C. 13C decoupling during t2 was achieved using WALTZ-16, and quadrature detection in ω1 was accomplished with States-TPPI. Before Fourier transformation, the time domain data were multiplied in t1 and t2 with sine-bell windows shifted by π/2. The digital resolution along ω1 after linear prediction and zero-filling was 1.23 Hz/point. The 13C–13C scalar coupling fine structures were extracted from the cross sections taken along the 13C axis in a HSQC spectrum by using the Bruker XWINNMR software. After manual baseline correction, the individual multiplet components of the scalar coupling fine structures were integrated to quantify the relative contributions of singlet, doublet, and quartet signals.
The assignments of metabolites in the HSQC spectra were made according to published data, by comparison with spectra of authentic compounds, by spiking the samples and by observing the 13C–13C scalar coupling fine structures. Details and chemical shifts are listed in Supplement Table S1. In all, 22 compounds could be identified in extracts of the MCF10 series of cells.
The concentrations of metabolites were determined by integrating the cross peaks in the HSQC spectra using the NMRPipe and Sparky software packages, comparing with the integral of resonances peaks of the l-methionine that was treated as an internal standard, and normalizing to the amount of total cellular protein (Supplement Table S2). For each sample, the spectra were recorded twice using the same acquisition parameters to determine the intra-individual variation in the determination of metabolite concentrations. The significance of the various concentration values was analyzed using Student’s t-test. Probability values (5%) were taken as significant results (P < 0.05). These values are shown throughout the “Results” section as well as Supplement Table S3.
Isotopomer analysis and modeling
A detailed presentation of the methodology used in the flux analysis for the pentose phosphate pathway, TCA cycle, anaplerosis and glutamate–glutathione local network is presented in the Supplement.
Results
We quantified key metabolites and carbon fluxes in four breast epithelial cell lines, allowing us to identify shifts in central metabolism that coincide with changes in cellular phenotype. Three types of measurements were made in this study. First, the basic metabolic parameters (glucose utilization and lactate production) and growth rates were measured. Second, the relative levels of 22 key metabolites were determined. These levels are presented as a ratio of the pool size in one type of cell type to the pool size in another type of cell. Third, the relative flux of carbon through 15 different metabolic steps was quantified (Figs. 1, 2). Flux was quantified by comparison of the isoptopic signature of metabolites that are indicators of given metabolic hubs [15]. All of the values in this study were normalized to the cells total protein content; therefore, increased flux or pool size represent true shifts in the way the cell is using its resources and are not just a reflection of the rate of proliferation.
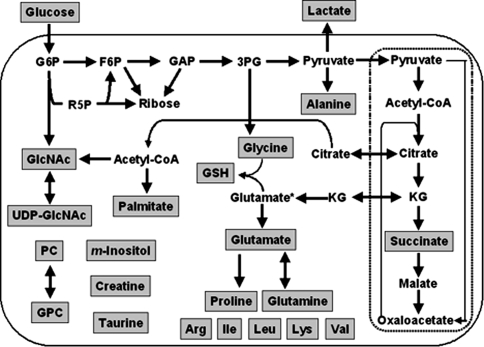
Fig. 1.
Metabolites, fluxes and pathways quantified in the MCF10 tumor progression model. Metabolites presented in the shaded boxes were observed by NMR, GCMS or enzymatic assay. Abbreviations are G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; R5P, ribose-5-phosphate; GAP, glyceraldehydes-3-phosphate; 3PG, 3-phosphoglycerate; KG, α-ketoglutarate; GSH, glutathione; GlcNAc, N-acetylglucosamine; PC, phosphocholine; GPC, glycerophosphocholine
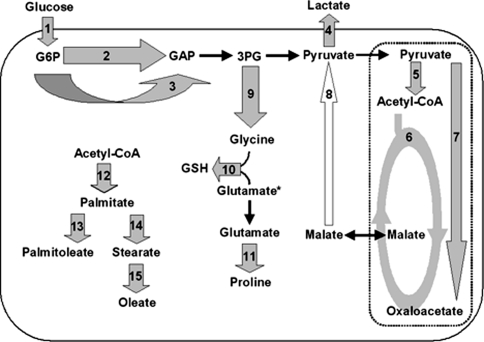
Fig. 2.
Metabolic fluxes and pathway activities determined by isotopomer analysis or enzymatic assay. Flux 1: glucose import; 2: glycolysis; 3: pentose phosphate pathway; 4: lactate excretion; 5: pyruvate dehydrogenase flux; 6: TCA cycle flux; 7: pyruvate carboxylase flux; 8: gluconeogenic flux; 9: glycine biosynthesis; 10: glutathione biosynthesis; 11: proline biosynthesis; 12: palmitate biosynthesis (fatty acid synthase activity); 13: desaturation of palmitate; 14: elongation of palmitate; 15: desaturation of stearate
Cellular transformation is associated with increased flux through the pentose phosphate pathway
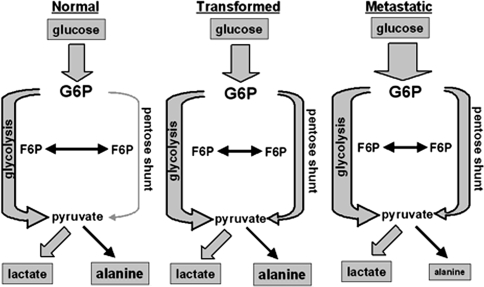
The early catabolism of glucose takes place in glycolysis or the pentose phosphate pathway (or both). Consequently, comparing the relative flux of glucose through each of these shunts is an important first step in tracking glucose metabolism. To determine the relative flux of each of these pathways the labeling state of pyruvate must be determined. However, pyruvate is in relatively low abundance within human cells and thus is not easily observable by HSQC in a complex mixture. Fortunately, pyruvate is in rapid equilibrium with alanine (which is present in much greater abundance). Therefore we observe the C2 of alanine in order to determine the labeling state of pyruvate and from these values determined the flux of glucose through the PPP and glycolysis. Surprisingly, the percentage of glucose metabolized through glycolysis is decreased in the transformed MCF10 cells (MCF10-AT, MCF10-AT1, and MCF10-CA1a) when compared to the non-transformed parental MCF10-A cells (Table 1). Nearly all of the pyruvate is derived from glycolysis in the MCF10-A cells, but in the transformed cells this value drops to between 65% and 70%.
Table 1.
Metabolic parameters quantified in the MCF10 progression series of breast cancer cell lines
| Parameter | MCF10-A | MCF10-AT | MCF10-AT1 | MCF10-CA1a | |
|---|---|---|---|---|---|
| Glucose-pyruvate (Fig. 2) | Growth rate (mg/g/h)a | 10 ± 2 | 20 ± 1** | 20 ± 1** | 18 ± 1*** |
| Glucose uptake rate (mmol/h/g protein)a | 0.8 ± 0.1 | 1.0 ± 0.2 | 1.1 ± 0.2 | 1.7 ± 0.1*** | |
| Pyruvate from glycolysis (%) | 98 ± 2 | 68 ± 7** | 65 ± 7** | 70 ± 6** | |
| Pyruvate from PPPb (%) | 2 ± 2 | 32 ± 7** | 35 ± 7** | 30 ± 6** | |
| Ribose synthesized form oxidative PPPb (%) | 12 ± 3 | 21 ± 4* | 21 ± 4* | 24 ± 4* | |
| Lactate excretion rate (mmol/g protein)a | 3.0 ± 0.6 | 3.4 ± 0.4 | 2.9 ± 0.3 | 2.9 ± 0.1 | |
| Intracellular lactate (rpsc) | 1 | 2.33 ± 0.48* | 1.68 ± 0.34* | 1.96 ± 0.40* | |
| Alanine (rpsc) | 1 | 2.54 ± 0.38** | 1.89 ± 0.27** | 1.08 ± 0.10 | |
| TCA (Fig. 3) | Relative TCA cycle fluxa | 1 | 1.8 ± 0.2** | 1.7 ± 0.2** | 1.4 ± 0.1* |
| Relative anaplerotic fluxa | 1 | 2.0 ± 0.2** | 2.0 ± 0.2** | 1.8 ± 0.2** | |
| Succinate (rpsc) | 1 | >5.26** | >5.13** | >5.29** | |
| Fatty acids (de novo) | Palmitate (%) | 1.9 ± 2.0 | 51.7 ± 2.9*** | 43.4 ± 0.5*** | 40.1 ± 0.3*** |
| Palmitoleate (%) | 0 ± 2.0 | 35.5 ± 4.9** | 7.3 ± 0.7* | 0.6 ± 0.8 | |
| Stearate (%) | 1.7 ± 2.1 | 35.9 ± 4.5** | 34.2 ± 1.1*** | 29.2 ± 3.0** | |
| Oleate (%) | 0 ± 2.0 | 25.3 ± 2.6** | 13.0 ± 4.8** | 13.3 ± 1.2** | |
| Glutamate hub (Fig. 4) | Glutamate (rpsc) | 1 | 1.22 ± 0.24 | 1.37 ± 0.21 | 0.72 ± 0.11 |
| Glutamine (rpsc) | 1 | 0.65 ± 0.11* | 0.60 ± 0.10* | 0.19 ± 0.04** | |
| Glutathione (rpsc) | 1 | 1.00 ± 0.07 | 1.20 ± 0.10 | 0.78 ± 0.06* | |
| Glycine (rpsc) | 1 | 0.59 ± 0.08* | 0.62 ± 0.09* | 0.40 ± 0.06** | |
| Proline (rpsc) | 1 | 0.72 ± 0.14 | 0.82 ± 0.13 | 1.43 ± 0.26 | |
| Glutathione flux (relative)a | 1 | 4.2 ± 0.6** | 4.9 ± 0.8** | 2.5 ± 0.4** | |
| Glycine flux (%)a | 2 ± 2 | 12 ± 2* | 21 ± 4** | 24 ± 4** | |
| Proline flux (%)a | 0 ± 2 | 0 ± 2 | 0 ± 2 | 52 ± 6 |
The table contains information on the overall metabolic status of the cell, flux through metabolic pathways and the pool size of individual metabolites
aThe overall metabolic state of the cells including growth rate, glucose uptake and lactate excretion are calculated per gram of total protein per hour
bFlux toward a given metabolite is calculated as a percentage of the total production of that metabolite from 13C-glucose through the given pathway
cIn other cases information on the synthesis of a metabolites is presented relative to the same flux in the MCF10A cells. This distinction is made because the metabolite is predominantly generated from 13C-glucose through a single pathway (alternative routes of production are not evident)
dThe relative size of the cellular pool of these metabolites is shown as a ratio to the same metabolite in the MCF10A cells
eFatty acid composition was measured by GC-MS and is presented as a percentage of the de novo synthesis of each fatty acid from 13C glucose
fSee “Materials and methods” for calculations
gPPP = pentose phosphate pathway
hrps = relative pool size
The decrease in the fraction of glucose shunted through glycolysis is more than compensated for by an increase in net flux through the PPP to pyruvate (Fig. 3). The non-transformed MCF10-A cells catabolized only a minor fraction of glucose via the PPP, but the transformed MCF10-AT cells catabolized approximately 30% of the glucose through this pathway. Flux through the PPP to pyruvate is similarly elevated in the more aggressive the MCF10-AT1 and MCF10-CA1a cells (Table 1). However, the conversion of glucose to pyruvate is only one of the possible “directions” of the pentose phosphate pathway. The non-oxidative branch of the PPP converts the glycolytic intermediates fructose-6-phosphate and glyceraldehydes-3-phosphate to ribose-5-phosphate, complementing the oxidative branch in the production of this nucleotide precursor. The relative flux through each branch of the PPP was gauged by using the C2 of UDP as an indicator (see Supplement for explanation). In comparison to the non-transformed MCF10-A cells, all three transformed MCF10 cells show a pronounced increase in flux through the oxidative branch of the PPP. The proportion of ribose generated through this branch is almost double in the MCF10-AT cells compared to the MCF10-A cells (30% of the total flux through PPP versus 16% of the total).
Fig. 3.
Relative distribution of the glucose-pyruvate local network in MCF10 cells. Flux through the pentose phosphate pathway increases as cells become progressively more tumorigenic. Observed metabolites are boxed in grey and their relative sizes between tumorigenic states are roughly proportional to the observed pool sizes. Observed fluxes are shown as grey arrows and their widths are roughly proportional to the relative pathway flux. See Table 1 for values. Normal is the MCF10-A cells; transformed is the MCF10-AT cells; metastatic is the MCF10-CA1a cells. G6P = glucose-6-phosphate; F6P = fructose-6-phosphate
Flux through the TCA cycle is higher in tumorigenic cell lines
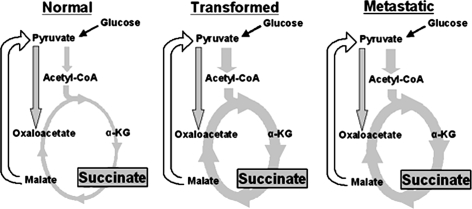
The relative flux through the TCA cycle in each cell line was measured using the isotopomer labeling of glutamate as an indicator (see Supplement II for explanation). Results showed the TCA cycle flux was increased approximately 1.8-fold in the transformed cells compared to the MCF-10A cells (Table 1). This flux, along with other TCA cycle-related results, is illustrated in Fig. 4. Additionally, our methodology can observe the gluconeogenic flux from malate to pyruvate via malic enzyme. This pathway was not active in any of the four cell lines however and is thus represented as an open arrow. A recent report by Forbes et al. reported a similar lack of malic enzyme activity in MCF7 breast cancer cells [16]. In our analysis of individual metabolites, we noticed a major change in succinate, a key metabolite in the TCA cycle. In the transformed cells, the pool of succinate is substantially increased (Fig. 4). The degree of the increase in succinate could not be precisely determined because the pool size is so low in the non-transformed MCF10-A cells. Nevertheless, the change was estimated to be at least 5-fold.
Fig. 4.
Relative distribution of the TCA cycle in MCF10 cells. Flux through the TCA cycle and the pool size of succinate are higher in tumorigenic cells than in untransformed cells. Observed metabolites are boxed in grey and their relative sizes between tumorigenic states are roughly proportional to the observed pool sizes. Observed fluxes are shown as grey arrows and their widths are roughly proportional to the relative pathway flux. See Table 1 for values. Normal is the MCF10-A cells; transformed is the MCF10-AT cells; metastatic is the MCF10-CA1a cells. α-KG = α-ketoglutarate
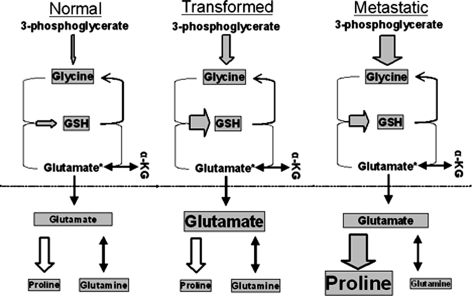
Flux through Glycine–Glutamine hub is dramatically increased in metastatic cells
The glycine–glutamine hub involves the inter-conversions of glycine, glutathione, glutamate, glutamine and proline. The entry point for carbon into this hub is the conversion of 3-phosphoglycerate to serine (an irreversible step), which is then converted to glycine. Flux through this first step was measured by direct assessment of the isotopic signature of glycine. Interestingly, flux toward glycine exhibits a striking step-wise increase in each of the MCF10 cells as they become more aggressive (Table 1). Flux into this hub step is 12-fold higher in the MCF10-AT cells than in the normal MCF10-A cells; in the metastatic MCF10-CA1a cells, the flux is 24-fold higher.
Interestingly, although the biosynthesis of glycine increased in these transformed cells, its pool size actually decreased because it is rapidly consumed for glutathione synthesis. Flux from glycine to glutathione is higher in all tumor cells compared to the MCF10-A line, but there is no corresponding increase in the size of the glutathione pool (Fig. 5). Rather, we observe increases in the pool sizes of glutamate or proline. The size of the glutamate pool is four to five times larger in the MCF10-AT and MCF10-AT1. However, in the metastatic MCF10-CA1a cells, the size of the glutamate pool is smaller than in the MCF10-A cells. The smaller size of this pool is probably the result of increased biosynthesis of proline, a step for which glutamate is the key precursor. Flux to proline is at least 50-fold higher in the MCF10-CA1a cells than in the MCF10-A cells. Despite the substantial differences in flux toward proline in all of these cells, there is only a slight increase in the size of the pool of proline, indicating that this amino acid is used rapidly.
Fig. 5.
Relative distribution of the glutamate–glutathione hub in MCF10 cells. Flux through the glycine–glutamine hub is higher in tumorigenic cells than in normal cells. Dramatic increases in the biosynthesis and pool size of proline coincide with the acquisition of the metastatic phenotype. Observed metabolites are boxed in grey and their relative sizes between tumorigenic states are roughly proportional to the pool sizes observed. Observed fluxes are shown as grey arrows and their widths are roughly proportional to the relative pathway flux. See Table 1 for values. Normal is the MCF10-A cells; transformed is the MCF10-AT cells; metastatic is the MCF10-CA1a cells; α-KG = α-ketoglutarate
Fatty acid synthesis is elevated in the transformed cell lines
GC/MS was used to quantify the 13C labeling of fatty acid species. From this value we can infer the percent of the total pool of each fatty acid that is 13C labeled, and thus the relative levels of de novo synthesis of each fatty acid. Four fatty acids were examined, palmitate (16:0), which is synthesized by fatty acid synthase, and palmitoleate (16:1), stearate (18:0) and oleate (18:1), which are derived from the desaturation and/or elongation of palmitate. The MCF10-A cells synthesized very little of the four fatty acid species (Table 1). However, de novo synthesis of palmitate increased 10-fold in the transformed MCF10-AT cells, and was elevated to similar levels in the other two tumorigenic cell lines. De novo synthesis of stearate and oleate were also elevated in all of the tumorigenic MCF10 lines in comparison to the MCF10A cells. The exception to this pattern was the de novo synthesis of palmitoleate, which was only substantially elevated in the transformed MCF10-AT cells. These observations show rather substantial changes to the fatty acid hub, and suggest that several enzymes in this pathway may be linked to breast tumor progression.
Discussion
The objective of this study was to identify changes in glucose metabolism that are associated with cellular transformation and acquisition of tumorigenic and metastatic potential in breast epithelial cells. The MCF10 panel of cell lines are an attractive system for such studies as they are a stable and well-characterized tissue culture model of mammary tumor progression. In order to minimize artifacts due to external components, a minimal common media was designed and used for all four cell lines investigated. The MCF10-A cell line requires growth factors to proliferate in two-dimensional culture, while the other MCF10 cell lines used here respond to such factors but do not require them for growth [5, 7]. However, the standard MCF10 media formulation contains a number of components which have the potential to alter cellular metabolism, including insulin [17]. We developed a MEM-based, glucose-centric media supplemented with EGF in which all four cell lines displayed continual proliferation over the 48 h 13C-glucose labeling period. We believe that growing each of the cell lines on this common background during log-phase growth has minimized any potential metabolic variation due to differential responses to media factors, basal proliferation rate and cell cycle stages.
Thus a comprehensive 2D HSQC NMR and GC-MS profiling strategy was used to characterize the metabolic program of the MCF10 model of mammary carcinoma and to determine if the findings support the Warburg effect or suggest another explanation for the uptake of glucose and increased lactate production in cancer cells. The transformed phenotype is accompanied by (1) increased catabolism of glucose by the pentose phosphate pathway (both oxidative and non-oxidative), (2) an increase in flux through the TCA cycle and a corresponding increase in the cellular pool of succinate, (3) increased flux through the glycine–glutamine hub and a corresponding increase in glutamate, and (4) a substantial increase in fatty acid synthesis and oxidation. These alterations are essentially stable through additional transitions in tumor cell phenotype and are evident in metastatic cells. Perhaps equally as significant though, the study reveals that acquisition of the metastatic phenotype is associated with a massive increase in the biosynthesis of proline. To our knowledge, this is the first study to take a comprehensive and comparative approach toward the links between breast tumor progression and metabolism, and the first study to characterize the metabolome of the metastatic cell.
Our results on glucose uptake and lactate excretion, the two parameters discussed by Warburg, are only partially consistent with his hypothesis. We do observe a doubling of glucose uptake in the metastatic MCF10-CA1a cell line when compared to the non-tumorigenic MCF10-A cells. Increases in glucose uptake were also observed in the less aggressive MCF-10AT and MCF-10AT1 cells, although the magnitude of these increases was small relative to the other tumor cell lines. However, we did not observe any consistent trend toward increased production of lactate by the tumor cells compared to the MCF10-A cells. This is most likely due to two factors: the immortalization of the MCF10-A cell line and the necessities of two-dimensional tissue culture. The MCF10-A cell line has a karyotypic gain of MYC, IL4 and IL12B concurrent with a homozygous loss of CDKN2BA and CDKN2B [10]. Cytokines have been shown to generally stimulate glycolysis (specifically, IL-3, IL-7 and IL-2) and increase the amount of lactate produced per glucose unit consumed [18]. Additionally, the MCF10A cell line requires EGF, which has also been shown to increase glycolysis [19]. Thus the relatively high lactate production in the MCF10-A cell line is most likely a result of the required tissue culture conditions.
However, our findings indicate that the distinctions between the tumorigenic and non-tumorigenic cell lines really center on how glucose is used and how carbon from glucose is shuttled through central carbon metabolism. The comprehensive strategy employed in the present study allowed an assessment of carbon flux through both the oxidative and non-oxidative branches of the PPP, each of which have two outputs—pyruvate and ribose. However, the oxidative branch has the additional role of synthesizing cellular NADPH, which is the primary source of reducing power of the cell. Flux through both branches of the PPP is increased in transformed cells, and the fact that flux through the non-oxidative branch is increased is consistent with the observations in different tumor cell lines [16, 20, 21]. Perhaps more interestingly, the magnitude of the increase in flux is higher for the oxidative branch. The increase in flux through the oxidative branch is probably a reflection of the amplified need of the transformed cells for the reducing power of NADPH, which is used for many biosynthetic reactions and is involved in protecting the rapidly proliferating cell from reactive oxygen species through glutathione. The increased flux through the oxidative branch also has potential implications for anti-tumor therapy. Many common chemotherapeutics act by inducing oxidative stress [22–26], which could be counteracted by the reducing power produced by the oxidative branch of the PPP. Therefore, drugs that independently inhibit the oxidative branch of the PPP may reduce the ability of the tumor cell to protect against oxidative stress brought on by chemotherapy.
The increased activity of the PPP also supports increased synthesis of nucleotides, which are in increased demand in highly proliferative tumor cells. Flux through each branch of the PPP can provide ribose-5-phosphate and then 5-phosphoribosylpyrophosphate, a key nucleotide precursor. The production of nucleotides also requires tetrahydrofolate, which is synthesized from glycine. Thus the reduction in the glycine pool size may relate to the fact that glycine donates methylene units to tetrahydrofolate. Glycine can also be converted to glutathione, which in turn can be converted to glutamate and then to proline. The increased flux through the PPP and the glycine–glutamine hub as cells become more tumorigenic is therefore critical because of the significant demand placed upon nucleotide synthesis during transformation.
Two key alterations in the TCA cycle were also observed in the H-ras transformed MCF10-AT cells; these alterations persisted in cells with increased hyperplasticity and metastatic potential. The changes include a near doubling of flux through the TCA cycle, and a corresponding increase in the size of the succinate pool. The exact size of the increase in the succinate pool could not be determined because the pool size is extremely low in the non-transformed MCF10-A cells. Nevertheless, we have determined the change to be at least 5-fold. This finding should be interpreted in the context of recently published work showing that high levels of cellular succinate can inhibit HIF-1α prolyl hydroxylase, and thereby cause a pseudo-hypoxic cellular response [27]. Such a response is expected to activate genes under the control of HIF-1α and thereby promote a tumorigenic phenotype. In fact, germline mutations in either fumarate hydratase or succinate dehydrogenase can drive this event and lead to a predisposition to tumors [28]. Therefore, the increased levels of succinate in the transformed MCF10-A cells may actually contribute to their tumorigenicity. The mechanisms underlying the increase in the succinate pool are not readily apparent. It is unlikely that inhibition of succinate dehydrogenase is involved in the transformed MCF10 cells because the total flux through the TCA cycle was increased. Thus, there is the distinct possibility that an increase in TCA cycle flux independently drives an increase in the succinate pool, which in turn could increase the level of HIF-1α. In other systems, such a connection between glycolysis and HIF-1α levels has been proposed, although succinate was not involved [29].
In this study two major alterations were observed in the glycine–glutamate hub. First, in the transformed cell lines displayed a 2- to 4-fold increase in the flux of carbons through GSH (indicating greater GSH turnover). This increase in flux through GSH is observed even as the size of the GSH pool decreases, indicating that increased demand for GSH in the metastatic cell is greater that the cell is able to replenish via de novo synthesis. Much like the results concerning the pentose phosphate pathway (which provides the NADPH needed for glutathione reduction), these findings demonstrate the significant demand placed upon cellular redox buffering during tumorigenic transformation. Second, we observed a substantial increase in the flux from glucose to proline in the MCF10-CA1a cell line. To our knowledge this is the first report of increased proline biosynthesis in metastatic cancer cells, which could be connected to increased turnover of the extracellular matrix in metastatic cells. Since tumor cell invasion is linked to increased degradation of collagen, one might also presume that as tumor cells invade they must replenish the digested extracellular matrix in order to adhere. This process would require increased synthesis of collagen and thus, since collagen is proline rich, a greater need for proline. There are few if any studies that examine the collagen synthesis by tumor cells, but the finding presented here indicate that this process should be the topic of future study.
Finally, these findings are consistent with our own observations for another pathway, the synthesis of fatty acids in breast tumor cells. There is a large body of work showing that fatty acid synthase (FAS) is up-regulated in a wide range of cancers [30] and that inhibition of FAS can halt tumor cell proliferation and induce tumor cell death (for review see [31]). In fact, we have reported that Orlistat, a drug approved for obesity, is an inhibitor of fatty acid synthase and that this drug elicits cell death in a number of breast tumor cell lines [32]. Not only are the findings of the present study consistent with these prior observations but they also add another dimension. Each of the fatty acids is derived from either elongation or desaturation (or both) of palmitate by the action of stearoyl-CoA desaturases and long chain fatty acid elongase. Both stearoyl-CoA desaturase [33] and long chain fatty acid elongase [34] are part of the SREBP-regulated lipogenic program that is often activated in solid tumor progression [35]. Increased stearoyl-CoA desaturase expression and activity results in increased mono-unsaturated fatty acid (MUFA) levels and this increase in MUFAs is associated with cancer risk [36] and mechanistically may help modulate membrane fluidity [37], and anchorage-independent growth [38] in tumor cells. The fact that tumor cells convert the newly synthesized palmitate to other fatty acids also suggests that stearoyl-CoA desaturases and fatty acid elongase should be explored as potential drug targets.
To our knowledge, this is the first study to take a comprehensive and comparative approach toward the links between breast tumor progression and metabolism, as well as the first study to characterize the metabolome of the metastatic cell. The results provide an essential foundation for subsequent interrogation of individual metabolic steps, and for gauging the requirement of each step in tumor cell progression. The analysis provides important information that will help guide the interpretation of studies of breast tumor metabolism under hypoxic conditions and studies of the metabolism of whole tumors in vivo, which are comprised of multiple cell types. It also provides a rationale for using metabolomics for cancer detection and diagnosis, as well as for the development of novel therapeutics that specifically target the tumor cell metabolome.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by grants from the National Cancer Institute (P30 CA030199) and from the National Institutes of Health (U54 RR020843 (JWS), R01 AI059146 (AO), and CA108959 (JWS)). Chen Yang was supported by a fellowship from the University of California Breast Cancer Research Program (12FB-0100).
Footnotes
Adam D. Richardson and Chen Yang contributed equally to this work.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-007-9732-3) contains supplementary material, which is available to authorized users.
References
- 1.Warburg O, Wind F, Negelein E (1927) The metabolism of tumors in the body. J Gen Physiol 8:519–530 [DOI] [PMC free article] [PubMed]
- 2.Kim JW, Dang CV (2006) Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res 66:8927–8930 [DOI] [PubMed]
- 3.Czernin J, Phelps ME (2002) Positron emission tomography scanning: current and future applications. Annu Rev Med 53:89–112 [DOI] [PubMed]
- 4.Zu XL, Guppy M (2004) Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun 313:459–465 [DOI] [PubMed]
- 5.Miller FR (2000) Xenograft models of premalignant breast disease. J Mammary Gland Biol Neoplasia 5:379–391 [DOI] [PubMed]
- 6.Soule HD, Maloney TM, Wolman SR et al (1990) Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res 50:6075–6086 [PubMed]
- 7.Basolo F, Elliott J, Tait L et al (1991) Transformation of human breast epithelial cells by c-Ha-ras oncogene. Mol Carcinog 4:25–35 [DOI] [PubMed]
- 8.Dawson PJ, Wolman SR, Tait L et al (1996) MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol 148:313–319 [PMC free article] [PubMed]
- 9.Santner SJ, Dawson PJ, Tait L et al (2001) Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat 65:101–110 [DOI] [PubMed]
- 10.Worsham MJ, Pals G, Schouten JP et al (2006) High-resolution mapping of molecular events associated with immortalization, transformation, and progression to breast cancer in the MCF10 model. Breast Cancer Res Treat 96:177–186 [DOI] [PubMed]
- 11.O’Neil KA, Miller FR, Barder TJ et al (2003) Profiling the progression of cancer: separation of microsomal proteins in MCF10 breast epithelial cell lines using nonporous chromatophoresis. Proteomics 3:1256–1269 [DOI] [PubMed]
- 12.Maharjan RP, Ferenci T (2003) Global metabolite analysis: the influence of extraction methodology on metabolome profiles of Escherichia coli. Anal Biochem 313:145–154 [DOI] [PubMed]
- 13.Lee WN, Bassilian S, Guo Z et al (1994) Measurement of fractional lipid synthesis using deuterated water (2H2O) and mass isotopomer analysis. Am J Physiol 266:E372–E383 [DOI] [PubMed]
- 14.Wittmann C, Heinzle E (1999) Mass spectrometry for metabolic flux analysis. Biotechnol Bioeng 62:739–750 [DOI] [PubMed]
- 15.Yang C, Richardson A, Smith J et al (2007) Comparative metabolomics of breast cancer. Pac Symp Biocomput 12:181–192 [PubMed]
- 16.Forbes NS, Meadows AL, Clark DS et al (2006) Estradiol stimulates the biosynthetic pathways of breast cancer cells: detection by metabolic flux analysis. Metab Eng 8:639–652 [DOI] [PubMed]
- 17.Marshall S (2006) Role of insulin, adipocyte hormones, and nutrient-sensing pathways in regulating fuel metabolism and energy homeostasis: a nutritional perspective of diabetes, obesity, and cancer. Sci STKE (2006):re7 [DOI] [PubMed]
- 18.Bauer DE, Harris MH, Plas DR et al (2004) Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. FASEB J 18:1303–1305 [DOI] [PMC free article] [PubMed]
- 19.Kaplan O, Jaroszewski JW, Faustino PJ et al (1990) Toxicity and effects of epidermal growth factor on glucose metabolism of MDA-468 human breast cancer cells. J Biol Chem 265:13641–13649 [PubMed]
- 20.Vizan P, Boros LG, Figueras A et al (2005) K-ras codon-specific mutations produce distinctive metabolic phenotypes in NIH3T3 mice [corrected] fibroblasts. Cancer Res 65:5512–5515 [DOI] [PubMed]
- 21.Lee WN, Boros LG, Puigjaner J et al (1998) Mass isotopomer study of the nonoxidative pathways of the pentose cycle with [1,2-13C2]glucose. Am J Physiol 274:E843–E851 [DOI] [PubMed]
- 22.Conklin KA (2004) Cancer chemotherapy and antioxidants. J Nutr 134:3201S–3204S [DOI] [PubMed]
- 23.Klaunig JE, Kamendulis LM (2004) The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol 44:239–267 [DOI] [PubMed]
- 24.Kurosu T, Fukuda T, Miki T et al (2003) BCL6 overexpression prevents increase in reactive oxygen species and inhibits apoptosis induced by chemotherapeutic reagents in B-cell lymphoma cells. Oncogene 22:4459–4468 [DOI] [PubMed]
- 25.Nohl H, Gille L, Staniek K (1998) The exogenous NADH dehydrogenase of heart mitochondria is the key enzyme responsible for selective cardiotoxicity of anthracyclines. Z Naturforsch [C] 53:279–285 [DOI] [PubMed]
- 26.Yokomizo A, Ono M, Nanri H et al (1995) Cellular levels of thioredoxin associated with drug sensitivity to cisplatin, mitomycin C, doxorubicin, and etoposide. Cancer Res 55:4293–4296 [PubMed]
- 27.Selak MA, Armour SM, MacKenzie ED et al (2005) Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 7:77–85 [DOI] [PubMed]
- 28.Pollard PJ, Briere JJ, Alam NA et al (2005) Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet 14:2231–2239 [DOI] [PubMed]
- 29.Lu H, Forbes RA, Verma A (2002) Hypoxia-inducible factor 1 activation by aerobic glycolysis implicates the Warburg effect in carcinogenesis. J Biol Chem 277:23111–23115 [DOI] [PubMed]
- 30.Kuhajda FP (2000) Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition 16:202–208 [DOI] [PubMed]
- 31.Kuhajda FP (2006) Fatty acid synthase and cancer: new application of an old pathway. Cancer Res 66:5977–5980 [DOI] [PubMed]
- 32.Kridel SJ, Axelrod F, Rozenkrantz N, Smith JW (2004) Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity. Cancer Res 64:2070–2075 [DOI] [PubMed]
- 33.Yang YA, Morin PJ, Han WF et al (2003) Regulation of fatty acid synthase expression in breast cancer by sterol regulatory element binding protein-1c. Exp Cell Res 282:132–137 [DOI] [PubMed]
- 34.Moon YA, Shah NA, Mohapatra S et al (2001) Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J Biol Chem 276:45358–45366 [DOI] [PubMed]
- 35.Swinnen JV, Brusselmans K, Verhoeven G (2006) Increased lipogenesis in cancer cells: new players, novel targets. Curr Opin Clin Nutr Metab Care 9:358–365 [DOI] [PubMed]
- 36.Pala V, Krogh V, Muti P et al (2001) Erythrocyte membrane fatty acids and subsequent breast cancer: a prospective Italian study. J Natl Cancer Inst 93:1088–1095 [DOI] [PubMed]
- 37.Aclimandos WA, Heinemann D, Kelly SB et al (1992) Erythrocyte stearic to oleic acid ratio in patients with ocular melanoma. Eye 6(Pt 4):416–419 [DOI] [PubMed]
- 38.Scaglia N, Igal RA (2005) Stearoyl-CoA desaturase is involved in the control of proliferation, anchorage-independent growth, and survival in human transformed cells. J Biol Chem 280:25339–25349 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.