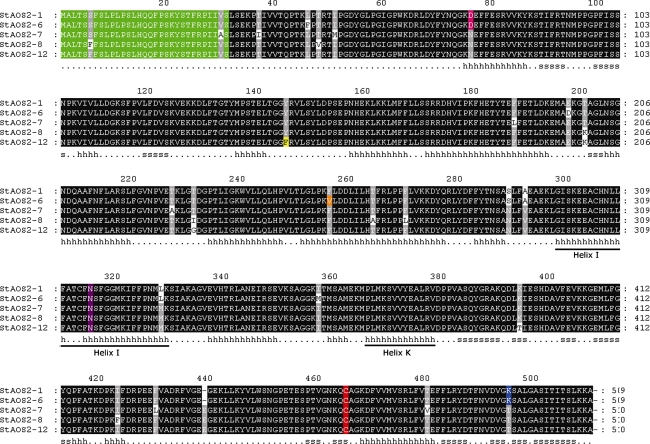
Fig. 1.
Deduced amino acid sequences alignment of the StAOS2 alleles. Black-shaded boxes represent conserved amino acid residues. Chloroplast targeting signal peptide is indicated in green color. Conserved heme-binding C464 and catalytic residue N315 are highlighted in red and purple, respectively. Amino acid substitutions close to the enzyme catalytic site, D76N, K494T, Y145F and F256V, are shown in pink, blue, yellow, and orange, respectively. The secondary structure based on the predicted alignment to Bacillus megaterium CYP102 (1jpza) is indicated below the sequences (h helix, s strand). Helix I (the oxygen-binding pocket; amino acids 298–329) and Helix K (bearing the EXXR motif; amino acids 366–378) are underlined