Short abstract
The glypican family of cell-surface heparan sulfate proteoglycans modulate the actions of many developmentally important signal proteins.
Abstract
Glypicans are heparan sulfate proteoglycans that are bound to the outer surface of the plasma membrane by a glycosyl-phosphatidylinositol anchor. Homologs of glypicans are found throughout the Eumetazoa. There are six family members in mammals (GPC1 to GPC6). Glypicans can be released from the cell surface by a lipase called Notum, and most of them are subjected to endoproteolytic cleavage by furin-like convertases. In vivo evidence published so far indicates that the main function of membrane-attached glypicans is to regulate the signaling of Wnts, Hedgehogs, fibroblast growth factors and bone morphogenetic proteins (BMPs). Depending on the context, glypicans may have a stimulatory or inhibitory activity on signaling. In the case of Wnt, it has been proposed that the stimulatory mechanism is based on the ability of glypicans to facilitate and/or stabilize the interaction of Wnts with their signaling receptors, the Frizzled proteins. On the other hand, GPC3 has recently been reported to inhibit Hedgehog protein signaling during development by competing with Patched, the Hedgehog receptor, for Hedgehog binding. Surprisingly, the regulatory activity of glypicans in the Wnt, Hedgehog and BMP signaling pathways is only partially dependent on the heparan sulfate chains.
Gene organization and evolutionary history
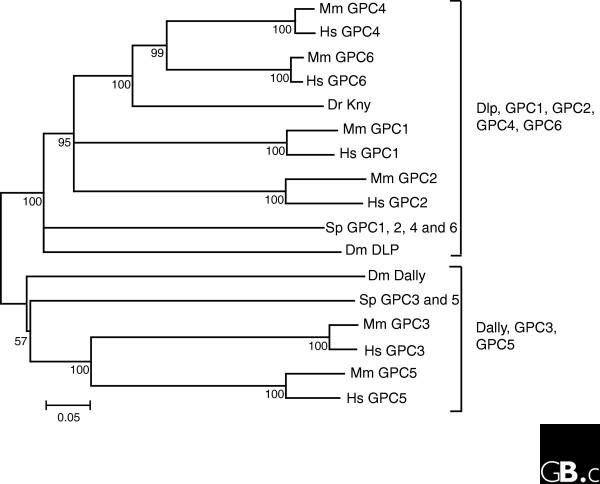
Glypicans are heparan sulfate proteoglycans that are bound to the external surface of the plasma membrane by a glycosyl-phosphatidylinositol (GPI) linkage [1,2]. Homologs of glypican are found throughout the Eumetazoa, with at least two genes in the starlet anemone Nematostella vectensis. Clear glypican homologs are not found outside the Metazoa. There are six glypican family members in the human genome (GPC1 to GPC6). The mouse genome also has six glypicans, which are identified by the same nomenclature (Table 1). Glypicans fall into two broad subfamilies: glypicans 1/2/4/6 and glypicans 3/5 (Figure 1), with approximately 25% amino-acid identity between groups. Within the first subfamily, glypicans 4 and 6 are relatively closely related (64% identity) and glypicans 1 and 2 form a more divergent clade. A single representative of each of the two subfamilies is present in Drosophila: Dally, an ortholog of the mammalian glypican 3/5 subfamily, and Dally-like protein, an ortholog of the glypican 1/2/4/6 subfamily. Basal deuterostomes such as the sea urchin also have one representative of each subfamily. Expansions of the multigene family in the lineage leading to mammals are thus characterized by an ancient gene duplication preceding the appearance of the common bilaterian (and possibly eumetazoan) ancestor giving rise to the two major subfamilies, followed by one or two rounds of duplication that probably took place in a vertebrate ancestor.
Table 1.
Glypicans in humans and Drosophila
| Gene name | Synonyms | Location | Gene accession number (GenBank) | Number of amino acids | Reference |
| Human | |||||
| GPC1 | Glypican | 2q35-37 | NM_002081 | 558 | [40] |
| GPC2 | Cerebroglycan | 7q22.1 | NM_152742 | 579 | [41] |
| GPC3 | OCI-5, MXR7 | Xq26 | NM_004484 | 580 | [42] |
| GPC4 | K-glypican | Xq26.1 | NM_001448 | 556 | [9] |
| GPC5 | 13q32 | NM_004466.3 | 572 | [43] | |
| GPC6 | 13q32 | NM_005708.2 | 555 | [44] | |
| Drosophila | |||||
| Dally | 3L,66E1-66E3 | NM_079259.2 | 626 | [45] | |
| Dally-like protein (Dlp) | 3L,70E5-70E7 | NM_206353.1 | 939 | [46] |
Figure 1.
Interrelationships among glypican proteins. The phylogeny was inferred using the neighbor-joining method. The tree is a bootstrap consensus generated from 1,000 replicates using the MEGA4 program suite [47]. The percentage of replicates in which the associated sequences cluster is shown next to branches. All positions containing gaps were eliminated from the dataset. The bar at the bottom indicates proportion of amino-acid differences. The species used are human (Hs), mouse (Mm), zebrafish (Dr), purple sea urchin (Sp), and fruit fly (Dm). Dlp, Dally-like protein. NCBI accession numbers for the sequences used in the analysis are as follows: HsGPC1, NP_002072.2; HsGPC2, NP_689955.1; HsGPC3, NP_004475.1; HsGPC4, NP_001439.2; HsGPC5, NP_004457.1; HsGPC6, NP_005699.1; MmGPC1, NP_057905.1; MmGPC2, NP_766000.1; MmGPC3, NP_057906.2; MmGPC4, NP_032176; MmGPC5, NP_780709.1; MmGPC6, NP_001073313.1; DrKNY, NP_571935; DmDally, AAA97401.1; DmDlp, AAG38110.1. Sea urchin sequences obtained from models generated in the Sea Urchin Genome Project [48] are as follows: SpGPC1/2/4/6, GLEAN3_03084; SpGPC3/5, GLEAN3_13086. A scan of the zebrafish genome reveals additional GPC family members, but complete transcript sequences are not available. The full complement of GPC genes is shown for the other species.
A notable genomic feature in the mouse and human genome is the presence of closely linked genes that form two glypican clusters: glypicans 3/4 on the X chromosome, and glypicans 5/6 on human chromosome 13 (mouse chromosome 14). Both of these clusters comprise one member of each of the two major glypican subfamilies, suggesting that their linkage may be ancient. Five glypican-like genes are present in the zebrafish genome (Ensembl [3]). Four of these zebrafish genes are linked in two clusters: a GPC3/Kny cluster and a GPC5/GPC1 cluster. Drosophila Dally and Dally-like protein are encoded on the same chromosome, but are far more distantly linked than are the mammalian clusters.
Glypican proteins are between 555 and 580 amino acids in length, and are encoded in eight to ten exons in human. The size of these genes can extend from a very compact 7.7 kb for human GPC2 to an expansive 1.5 Mb for human GPC5. This remarkable divergence in gene size begs the question of whether the small glypicans (GPC1 and 2) differ in some essential way from the much larger relatives in terms of complexity of gene usage or other regulatory characteristics.
Characteristic structural features
Because there are no reports on the analysis of glypicans by X-ray crystallography or other imaging techniques, our knowledge of the three-dimensional structure of glypicans is very limited. Furthermore, glypicans do not seem to have domains with significant homology to characterized structures. It is clear, however, that the three-dimensional structure of glypicans is highly conserved across the family, as the localization of 14 cysteine residues is preserved in all family members [4]. A weak identity between a fragment that extends approximately from residue 200 to residue 300 of glypicans and the cysteine-rich domain of Frizzled proteins has been reported [5]. Whether this has functional implications is still unknown, however. Another interesting structural feature shared by all glypicans is the insertion sites for the heparan sulfate (HS) chains, which are located close to the carboxyl terminus. This places the HS chains close to the cell surface, suggesting that these chains could mediate the interaction of glypicans with other cell-surface molecules, including growth factor receptors.
Most glypicans, including those of Drosophila [6], are subjected to endoproteolytic cleavage by a furin-like convertase [7]. This cleavage has been observed in vivo [8], and in many types of cultured cells [7,9]. The cleavage site is located at the carboxy-terminal end of the CRD domain, and generates two subunits that remain attached to each other by one or more disulfide bonds [7]. Whether the convertase-induced cleavage of glypicans is complete, and whether it occurs in all cell types, is still unknown. It should be noted, however, that this cleavage is not required for all glypican functions [10].
GPC5 displays a mixture of HS and chondroitin sulfate when transiently transfected into Cos-7 cells [11]. It remains to be seen whether the unexpected presence of chondroitin sulfate chains in a glypican is just a peculiarity of transiently transfected Cos-7 cells, or whether endogenous GPC5 can also display such chains at least in specific tissues.
Localization and function
As expected for proteins that carry GPI anchors, glypicans are mostly found at the cell membrane. In polarized cells, GPI-anchored proteins are usually located at the apical membrane. It is thought that apical sorting is due to their association with lipid rafts [12]. These are cell-membrane subdomains that are glycolipid-enriched and detergent-resistant. It has been proposed that these domains facilitate selective protein-protein interactions that establish transient cell-signaling platforms [13]. Unlike other GPI-anchored proteins, however, significant amounts of glypicans can be found outside lipid rafts, and at the basolateral membranes of polarized cells [14]. Interestingly, the HS chains seem to play a critical role in this unexpected localization, since non-glycanated glypicans are sorted apically [14]. Most of the in vivo evidence published so far indicates that the main function of membrane-attached glypicans is to regulate the signaling of Wnts, Hedgehogs (Hhs), fibroblast growth factors (FGFs), and bone morphogenetic proteins (BMPs) [5,15-18]. For example, GPC3-null mice display alterations in Wnt and Hh signaling [16,19], and Drosophila glypican mutants have defective Hh, Wnt, BMP and FGF signaling in specific tissues [15,18,20,21]. Furthermore, GPC3 promotes the growth of hepatocellular carcinoma cells by stimulating Wnt signaling [22]. The function of glypicans is not limited to the regulation of growth factor activity. For example, Dally-like protein, a Drosophila glypican, has been shown to play a role in synapse morphogenesis and function by binding and inhibiting the receptor phosphatase LAR [23]. In addition, it has been proposed that glypicans can be involved in the uptake of polyamines [24].
Glypicans can also be shed into the extracellular environment. This shedding is generated, at least in part, by Notum, an extracellular lipase that releases glypicans by cleaving the GPI anchor [25,26]. Studies in Drosophila have demonstrated that shed glypicans play a role in the transport of Wnts, Hhs and BMPs for the purpose of morphogen gradient formation [27-32]. Interestingly, glypicans have been found in lipophorins, the Drosophila lipoproteins. These particles are critical for the long-range activity of Wnts and Hhs [6,33]. In the particular case of Hh, it has been proposed that the glypicans in lipophorins may promote the formation of ligand-receptor complexes in the target cells [6].
In addition to their localization on the cell membrane and in the extracellular environment, glypicans can also be found in the cytoplasm. In particular, there have been several studies reporting the presence of GPC3 in the cytoplasm of liver cancer cells [34,35]. Whether cytoplasmic GPC3 plays a specific role is unknown.
Mechanism of action
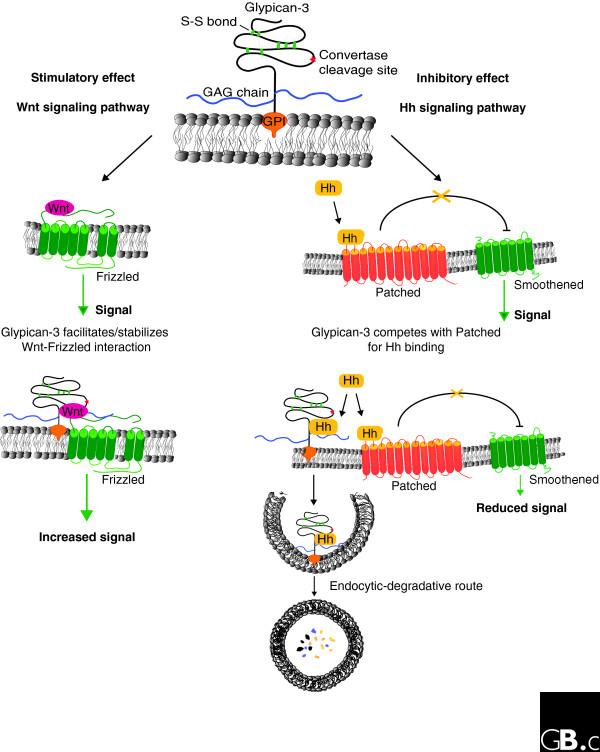
Depending on the biological context, glypicans can either stimulate or inhibit signaling activity. In the case of the stimulation of Wnt signaling, it has been proposed that the stimulatory mechanism is based on the ability of glypicans to facilitate and/or stabilize the interaction of Wnts with their signaling receptors, the Frizzled proteins (Figure 2) [22].
Figure 2.
Positive and negative effects of GPC3 on cell signaling. In the Wnt signaling pathway (left), GPC3 exerts a positive effect. Wnt binds to the receptor Frizzled to induce signaling (green arrow). GPC3 facilitates and/or stabilizes the interaction between Wnt and Frizzled with the consequent increment on signaling. In the Hedgehog (Hh) signaling pathway (right), GPC3 exerts an inhibitory effect. The binding of Hh to the receptor Patched (Ptc) triggers the signaling pathway by blocking the inhibitory activity of Ptc on Smoothened. GPC3 competes with Ptc for Hh binding. The interaction of Hh with GPC3 triggers the endocytosis and degradation of the complex with the consequent reduction of Hh available for binding to Ptc.
This hypothesis is based on the finding that glypicans can bind to Wnts and to Frizzleds [16,18,22,36], and that transfection of glypicans increases the Wnt-binding capacity of the transfected cells [22]. In the case of Hhs, it has been recently reported that GPC3 inhibits their signaling during development by competing with Patched, the Hh receptor, for Hh binding (Figure 2) [19]. The binding of Hh to GPC3 triggers its endocytosis and degradation. On the other hand, it has been shown that the Drosophila glypican Dally-like protein stimulates Hh signaling, although the mechanism of this stimulatory activity remains unknown [37].
Because the HS chains have a strong negative charge, HS proteoglycans can interact in a rather promiscuous way with proteins that display positively charged domains. On this basis it was originally thought that the HS chains were essential for glypican activity. Indeed, this seems to be the case for the glypican-induced stimulation of FGF activity [38]. However, recent experimental evidence has demonstrated that the HS chains are only partially required for the regulatory activity of glypicans in Hh, Wnt and BMP signaling [16,19,39]. Furthermore, Hh has been shown to bind to the core protein of GPC3 with high affinity [19].
Frontiers
One of the main issues that requires attention in the near future is the cellular and molecular basis of the context specificity that characterizes glypican activity. For example, what is the reason for the opposite effects of GPC3 and Dally-like protein on Hh signaling? Equally important will be a detailed characterization of the interaction of glypicans with Hhs, Wnts, and BMPs. Some of the questions to be answered in this regard are: Do all glypican core proteins interact with Hhs, Wnts and BMPs? What are the domains involved in these interactions? Do glypicans interact with the corresponding signaling receptors?
A further important topic of investigation will be the role of glypicans in morphogen gradient formation. We still do not understand the precise role of these proteins in regulating morphogen movement. Furthermore, whether glypicans are involved in this process in mammals remains to be investigated.
It is obvious that our knowledge of glypican functions is still very limited despite the recent advances. A better understanding of these functions will make a significant contribution to the study of signaling pathways that play a very important role in developmental morphogenesis and several human diseases, including cancer.
Acknowledgments
Acknowledgements
JF and JR thank the Canadian Institute of Health Research for funding (MOP 62815 and MOP74667, respectively).
References
- Filmus J, Selleck SB. Glypicans: proteoglycans with a surprise. J Clin Invest. 2001;108:497–501. doi: 10.1172/JCI13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fico A, Maina F, Dono R. Fine-tuning of cell signalling by glypicans. Cell Mol Life Sci doi: 10.1007/s00018-007-7471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensembl http://www.ensembl.org/index.html
- Veugelers M, De Cat B, Ceulemans H, Bruystens AM, Coomans C, Durr J, Vermeesch J, Marynen P, David G. Glypican-6, a new member of the glypican family of cell surface proteoglycans. J Biol Chem. 1999;274:26968–26977. doi: 10.1074/jbc.274.38.26968. [DOI] [PubMed] [Google Scholar]
- Topczewsky J, Sepich DS, Myers DC, Walker C, Amores A, Lele Z, Hammerschmidt M, Postlethwait J, Solnica-Krezel L. The zebrafish glypican Knypek controls cell polarity during gastrulation movements of convergent extension. Dev Cell. 2001;1:251–264. doi: 10.1016/S1534-5807(01)00005-3. [DOI] [PubMed] [Google Scholar]
- Eugster C, Panakova D, Mahmoud A, Eaton S. Lipoprotein-heparan sulfate interactions in the Hh pathway. Dev Cell. 2007;13:57–71. doi: 10.1016/j.devcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- De Cat B, Muyldermans SY, Coomans C, Degeest G, Vander-schueren B, Creemers J, Biemar F, Peers B, David G. Processing by proprotein convertases is required for glypican-3 modulation of cell survival, Wnt signaling, and gastrulation movements. J Cell Biol. 2003;163:625–635. doi: 10.1083/jcb.200302152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara K, Watanabe K, Chun J, Yamaguchi Y. Glypican-4 is an FGF2-binding heparan sulfate proteoglycan expressed in neural precursor cells. Dev Dyn. 2000;219:353–367. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1059>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Yamada H, Yamaguchi Y. K-glypican: a novel GPI-linked heparan sulfate proteoglycan that is highly expressed in developing brain and kidney. J Cell Biol. 1995;130:1207–1218. doi: 10.1083/jcb.130.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurro MI, Shi W, Sandal S, Filmus J. Processing by convertases is not required for glypican-3-induced stimulation of hepatocellular carcinoma. J Biol Chem. 2005;280:41201–41206. doi: 10.1074/jbc.M507004200. [DOI] [PubMed] [Google Scholar]
- Saunders S, Paine-Saunders S, Lander AD. Expression of the cell surface proteoglycan glypican-5 is developmentally regulated in kidney, limb, and brain. Dev Biol. 1997;190:78–93. doi: 10.1006/dbio.1997.8690. [DOI] [PubMed] [Google Scholar]
- Mayor S, Riezman H. Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- Hancock JF. Lipid rafts: contentious only from simplistic standpoints. Nat Rev Mol Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens G, Schueren B Van den, Berghe H Van Den, David G. Heparan sulfate expression in polarized epithelial cells: the apical sorting of glypican (GPI-anchored proteoglycan) is inversely related to its heparan sulfate content. J Cell Biol. 1996;132:487–497. doi: 10.1083/jcb.132.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Perrimon N. Dally cooperates with Drosophila Frizzled 2 to transduce Wingless signaling. Nature. 1999;400:281–284. doi: 10.1038/22343. [DOI] [PubMed] [Google Scholar]
- Song HH, Shi W, Xiang Y, Filmus J. The loss of Glypican-3 induces alterations in Wnt signaling. J Biol Chem. 2005;280:2116–2125. doi: 10.1074/jbc.M410090200. [DOI] [PubMed] [Google Scholar]
- Yan D, Lin X. Drosophila glypican Dally-like acts in FGF-receiving cells to modulate FGF signaling during tracheal morphogenesis. Dev Biol. 2007;312:203–216. doi: 10.1016/j.ydbio.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawara B, Yamamoto TS, Tada M, Ueno N. Role of glypican 4 in the regulation of convergent extension movements during gastrulation in Xenopus laevis. Development. 2003;130:2129–2138. doi: 10.1242/dev.00435. [DOI] [PubMed] [Google Scholar]
- Capurro MI, Xu P, Shi W, Li F, Jia A, Filmus J. Glypican-3 inhibits hedgehog signaling during development by competing with Patched for Hedgehog binding. Dev Cell. 2008;14:700–711. doi: 10.1016/j.devcel.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Desbordes SC, Sanson B. The glypican Dally-like is required for hedgehog signalling in the embryonic epidermis of Drosophila. Development. 2003;130:6245–6255. doi: 10.1242/dev.00874. [DOI] [PubMed] [Google Scholar]
- Jackson SM, Nakato H, Sugiura M, Jannuzi A, Oakes R, Kaluza V, Golden C, Selleck SB. dally, a Drosophila glypican, controls celular responses to the TGF-beta-related morphogen Dpp. Development. 1997;124:4113–4120. doi: 10.1242/dev.124.20.4113. [DOI] [PubMed] [Google Scholar]
- Capurro M, Xiang YY, Lobe C, Filmus J. Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 2005;65:6245–6254. doi: 10.1158/0008-5472.CAN-04-4244. [DOI] [PubMed] [Google Scholar]
- Johnson KG, Tenney AP, Ghose A, Duckworth AM, Higashi ME, Parfitt K, Marcu O, Heslip TR, Marsh JL, Schwarz TL, Flanagan JG, Van Vactor D. The HSPGs syndecan and Dallylike bind the receptor phophatase LAR and exert distinct effects on synaptic development. Neuron. 2006;49:517–531. doi: 10.1016/j.neuron.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Fransson LA. Glypicans. Int J Biochem Cell Biol. 2003;35:125–129. doi: 10.1016/S1357-2725(02)00095-X. [DOI] [PubMed] [Google Scholar]
- Kreuger J, Perez L, Giraldez AJ, Cohen SM. Opposing activities of Dally-like glypican at high and low levels of Wingless morphogen activity. Dev Cell. 2004;7:503–512. doi: 10.1016/j.devcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Traister A, Shi W, Filmus J. Mammalian Notum induces the release of glypicans and other GPI-anchored proteins from the cell surface. Biochem J. 2008;410:503–511. doi: 10.1042/BJ20070511. [DOI] [PubMed] [Google Scholar]
- Fujise M, Takeo S, Kamimura K, Matsuo T, Aigaki T, Izumi S, Nakato H. Dally regulates Dpp morphogen gradient formation in the Drosophila wing. Development. 2003;130:1515–1522. doi: 10.1242/dev.00379. [DOI] [PubMed] [Google Scholar]
- Han C, Belenkaya TY, Wang B, Lin X. Drosophila glypicans control the cell-to-cell movement of hedgehog by a dynamin-independent process. Development. 2004;131:601–611. doi: 10.1242/dev.00958. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CA, Dimitroff BD, Rawson JM, Selleck SB. Spatial regulation of wingless morphogen distribution and signaling by Dally-like protein. Dev Cell. 2004;7:513–523. doi: 10.1016/j.devcel.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Belenkaya TY, Han C, Yan D, Opoka RJ, Khodoun M, Liu H, Lin X. Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell. 2004;119:231–244. doi: 10.1016/j.cell.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Han C, Yan D, Belenkaya TY, Lin X. Drosophila glypicans Dally and Dally-like shape the extracellular Wingless morphogen gradient in the wing disk. Development. 2005;132:667–679. doi: 10.1242/dev.01636. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Kamimura K, Firkus C, Takeo S, Shimmi O, Nakato H. Dally regulates Dpp morphogen gradient formation by stabilizing Dpp on the cell surface. Dev Biol. 2008;313:408–419. doi: 10.1016/j.ydbio.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willnow TE, Hammes A, Eaton S. Lipoproteins and their receptors in embryonic development: more than cholesterol clearance. Development. 2007;134:3239–3249. doi: 10.1242/dev.004408. [DOI] [PubMed] [Google Scholar]
- Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125:81–90. doi: 10.1016/S0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- Ligato S, Mandich D, Cartun RW. Utility of glypican-3 in differentiating hepatocellular carcinoma from other primary and metastatic lesions in FNA of the liver: an imunocytochemical study. Modern Pathol. 2008;21:626–631. doi: 10.1038/modpathol.2008.26. [DOI] [PubMed] [Google Scholar]
- Ai X, Do AT, Lozynska O, Kusche-Gullberg M, Lindhal U, Emerson CP. QSulf1 remodels the 6-O sulfation states of cell surface proteoglycans to promote Wnt signaling. J Cell Biol. 2003;162:341–351. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Identification of hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299:2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- Song HH, Shi W, Filmus J. OCI-5/rat glypican-3 binds to fibroblast growth factor-2 but not to insulin-like growth factor-2. J Biol Chem. 1997;272:7574–7577. doi: 10.1074/jbc.272.12.7574. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CA, Knox SM, Staatz WD, Fox B, Lercher DM, Selleck SB. The function of a Drosophila glypican does not depend entirely on heparan sulfate modification. Dev Biol. 2006;300:570–582. doi: 10.1016/j.ydbio.2006.09.011. [DOI] [PubMed] [Google Scholar]
- David G, Lories V, Decock B, Marynen P, Cassiman J, Berghe H Van Den. Molecular cloning of a phosphatidylinositol-anchored membrane heparan sulfate proteoglycan from human lung fibroblasts. J Cell Biol. 1990;111:3165–3176. doi: 10.1083/jcb.111.6.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp CS, Litwac ED, Lander AD. Cerebroglycan: an integral membrane heparan sulfate proteoglycan that is unique to the developing nervous system and expressed specifically during neuronal differentiation. J Cell Biol. 1994;124:149–160. doi: 10.1083/jcb.124.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filmus J, Church J, Buick RN. Isolation of a cDNA corresponding to a developmentally regulated transcript in rat intestine. Mol Cell Biol. 1988;8:4243–4249. doi: 10.1128/mcb.8.10.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veugelers M, Vermeesch J, Reekmans G, Steinfeld R, Marynen P, David G. Characterization of glypican-5 and chromosomal localization of human GPC5, a new member of the glypican gene family. Genomics. 1997;40:24–30. doi: 10.1006/geno.1996.4518. [DOI] [PubMed] [Google Scholar]
- Paine-Saunders S, Viviano BL, Saunders S. GPC6, a novel member of the glypican gene family, encodes a product structurally related to GPC4 and is colocalized with GPC5 on human chromosome 13. Genomics. 1999;57:455–458. doi: 10.1006/geno.1999.5793. [DOI] [PubMed] [Google Scholar]
- Nakato H, Futch TA, Selleck SB. The division abnormally delayed (dally) gene: a putative integral membrane proteoglycan required for cell division patterning during postembryonic development of the nervous system in Drosophila. Development. 1995;121:3687–3702. doi: 10.1242/dev.121.11.3687. [DOI] [PubMed] [Google Scholar]
- Baeg GH, Lin X, Khare N, Baumgartner S, Perrimon N. Heparan sulfate proteoglycans are critical for the organization of the extracellular distribution of Wingless. Development. 2001;128:87–94. doi: 10.1242/dev.128.1.87. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Sea Urchin Genome Project http://www.hgsc.bcm.tmc.edu/projects/seaurchin